Abstract
A novel X-linked mental retardation (XLMR) syndrome was recently identified, resulting from creatine deficiency in the brain caused by mutations in the creatine transporter gene, SLC6A8. We have studied the prevalence of SLC6A8 mutations in a panel of 290 patients with nonsyndromic XLMR archived by the European XLMR Consortium. The full-length open reading frame and splice sites of the SLC6A8 gene were investigated by DNA sequence analysis. Six pathogenic mutations, of which five were novel, were identified in a total of 288 patients with XLMR, showing a prevalence of at least 2.1% (6/288). The novel pathogenic mutations are a nonsense mutation (p.Y317X) and four missense mutations. Three missense mutations (p.G87R, p.P390L, and p.P554L) were concluded to be pathogenic on the basis of conservation, segregation, chemical properties of the residues involved, as well as the absence of these and any other missense mutation in 276 controls. For the p.C337W mutation, additional material was available to biochemically prove (i.e., by increased urinary creatine:creatinine ratio) pathogenicity. In addition, we found nine novel polymorphisms (IVS1+26G→A, IVS7+37G→A, IVS7+87A→G, IVS7-35G→A, IVS12-3C→T, IVS2+88G→C, IVS9-36G→A, IVS12-82G→C, and p.Y498) that were present in the XLMR panel and/or in the control panel. Two missense variants (p.V629I and p.M560V) that were not highly conserved and were not associated with increased creatine:creatinine ratio, one translational silent variant (p.L472), and 10 intervening sequence variants or untranslated region variants (IVS6+9C→T, IVS7-151_152delGA, IVS7-99C→A, IVS8-35G→A, IVS8+28C→T, IVS10-18C→T, IVS11+21G→A, IVS12+15C→T, *207G→C, IVS12+32C→A) were found only in the XLMR panel but should be considered as unclassified variants or as a polymorphism (p.M560V). Our data indicate that the frequency of SLC6A8 mutations in the XLMR population is close to that of CGG expansions in FMR1, the gene responsible for fragile-X syndrome.
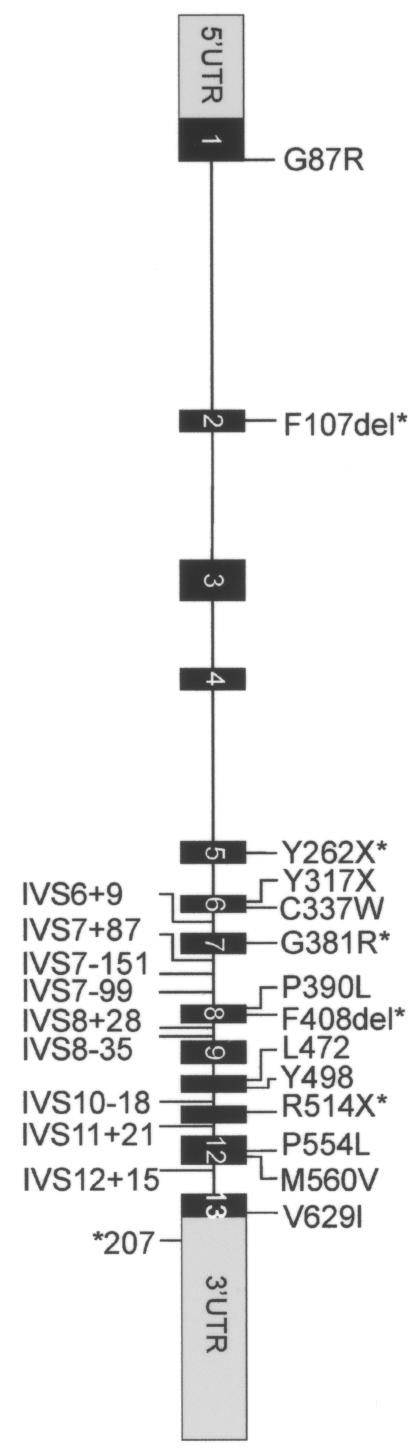
Creatine and phosphocreatine play essential roles in the storage and transmission of phosphate-bound energy (Walker 1979; Wyss and Kaddurah-Daouk 2000). Humans maintain their creatine pool by creatine biosynthesis, which involves two enzymes—L-arginine:glycine amidinotransferase (AGAT; Enzyme Commission [EC] number 2.1.4.1) and guanidinoacetate methyltransferase (GAMT; EC 2.1.1.2)—as well as nutritional uptake. Cellular transport is of fundamental importance for creatine homeostasis in tissues void of creatine biosynthesis. The creatine transporter gene (SLC6A8/CT1/CRTR1 [MIM 300036]) has been mapped to Xq28 (Gregor et al. 1995) and is a member of the solute-carrier family 6 (neurotransmitter transporters). The SLC6A8 gene spans ∼8.4 kb; it consists of 13 exons (GenBank accession number Z66539) and encodes a protein of 635 amino acids with a predicted molecular weight of 70 kDa (Sandoval et al. 1996) (fig. 1).
Figure 1.
Mutations and polymorphisms in patients with XLMR. Primer sequences were designed specifically to amplify all exons of the SLC6A8 gene (and not the SLC6A10 gene, a presumed creatine transporter pseudogene mapped at chromosome 16 [Eichler et al. 1996]), including short fragments of the flanking intronic sequences. By comparing the SLC6A8 and SLC6A10 sequences, we have found that at least two nucleotide variations were present in all amplicons, confirming selective amplification of the SLC6A8 sequences (data not shown). PCR reactions were performed with HotStar Taq (Qiagen) in a PE Applied Biosystems model 9700. Direct sequence analysis was performed on purified PCR products (Millipore vacufold) by use of BigDye v3.1 terminators and an ABI 3100 sequence machine (PE Applied Biosystems). The obtained electropherograms were assembled and analyzed to identify potential genomic alterations by use of the Mutation Surveyor software package (SoftGenetics). Sequence variants were annotated according the guidelines of den Dunnen and Antonarakis (2001). On the basis of impaired uptake in fibroblasts, five alterations (asterisks [*]) have been proven elsewhere to be mutations. p.F107del was also found in our XLMR cohort. One novel nonsense mutation (p.Y317X) is strongly predictive of impaired creatine uptake. p.M560V is a rare polymorphism, and p.V629I is an unclassified variant. The implications of the translational silent variant, the IVS variant, and the 3′ UTR variant could not yet be investigated, and these variants are therefore assigned as “unclassified.” IVS variants in introns 1, 2, 3, and 4 may have been missed, since only small exon-flanking parts were included.
The use of proton magnetic resonance spectroscopy (H-MRS) resulted in the identification of three inborn errors of metabolism: two creatine biosynthesis errors (AGAT deficiency [MIM 602360] and GAMT deficiency [MIM 601240]) and one creatine transport error (SLC6A8 deficiency [MIM 300036]) (Stockler et al. 1994, 1996a, 1996b; Schulze et al. 1997; Bianchi et al. 2000; van der Knaap et al. 2000; Cecil et al. 2001; Item et al. 2001; Salomons et al. 2001; Bizzi et al. 2002; deGrauw et al. 2002; Hahn et al. 2002; Salomons et al. 2003). These creatine-deficiency syndromes (CDS) share the almost-complete lack of creatine/phosphocreatine in the brain, as measured by in vivo H-MRS. Metabolite measurements in urine and plasma are indicative of the specific disorders (e.g., in SLC6A8-deficient males, the creatine:creatinine [Cr:Crn] ratio is increased in urine, and guanidinoacetate is increased in urine and plasma of GAMT-deficient patients). Characteristic in the clinical presentation of all CDS are mental retardation, expressive speech and language delay, and epilepsy (varying from intractable seizures in GAMT-deficient patients to mild epileptic or febrile seizures in AGAT-deficient and transporter-deficient patients). GAMT-deficient patients and transporter-deficient patients may show autistic behavior; in GAMT-deficient patients with a severe phenotype, (extra)pyramidal symptoms are present (Salomons et al. 2003; Stromberger et al. 2003). Female carriers of SLC6A8 mutations may have learning disabilities and/or behavioral problems.
In Western countries, mental retardation (MR) affects 2%–3% of the general population (Chelly and Mandel 2001). For the majority of the cases of inherited MR, the genetic cause has not yet been elucidated. The larger number of males than females in the MR population (∼70%) suggests a high contribution of X-linked disorders. X-linked mental retardation (XLMR) is estimated to account for 5%–12% of all cases of MR, with a relatively large (2%) contribution of fragile-X syndrome (fraX) (Herbst and Miller 1980; de Vries et al. 1997). The fact that >15 SLC6A8-deficient families (five families in one metropolitan area) have been identified in the 2 years after the first recognition of this disorder suggests a high incidence of creatine transporter deficiency in the Western population. Two prominent features of SLC6A8 deficiency are MR and X-linked inheritance of the disease. We therefore investigated by DNA sequence analysis the prevalence of SLC6A8 defects in a panel of 290 unrelated families of the European XLMR Consortium (European XLMR Consortium Web site). Families are included in this panel if at least two males in the family are affected. Informed consent has been obtained from the parents or caretakers of the affected males. Of the 290 cases, 2 were excluded from this study, since we were unable to amplify, by PCR, DNA sequences of the SLC6A8 gene and of two autosomal genes, because of the poor quality of the DNA. In addition, DNAs of 276 healthy controls were similarly analyzed.
In the European XLMR panel, seven novel nucleotide substitutions in the ORF of SLC6A8 were encountered: six missense variations/mutations and one insertion resulting in an immediate nonsense codon. In addition, one known pathogenic single–amino acid deletion was found. Furthermore, one translational silent change, eight intervening sequence (IVS) variants, and one substitution in the 3′ UTR were detected—none of which were found in either the 276 controls or the human EST database (table 1; fig. 1).
Table 1.
Nucleotide Changes in SLC6A8 in the XLMR Panel[Note]
| Genomic Mutation | cDNA Mutation | Deduced Effect | Family |
| g.2933G→A | c.259G→A | p.G87Ra | T31 |
| g.4533_4535delCTT | c.319_321delCTT | p.F107dela | T115 |
| g.7400_7401insA | c.950_951insA | p.Y317stopa | P66 |
| g.7461C→G | c.1011C→G | p.C337Wa | N87 |
| g.8032C→T | c.1169C→T | p.P390La | T132 |
| g.8883C→T | c.1661C→T | p.P554La | D11 |
| g.8900A→G | c.1678A→G | p.M560Vb | P18 |
| g.9291G→A | c.1885G→A | p.V629Ic | N67 |
| g.8467G→A | c.1416G→A | p.L472c | ⩾1 family |
| IVS6+9C→T | ND | ND | ⩾1 family |
| IVS7-151_152delGA | ND | ND | ⩾1 family |
| IVS7-99C→A | ND | ND | ⩾1 family |
| IVS8+28C→T | ND | ND | ⩾1 family |
| IVS8-35G→A | ND | ND | ⩾1 family |
| IVS10-18C→T | ND | ND | ⩾1 family |
| IVS11+21G→A | ND | ND | ⩾1 family |
| IVS12+15C→T | ND | ND | ⩾1 family |
| *207G→C | ND | ND | ⩾1 family |
| IVS12+32C→A | ND | ND | ⩾1 family |
Note.— The SLC6A8 nucleotide changes shown in this table were not found in the 276 controls or in the human EST database. ND = not determined.
Pathogenic mutation.
Polymorphism.
Unclassified variant.
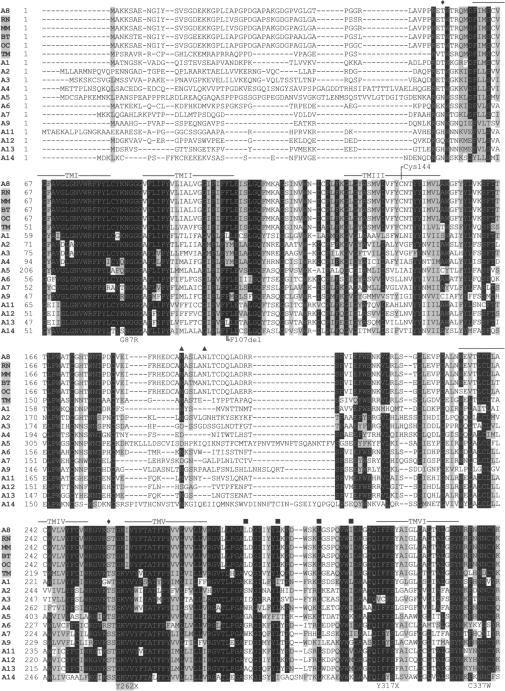
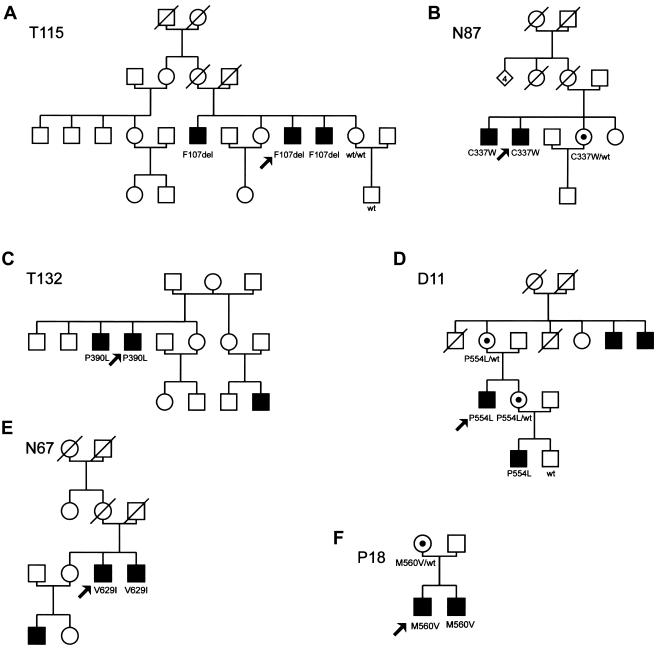
In DNA of the index patient of family T115, a deletion of 3 bp (c.319_321delCTT) in SLC6A8 was detected, which results in the loss of phenylalanine at position p.107 (p.F107del). The mutation is located in a short repeat of two phenylalanines in exon 2. This repeat is part of transmembrane-spanning domain (TM) II, which is highly conserved among all known creatine transporters (i.e., bovine, rat, rabbit, marbled electric ray, and mouse [see fig. 2]) and within the Na+- and Cl−-dependent neurotransmitter transporter family. The mutation segregates within the family (fig. 3A). SLC6A8 F107del was previously reported in an unrelated family (deGrauw et al. 2002) (fig. 1). The index male of that family showed impairment of creatine uptake in fibroblasts. The following observations indicate that p.F107del is pathogenic: (1) p.F107 is highly conserved between species; (2) p.F107 is located in a conserved area within its solute carrier family; (3) F107del segregates with the XLMR phenotype; (4) the mutation was detected in an unrelated family with SLC6A8 deficiency; (5) urine of affected males with this mutation showed an increased Cr:Crn ratio, a hallmark of this disease; and (6) fibroblasts with this deletion are impaired in creatine uptake.
Figure 2.
Multiple-sequence alignment with hierarchical clustering among the SLC6A8 proteins of different species and the superfamily of neurotransmitter transporters. The SLC6A8 protein sequence is shown for the following species: Homo sapiens (A8), Rattus norvegicus (RN), Mus musculus (MM), Bos taurus (BT), Oryctolagus cuniculus (OC), and Torpedo marmorata (TM). SLC6A8 genes were identified by a protein blast (BLASTP). Alignment was determined by the ClustalW program; the SLC6A8 proteins that were used for this analysis were the ones identified by the BLASTP search to be most related to the Homo sapiens SLC6A8 protein. All were saved in FASTA format. The BOXSHADE program was used to visualize identical amino acids (highlighted in black) and functionally conserved amino acids (gray). Functionally conserved amino acids are classified as follows: V, I, L, and M; D, E, Q, and N; F, Y, and W; G, S, T, P, and A; and K, R, and H. The codes A1–A14 represent neurotransmitter transporters SLC6A1–SLC6A14 (A1 = GABA1, A2 = noradrenaline, A3 = dopamine, A4 = serotonine, A5 = glycine2, A6 = taurine, A7 = proline, A9 = glycine 1, A11 = GABA3, A12 = betaine/GABA, A13 = GABA2, and A14 = ATB0+). In the protein sequence of A5, certain amino acids (20–46, 51–93, 116–127, 143–152, and 168–184) were cut out, because these stretches occur only in SLC6A5 and would make the figure disorganized. TMI–TMXII = putative transmembrane domain, predicted by ExPASy (Swiss-Prot S6A8_Human P48029); ♦ = putative cAMP-PK phosphorylation site (2×); ▴ = putative N-glycosylation sites (3×); ▪ = putative Leu zipper motif (4×).
Figure 3.
Pedigree charts of families in which segregation could be studied. (For designation of the variants, see table 1.)
In DNA of the index patient of family P66, an insertion of adenosine at position c.951 in exon 6 (c.950_951insA) of the SLC6A8 gene was found. This mutation predicts a premature stop (p.Y317X) that results in a truncated protein of 316 amino acids that lacks 319 amino acids of the C-terminus, including the putative TM VII–XII of SLC6A8 (fig. 2). The mutation most likely results in an unstable and/or inappropriately folded protein that is completely inactive. Indeed, two other nonsense mutations in the SLC6A8 gene (p.R514X and p.Y262X) (fig. 1) have been shown to impair creatine uptake and are pathogenic.
In DNA of the index patient of family T31, the transition G→A at position c.259 in the SLC6A8 gene was found. This transition results in the substitution p.G87R. The mutation is located in a short repeat of three glycines in exon 1. This repeat is located between the putative TM I and II in a small intracellular loop that is highly conserved among all known creatine transporters and within the neurotransmitter transporter family SLC6 (fig. 2). The mutation results in the substitution of the nonpolar and neutral glycine for the polar and basic arginine. The highly conserved nature of p.G87 and the clear difference in the chemical properties of the wild-type and mutant amino acids indicate that the p.G87R mutation is pathogenic. Because no additional biological materials (e.g., fibroblasts or urine) were available, the pathogenicity could not be studied at the biochemical level or by H-MRS of the brain.
In two affected males of family N87, the transversion C→G at SLC6A8 c.1011 was found, resulting in the substitution p.C337W. C337 is located in the intracellular loop between TM VI and VII and is highly conserved (fig. 2). The substitution of cysteine by tryptophan is highly significant: cysteine is polar and weakly acidic, and the sulfhydryl group is often involved in protein folding by forming disulfide links, whereas tryptophan is a nonpolar and neutral amino acid, with a very rigid cyclic structure. The patients' sister was a carrier of the p.C337W mutation (fig. 3B). In urine of the two affected males, an increased Cr:Crn ratio was found, a biochemical hallmark of SLC6A8 deficiency. These data indicate that the p.C337W is pathogenic.
In DNA of the index patient of family T132, a transition of c.1169C→T was detected, resulting in the substitution p.P390L. The residue P390 is located in the extracellular loop between TM VII and VIII and is conserved in all known SLC6 family members, except for the serotonine transporter (fig. 2). The p.P390L substitution may have a significant effect on correct protein folding. An affected brother of the index patient also had the p.P390L substitution (fig. 3C). Therefore, we consider p.P390L a pathogenic mutation. It is unfortunate that, because of the unavailability of fibroblasts or urine, the pathogenicity could not be studied at the biochemical level or by H-MRS of the brain.
In DNA of the index patient of family D11, a transition of c.1661C→T was detected in exon 12 of SLC6A8, which resulted in the substitution p.P554L. P554 is a residue of the extracellular loop between TM XI and XII. The region of TM IX–XI is expected to harbor the determinants of substrate specificity. The residue p.P554 is highly conserved (see fig. 2). The effect of a substitution of proline by leucine was discussed above. The p.P554L mutation segregates with the clinical phenotype within the family (fig. 3D). Thus, p.P554L is pathogenic. As discussed above, because of the lack of appropriate biological material from the affected males, the pathogenicity could not be studied at the biochemical or clinical level.
In DNA of the index patient of family N67, a G→A transition was detected at SLC6A8 c.1885, which resulted in the substitution p.V629I in exon 13. The nonsynonymous substitution is located in a short repeat of four valines in the intracellular C-terminus of the creatine transporter protein (fig. 2). This residue is conserved in the creatine transporters between species but not among the SLC6 family members. Moreover, both valine and isoleucine are members of the same group of aliphatic amino acids, and, therefore, no change of chemical properties is predicted. The p.V629I mutation was also present in the affected brother (fig. 3E). Cr:Crn ratios in urine were within the low normal range (index patient and brother 0.001; control values 0–0.3). It is not known whether mutations in specific SLC6A8 regions could alter the function, stability, or trafficking of the protein, as is described for SLC6A2 (Bauman and Blakely 2002) and SLC6A13 (Brown et al. 2003). The c.1885G→A transition was not found in 276 controls and 95 ESTs. p.V629I is currently considered to be an unclassified variant.
In DNA of the index patient of family P18, an A→G transition was found in SLC6A8 at c.1678 in exon 12, resulting in the substitution p.M560V. The substitution was also present in the affected brother of the index patient. The mother is a carrier of the transition (fig. 3F). p.M560 is conserved in rat, rabbit, and mouse creatine transporter but not in bovine (Val), marbled electric ray (Ile), or other members of the SLC6 family (fig. 2). The p.M560V mutation was not found in 276 control chromosomes. However, fibroblasts of the patient are not deficient in creatine uptake, and, in the H-MRS of the brain, the creatine signal was present. Therefore, we conclude that p.M560V is a rare polymorphism. Whether any of the above-mentioned silent, IVS, and 3′ UTR variants (table 1) are disease causing or just represent rare polymorphisms remains to be investigated.
The absence of missense mutations in 276 control chromosomes (appendix A) reduces the chance to <1% for a missense mutation to be a polymorphism with 80% power (Collins and Schwartz 2002). The size of the control population is rather small to provide conclusive information on the nature of a specific missense mutation found in the equal-sized XLMR population. On the other hand, the number of missense mutations differs significantly between the patient group and control groups (6/288 versus 0/276; Fisher's exact test P value = .03).
We have identified 6 pathogenic mutations in 288 patients. This means a prevalence of 2.1% SLC6A8-deficient patients in this XLMR panel. This may be an underestimate, since we were able to investigate only 93% of the coding sequence in 288 patients (the complete coding sequence in 180 patients and, overall, 80% of the coding sequence in the remaining 108 patients). In addition, some of the unclassified variants (n=11) may prove to be pathogenic.
The presented results were calculated on the basis of our findings in a group of 288 patients who were preselected for suspected X-linked inheritance of MR. Currently, collaboration is being initiated to study the contribution of SLC6A8 deficiency in males with MR but without molecular defects in the FMR1 gene (i.e., fraX negative).
Throughout exons 1–13 of the coding region of the SLC6A8 gene, 14 previously reported and novel sequence variants have been identified. Mutation types include three nonsense mutations, two single–amino acid deletions in four families, one large deletion, four missense mutations, one combined missense/splice-site mutation, one unclassified variant (missense type), one translational silent mutation with unknown consequence, and one missense-type polymorphism. Although several mutations have been identified already, there is no indication of a recurrent mutation or a mutation-sensitive/-prone site (fig. 1).
Knockout-mouse models and clinical trials may result in a better understanding of the disorder and in appropriate treatment protocols for both affected males and females. The authors suggest that male patients with MR, autistic behavior, epilepsy, and/or expressive speech and language delay should be tested for creatine-deficiency disorders. Screening could be based on metabolite measurement of urine, H-MRS of the brain, and/or mutational analysis by direct sequencing of the gene(s). Functional tests could prove the biochemical defect and should include tests for AGAT and GAMT enzyme activity or creatine uptake into lymphoblasts or fibroblasts, respectively. SLC6A8 deficiency may contribute, together with fraX, to MR in an extensive proportion of males with nonsyndromic XLMR of unknown cause.
Acknowledgments
We are grateful to Patricia S. Darmin, Silvy J. M. van Dooren, and Yodit Jakob for their assistance with DNA sequence analysis. We thank Astrid R. Oudakker for assistance to T.K. and H.G.Y., and we acknowledge Michel Willemsen, M.D., Ph.D., for his valuable contribution prior to the initiation of the present study. We are thankful to the human genetics departments of the VU University Medical Center in Amsterdam, the University Medical Center in Nijmegen, and the University Medical Center in Leiden for providing the control DNA samples. This work was supported by ZonMw grant 940-37-031 and by European Union grant QLG3-CT-2002-01810 EURO-MRX.
Appendix A: Polymorphisms in the SLC6A8 Gene
The anonymous DNAs of unaffected males were kindly provided by the Departments of Human Genetics of the VU University Medical Center in Amsterdam, the University Medical Center in Nijmegen, and the University Medical Center in Leiden.
SLC6A8 Polymorphisms in Controls and Patients with XLMR
IVS1+26G→A
IVS7+37G→A
IVS7+87A→G
IVS7-35G→A
IVS12-3C→T
c.1494C→T: p.Y498
SLC6A8 Polymorphisms in Controls Only
IVS2+88G→C
IVS9-36G→A
IVS12-82G→C
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- European XLMR Consortium, http://www.euromrx.com/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SLC6A8 [accession number Z66539])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SLC6A8, AGAT deficiency, GAMT deficiency, and SLC6A8 deficiency)
References
- Bauman PA, Blakely RD (2002) Determinants within the C-terminus of the human norepinephrine transporter dictate transporter trafficking, stability, and activity. Arch Biochem Biophys 404:80–91 10.1016/S0003-9861(02)00232-1 [DOI] [PubMed] [Google Scholar]
- Bianchi MC, Tosetti M, Fornai F, Alessandri’ MG, Cipriani P, De Vito G, Canapicchi R (2000) Reversible brain creatine deficiency in two sisters with normal blood creatine level. Ann Neurol 47:511–513 [DOI] [PubMed] [Google Scholar]
- Bizzi A, Bugiani M, Salomons GS, Hunneman DH, Moroni I, Estienne M, Danesi U, Jakobs C, Uziel G (2002) X-linked creatine deficiency syndrome: a novel mutation in creatine transporter gene SLC6A8. Ann Neurol 52:227–231 10.1002/ana.10246 [DOI] [PubMed] [Google Scholar]
- Brown AN, Muth TR, Caplan MJ (2003) The C-terminal tail of the GAT-2 GABA transporter contains a novel motif that plays a role in basolateral targeting. Am J Physiol Cell Physiol 286:c1071–c1077 [DOI] [PubMed] [Google Scholar]
- Cecil KM, Salomons GS, Ball WS Jr, Wong B, Chuck G, Verhoeven NM, Jakobs C, deGrauw TJ (2001) Irreversible brain creatine deficiency with elevated serum and urine creatine: a creatine transporter defect? Ann Neurol 49:401–404 10.1002/ana.79 [DOI] [PubMed] [Google Scholar]
- Chelly J, Mandel JL (2001) Monogenic causes of X-linked mental retardation. Nat Rev Genet 2:669–680 10.1038/35088558 [DOI] [PubMed] [Google Scholar]
- Collins JS, Schwartz CE (2002) Detecting polymorphisms and mutations in candidate genes. Am J Hum Genet 71:1251–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deGrauw TJ, Salomons GS, Cecil KM, Chuck G, Newmeyer A, Schapiro MB, Jakobs C (2002) Congenital creatine transporter deficiency. Neuropediatrics 33:232–238 10.1055/s-2002-36743 [DOI] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE (2001) Nomenclature for the description of human sequence variations. Hum Genet 109:121–124 10.1007/s004390100505 [DOI] [PubMed] [Google Scholar]
- de Vries BB, van den Ouweland AM, Mohkamsing S, Duivenvoorden HJ, Mol E, Gelsema K, van Rijn M, Halley DJ, Sandkuijl LA, Oostra BA, Tibben A, Niermeijer MF, Collaborative Fragile X Study Group (1997) Screening and diagnosis for the fragile X syndrome among the mentally retarded: an epidemiological and psychological survey. Am J Hum Genet 61:660–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Lu F, Shen Y, Antonacci R, Jurecic V, Doggett NA, Moyzis RK, Baldini A, Gibbs RA, Nelson DL (1996) Duplication of a gene-rich cluster between 16p11.1 and Xq28: a novel pericentromeric-directed mechanism for paralogous genome evolution. Hum Mol Genet 5:899–912 10.1093/hmg/5.7.899 [DOI] [PubMed] [Google Scholar]
- Gregor P, Nash SR, Caron MG, Seldin MF, Warren ST (1995) Assignment of the creatine transporter gene (SLC6A8) to human chromosome Xq28 telomeric to G6PD. Genomics 25:332–333 10.1016/0888-7543(95)80155-F [DOI] [PubMed] [Google Scholar]
- Hahn KA, Salomons GS, Tackels-Horne D, Wood TC, Taylor HA, Schroer RJ, Lubs HA, Jakobs C, Olson RL, Holden KR, Stevenson RE, Schwartz CE (2002) X-linked mental retardation with seizures and carrier manifestations is caused by a mutation in the creatine-transporter gene (SLC6A8) located in Xq28. Am J Hum Genet 70:1349–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst DS, Miller JR (1980) Nonspecific X-linked mental retardation II: the frequency in British Columbia. Am J Med Genet 7:461–469 [DOI] [PubMed] [Google Scholar]
- Item CB, Stockler-Ipsiroglu S, Stromberger C, Muhl A, Alessandri MG, Bianchi MC, Tosetti M, Fornai F, Cioni G (2001) Arginine:glycine amidinotransferase deficiency: the third inborn error of creatine metabolism in humans. Am J Hum Genet 69:1127–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomons GS, van Dooren SJ, Verhoeven NM, Cecil KM, Ball WS, deGrauw TJ, Jakobs C (2001) X-linked creatine-transporter gene (SLC6A8) defect: a new creatine-deficiency syndrome. Am J Hum Genet 68:1497–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomons GS, van Dooren SJ, Verhoeven NM, Marsden D, Schwartz C, Cecil KM, deGrauw TJ, Jakobs C (2003) X-linked creatine transporter defect: an overview. J Inherit Metab Dis 26:309–318 10.1023/A:1024405821638 [DOI] [PubMed] [Google Scholar]
- Sandoval N, Bauer D, Brenner V, Coy JF, Drescher B, Kioschis P, Korn B, Nyakatura G, Poustka A, Reichwald K, Rosenthal A, Platzer M (1996) The genomic organization of a human creatine transporter (CRTR) gene located in Xq28. Genomics 35:383–385 10.1006/geno.1996.0373 [DOI] [PubMed] [Google Scholar]
- Schulze A, Hess T, Wevers R, Mayatepek E, Bachert P, Marescau B, Knopp MV, De Deyn PP, Bremer HJ, Rating D (1997) Creatine deficiency syndrome caused by guanidinoacetate methyltransferase deficiency: diagnostic tools for a new inborn error of metabolism. J Pediatr 131:626–631 [DOI] [PubMed] [Google Scholar]
- Stockler S, Hanefeld F, Frahm J (1996a) Creatine replacement therapy in guanidinoacetate methyltransferase deficiency: a novel inborn error of metabolism. Lancet 348:789–790 10.1016/S0140-6736(96)04116-5 [DOI] [PubMed] [Google Scholar]
- Stockler S, Holzbach U, Hanefeld F, Marquardt I, Helms G, Requart M, Hanicke W, Frahm J (1994) Creatine deficiency in the brain: a new, treatable inborn error of metabolism. Pediatr Res 36:409–413 [DOI] [PubMed] [Google Scholar]
- Stockler S, Isbrandt D, Hanefeld F, Schmidt B, von Figura K (1996b) Guanidinoacetate methyltransferase deficiency: the first inborn error of creatine metabolism in man. Am J Hum Genet 58:914–922 [PMC free article] [PubMed] [Google Scholar]
- Stromberger C, Bodamer OA, Stockler-Ipsiroglu S (2003) Clinical characteristics and diagnostic clues in inborn errors of creatine metabolism. J Inherit Metab Dis 26:299–308 10.1023/A:1024453704800 [DOI] [PubMed] [Google Scholar]
- van der Knaap MS, Verhoeven NM, Maaswinkel-Mooij P, Pouwels PJ, Onkenhout W, Peeters EA, Stockler-Ipsiroglu S, Jakobs C (2000) Mental retardation and behavioral problems as presenting signs of a creatine synthesis defect. Ann Neurol 47:540–543 [DOI] [PubMed] [Google Scholar]
- Walker JB (1979) Creatine: biosynthesis, regulation, and function. Adv Enzymol Relat Areas Mol Biol 50:177–242 [DOI] [PubMed] [Google Scholar]
- Wyss M, Kaddurah-Daouk R (2000) Creatine and creatinine metabolism. Physiol Rev 80:1107–1213 [DOI] [PubMed] [Google Scholar]