Abstract
A family history of coronary artery disease (CAD), especially when the disease occurs at a young age, is a potent risk factor for CAD. DNA collection in families in which two or more siblings are affected at an early age allows identification of genetic factors for CAD by linkage analysis. We performed a genomewide scan in 1,168 individuals from 438 families, including 493 affected sibling pairs with documented onset of CAD before 51 years of age in men and before 56 years of age in women. We prospectively defined three phenotypic subsets of families: (1) acute coronary syndrome in two or more siblings; (2) absence of type 2 diabetes in all affected siblings; and (3) atherogenic dyslipidemia in any one sibling. Genotypes were analyzed for 395 microsatellite markers. Regions were defined as providing evidence for linkage if they provided parametric two-point LOD scores >1.5, together with nonparametric multipoint LOD scores >1.0. Regions on chromosomes 3q13 (multipoint LOD = 3.3; empirical P value <.001) and 5q31 (multipoint LOD = 1.4; empirical P value <.081) met these criteria in the entire data set, and regions on chromosomes 1q25, 3q13, 7p14, and 19p13 met these criteria in one or more of the subsets. Two regions, 3q13 and 1q25, met the criteria for genomewide significance. We have identified a region on chromosome 3q13 that is linked to early-onset CAD, as well as additional regions of interest that will require further analysis. These data provide initial areas of the human genome where further investigation may reveal susceptibility genes for early-onset CAD.
Introduction
More than 13 million Americans are afflicted with clinically significant coronary artery disease (CAD) (American Heart Association 2004), and the care of these patients costs >$133 billion annually. Of these Americans with CAD, ∼10% are <54 years of age. Although a minority of the patient base, this 10% represents a fertile group for the investigation of the genetics underlying cardiac risk, since a family history of CAD is one of the most robust risk factors for the disease, even after adjustment for environmental risks that may be shared within families (Shea et al. 1984). Additionally, the disease inflicts a particularly high economic impact on this group of patients with early-onset CAD. The identification of genetic markers for the predisposition to develop CAD is likely to lead to new insights into the pathophysiology of the disease and the development of therapeutic agents for the prevention and treatment of CAD. There are two possible advantages to studying families with more than one member affected at an early age: (1) a more etiologically homogeneous sample will be ascertained, and (2) the genetic effect should be stronger in these families.
Several studies have estimated that the relative risk of developing early-onset CAD in a first-degree relative (sib) is between 3.8 and 12.1, depending on the age at onset of the proband (Rissanen 1979; Shea et al. 1984). Twin studies have shown increased risk to MZ and DZ twins, with elevated risk to the co-twin when the index twin was <65 years of age (Marenberg et al. 1994). Although family history has been consistently identified as an important risk factor in many independent studies, there are many other individual-specific factors that increase the risk of CAD, including lipid abnormalities of several kinds, smoking, hypertension, diabetes, physical inactivity, and obesity. These risk factors exhibit strong concordance in families and may add to the familial mechanism of CAD. In a population-based study of 503 Australian families, Harrap et al. (2000) identified shared environmental effects that persisted into adulthood, particularly body mass index. Furthermore, some risk factors themselves may exhibit significant familial clustering, including dyslipidemia, obesity, and diabetes (Perusse et al. 1997; Rice et al. 1997; Katzmarzyk et al. 2000; Watanabe et al. 2000). The importance of these risk factors in the etiology of CAD suggests that they must be considered in studies identifying genes for CAD.
The list of previously implicated candidate genes for CAD is long and includes polymorphisms in various genes plausibly associated with CAD (Lusis et al. 1992; Breslow 2000; Lusis 2000; Yamada et al. 2002). Recently, Yamada et al. (2002) genotyped thousands of individuals with CAD for 71 genes, and Ozaki et al. (2002) genotyped hundreds of individuals for thousands of SNPs. In both of these large-scale candidate-gene studies, only a few SNPs were shown to be associated in a second set of patients. Despite the considerable resources devoted to the study of candidate genes as risk factors for CAD, none have been shown to account for even a modest fraction of the familial risk of CAD. Thus, there is a need for a complementary genomewide approach that is unbiased with respect to gene function. Here we report the results of a genomewide scan for genetic regions linked to early-onset CAD by use of data from 438 families with two siblings who had CAD diagnosed before the age of 51 years in men and 56 years in women.
Material and Methods
Clinical Data Collection
The GENECARD study design has been described elsewhere (Jomini et al. 2002; Hauser et al. 2003). In brief, probands were ascertained by investigators affiliated with six international clinical centers: Duke University Medical Center (United States), University of Sheffield (United Kingdom), University of Wales College of Medicine (United Kingdom), University of Lausanne (Switzerland), Ludwigshafen Heart Center (Germany), and Vanderbilt University (United States). Study protocol and data-sharing agreements were approved by the institutional review boards of each participating institution. Informed consent was obtained from each participant.
Individuals were recruited if they met the diagnostic criteria for CAD and if the disease had been diagnosed before the age of 51 years in men and 56 years in women. These age limits were based on the incidence curves of CAD and the estimates of genetic effect at various ages (Rissanen 1979; Shea et al. 1984; Marenberg et al. 1994). To be eligible for participation in the GENECARD study, families were required to have at least two living siblings who met the robust diagnostic criteria. Living parents of the affected sibling pair (ASP) and additional affected siblings were ascertained when available. Family members (and connecting relatives) outside the proband’s nuclear family were also considered to be affected if they met the diagnostic and age criteria for inclusion in GENECARD. Any sample with a sibling with an age at onset older than the age criteria was included for the purpose of estimating identity-by-descent sharing but was not considered to be affected. Blood samples were obtained by study staff at the medical center or clinic, by visiting participants’ homes, or, in a few cases, by mail. Medical history and risk-factor information were obtained by interviewing patients and by abstracting information from medical records. Table 1 shows the number of families, individuals, and ASPs included in the GENECARD sample. There were 493 independent ASPs in the 438 families: 395 families with two affected siblings, 36 families with three affected siblings, and 7 families with four or more affected siblings. As with other adult-onset diseases, a significant number of parents could not be ascertained; only 19 fathers and 62 mothers of the ASPs are included in the genome screen.
Table 1.
Distribution of Families Included in the GENECARD Genome Screen across the Six International Ascertainment Sites[Note]
| Ascertainment Site | No. of Families | No. of ASPs | No. of Individuals | No. of Affected Individuals |
| Princess of Wales Hospital (Cardiff, United Kingdom) | 62 (39, 21, 24) | 64 | 135 | 128 |
| Duke University Medical Center (Durham, NC) | 194 (95, 78, 46) | 223 | 599 | 462 |
| Ludwigshafen Heart Center (Ludwigshafen, Germany) | 24 (12, 5, 6) | 24 | 48 | 48 |
| Lausanne University Hospital (Lausanne, Switzerland) | 50 (25, 13, 20) | 51 | 103 | 103 |
| Vanderbilt University (Nashville, TN) | 45 (25, 18, 22) | 67 | 144 | 117 |
| University of Sheffield (Sheffield, United Kingdom) | 63 (32, 17, 28) | 64 | 139 | 127 |
| Total | 438 | 493 | 1,168 | 985 |
Note.— Numbers in parentheses reflect the number of families in the subsets of patients with ACS, NDIA, and ADL, respectively.
We established a stringent definition of CAD to ensure accurate diagnosis and to reduce possible misclassification, as described by Hauser et al. (2003). In addition to meeting the GENECARD sex-specific age criteria, patients were required to have diagnoses accompanied by an index event—coronary angiography, myocardial infarction (MI), coronary artery bypass graft, or a functional test documenting reversible myocardial ischemia (Hauser et al. 2003)—verified through medical record evidence. A system of periodic review was implemented to establish quality control (QC) and to ensure consistency among all clinical sites. At 6-mo intervals during ascertainment, a random sample of 10% of the families was selected, and a complete review of the diagnosis and the information supporting that diagnosis was performed by the GENECARD cardiologists. As evidence of the success of this effort, the clinical characteristics of affected individuals were remarkably similar across all collection sites (Hauser et al. 2003).
Laboratory Methods
DNA was extracted by use of the Puregene system (Gentra Systems). The DNA samples from the study subjects were organized into genotyping lists, producing a standardized order of samples; QC samples were incorporated at specified slots in the list. The laboratory technicians were blinded to the identity of the QC samples and were also blinded to affection status and family composition of all samples. Genotyping was performed by use of the gel-based FAAST method (Vance and Ben Othmane 1998). Multiplex reactions were used to increase efficiency, lower the cost, and increase the speed of the genomic screens. Two-color fluorescence allowed for the combination of two multiplex sets of genomic markers in the same lane (Vance et al. 1996).
We implemented a series of QC checks to maximize data quality during genotyping. Two control individuals common to all gels, as well as six duplicated QC samples from the GENECARD study, were included for each set of 84 samples. QC mismatches identified technical difficulties or systematic marker-reading errors that were resolved prior to analysis. Each marker was tested for deviations from Hardy-Weinberg equilibrium (HWE) in the 438 probands, with particular attention given to deviations from the expected rate of homozygosity. Markers that showed deviations from HWE were examined for technical problems, such as the presence of a null allele. A total of 395 (98.3%) of 402 markers attempted passed the QC tests and were included in the genome screen analysis. The mean genotyping efficiency (proportion of nonzero genotypes over the 395 markers) was 97.6%. By use of data from several large studies performed at the Duke Center for Human Genetics, we estimated an error rate in sample processing and allocation of 0.14%, and we estimated the genotyping error rate to be ∼0.8%.
The program PedCheck (O’Connell and Weeks 1998) was used to identify Mendelian inconsistencies. If an inconsistency persisted after rereading of the gel, the genotypes for that marker were removed for the entire family. If the family had multiple pedigree inconsistencies involving ⩾10 markers, the family was removed from analysis. Since the majority of GENECARD families were ASPs without parents, pedigree error detection by use of Mendelian inheritance inconsistencies was limited. To overcome this limitation, we used the RELPAIR software package (Boehnke and Cox 1997; Epstein et al. 2000) on 395 markers. RELPAIR identified the likely biological relationship between pairs of individuals by use of the entirety of the marker data. A total of 24 families with misspecified relationships were identified in the GENECARD genome screen set. Of these families, 10 were also identified with Mendelian inconsistencies by use of PedCheck. The remaining 14 families (3.2%) were single ASPs that were identified as likely half-sibling pairs. After all pedigree QC checks, pedigrees were corrected on the basis of pedigree drawings for six families. Eighteen families were removed from the data set, leaving 420 families for the final analysis.
The families were collected from six sites in the United States and Europe. It is possible that they represent genetically distinct subpopulations. To test for population substructure, we used the methods implemented in Structure (Pritchard et al. 2000) and in Arlequin (Schneider et al. 2000), by use of an indicator for each site. There was no evidence from either analysis that the sites could be distinguished on the basis of allele frequencies at the 395 markers in the genome scan. On the basis of these results, we estimated allele frequencies from the family members in the entire sample, as suggested by Broman (2001). The majority of families were white (n=391 [93.1%]). Small numbers of families prevented analysis of each ethnic group separately, but we performed a genome screen with samples from only the white families. Results from this analysis were very similar to the results for all families, and thus we report the linkage results for all families combined.
The genetic complexity of CAD required a multi-analytic strategy that would capture all of the linkage information in the families. We used Genehunter-Plus (Kong and Cox 1997) for nonparametric nuclear-family analysis, and VITESSE (O’Connell and Weeks 1995) and HOMOG (Ott 1991) were used for two-point affected individuals–only parametric linkage analysis incorporating genetic heterogeneity (MLOD). Two parametric models were used: a recessive model with a disease-allele frequency of 0.20 and a rare dominant model with a disease-allele frequency of 0.01. The MLOD was defined as the maximum from these two models. Multipoint linkage analysis was performed by use of Genehunter-Plus (LOD*) (Kong and Cox 1997). The Marshfield genetic linkage maps were used to estimate intermarker distances (Broman et al. 1998). We compared results from all analyses to assess the consistency in linkage evidence. We defined our most interesting regions as those with parametric two-point MLOD results >1.5 and nonparametric multipoint LOD* results >1.0 (Hauser et al. 1996; Holmans and Craddock 1997). In addition, we required that the MLOD and LOD* results be within 5 cM of each other. The LOD score cut points we chose were lower than the LOD scores defined by Lander and Kruglyak (1995) as meeting a genomewide significance level (LOD>3.6; P<2×10-5), which is known to be conservative (Holmans and Craddock 1997). We chose lower LOD score cut points to avoid excluding any important loci in the first stage of the analysis, even though this meant accepting the risk of including some false-positive regions in the follow-up analyses. To further evaluate the significance of regions meeting our LOD score cutoffs, we used the pairs-score LOD score statistic implemented in Merlin (Abecasis et al. 2002), and we used SIMLA (Bass et al. 2004) with Genehunter-Plus (Kong and Cox 1997) to estimate empirical P values by use of the observed family structures, marker-allele frequencies, marker-map parameters, and missing-data patterns. Both methods of estimating the P values gave consistent results. However, since the use of SIMLA together with Genehunter-Plus for analysis is under development, we report the Merlin empirical P values. To evaluate genomewide significance, we used a method of locus counting that counts the number of independent linkage regions (ILR) above a specified LOD score threshold for each genome screen (Wiltshire et al. 2002). For the 1,000 data sets simulated by Merlin from the null hypothesis of no linkage, we estimated the LOD score at which 5% of the replicates had no more than 1 ILR.
To reduce etiologic heterogeneity due to phenotypic characteristics or the presence of other genetic risk factors and to be able to compare our results to other published genome screens, three phenotypic stratifications of our families were developed by the investigators prior to the linkage analysis. These stratifications were also chosen to isolate what may be major etiologic classifications of CAD (e.g., acute coronary syndrome vs. stable occlusive coronary disease, coronary disease not associated with diabetes, and CAD associated with atherogenic dyslipidemias). We defined three subgroups to attempt to isolate CAD-specific effects and to match other patient populations: (1) presence of acute coronary syndrome (ACS), defined as diagnosis of MI or unstable angina, in at least two affected siblings (228 families); (2) presence of atherogenic dyslipidemia (ADL) (Grundy 1995), defined as concomitant presence of plasma triglyceride levels above the 75th percentile for age and sex and high-density lipoprotein (HDL) cholesterol levels below the 25th percentile for age and sex, in at least one member of the nuclear family (146 families); and (3) no diabetes in any affected member of the family (NDIA) (274 families). There is overlap, in that some families appear in more than one group, with 50 families in all groups. However, there was no evidence for association of any pair of strata among the families (P>.34 for all comparisons). The linkage analysis of CAD was performed in each of these subsets, and the subset results were compared with the results for all families. As reported in the GENECARD study-design paper, all sites were remarkably similar with respect to the distribution of well-accepted CAD risk factors (Hauser et al. 2003). The distribution of families from each site contributing to each subset is included in table 1.
Results
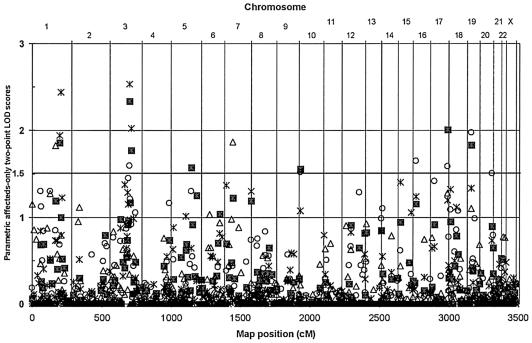
After the exclusions noted above, 420 families with early-onset CAD passed the pedigree QC screen and linkage analysis was performed by use of 395 microsatellite markers that passed the marker QC screen. Figure 1 shows the two-point MLOD results for all 395 markers arranged in marker-map order. Table 2 shows the maximum multipoint LOD* linkage and location estimates (in cM) for each chromosome. (Tables showing the two-point results and multipoint linkage plots for all chromosomes are available on the Duke Center for Human Genetics Web site.) In the overall GENECARD family sample, six regions were identified with a two-point MLOD score of >1.5 (chromosomes 1q25, 3q13, 5q31, 10p15, 17q25, and 19p13) (see fig. 1), and four regions were identified with a multipoint LOD* score >1.0 (chromosomes 3q13, 5q31, 6q24, and 7p15) (see table 2). By use of the criteria for follow-up described above, two regions were identified. Chromosome 3q13 achieved an MLOD score of 2.3 at D3S2460 and a LOD* score of 3.5 near D3S2460 at 140 cM (fig. 2A). The pairs-score LOD score was 3.28, and the empirical P value for this result was <.001. The genomewide significant LOD score (P<.05) under the null hypothesis of no linkage for the set of all families was 2.70, and thus the chromosome 3q13 region meets the criteria for genomewide significance. Chromosome 5q31 achieved an MLOD score of 1.57 at D5S816 and a LOD* score of 1.4 at 154.5 cM (fig. 2B). The pairs-score LOD score was 1.37, with an empirical P value of .081. The chromosome 5q31 region did not achieve genomewide significance. For the other regions with MLOD scores >1.5, the multipoint LOD* results did not reach the cut point of 1.0, and, for the regions on chromosomes 6 and 7 with LOD* scores >1.0, the MLOD result did not reach 1.5 (tables 2 and 3).
Figure 1.
Parametric LOD scores for affected subjects, allowing for genetic heterogeneity (MLOD). Results for all 395 markers typed in the GENECARD study are shown for four groups: all families (squares), families with ACS (stars), families with ADL (triangles), and families with NDIA (circles).
Table 2.
Multipoint Maximum LOD Scores (and Location of Maximum in cM) on Each Chromosome for GENECARD Families [Note]
|
Maximum LOD Score (and Location in cM) for |
||||
| Chromosome | All Families(n = 420) | Families with ACS(n = 228) | Families with ADL(n = 146) | Families with NDIA (n = 274) |
| 1 | .95 (198) | 2.17 (202) | 1.20 (3) | 1.11 (62) |
| 2 | .31 (229) | .39 (46) | .77 (44) | .54 (245) |
| 3 | 3.50 (140) | 3.16 (140) | .54 (121) | 2.42 (140) |
| 4 | .36 (189) | .56 (180) | .31 (189) | .95 (189) |
| 5 | 1.39 (154) | .82 (16) | .31 (98) | .98 (153) |
| 6 | 1.00 (142) | .84 (160) | 1.08 (69) | .33 (140) |
| 7 | 1.04 (32) | .77 (194) | 1.61 (55) | 1.00 (32) |
| 8 | .59 (114) | .42 (119) | .36 (116) | .53 (108) |
| 9 | .04 (106) | .56 (106) | .04 (115) | .07 (80) |
| 10 | .09 (7.9) | .13 (5) | .30 (152) | .26 (8) |
| 11 | .00 (…) | .23 (12) | .45 (50) | .02 (12) |
| 12 | .25 (163) | .27 (163) | .89 (32) | .45 (140) |
| 13 | .01 (114) | .12 (114) | .03 (103) | .00 (…) |
| 14 | .62 (0) | .38 (0) | .70 (71) | 1.06 (4) |
| 15 | .06 (16.7) | .13 (17) | .03 (17) | .00 (…) |
| 16 | .20 (21) | .79 (21) | .17 (82) | .37 (21) |
| 17 | .97 (123) | .67 (123) | 1.17 (123) | .76 (123) |
| 18 | .97 (51) | 1.01 (51) | 1.14 (75) | 1.20 (54) |
| 19 | .86 (44) | .35 (44) | .66 (44) | 1.72 (44) |
| 20 | .67 (102) | .42 (90) | .23 (102) | 1.08 (102) |
| 21 | .18 (58) | .23 (55) | .00 (…) | .05 (35) |
| 22 | .05 (14) | .21 (36) | .38 (24) | .31 (0) |
| X | .04 (60) | .14 (60) | .00 (…) | .00 (…) |
Note.— LOD* values >1.0 are in bold italics.
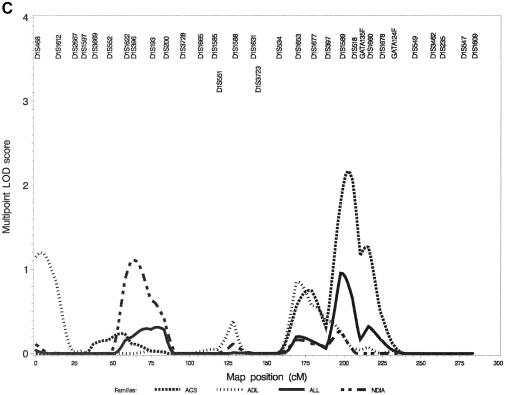
Figure 2.
Multipoint nonparametric LOD score (LOD*) curves for all of the GENECARD families (solid lines) and for the three phenotypic subsets (dashed lines [see legend]). Marker names and locations are indicated across the top. A, Chromosome 3. B, Chromosome 5. C, Chromosome 1.
Table 3.
Support Intervals for Linkage Peaks Identified in the GENECARD Study for GENECARD Families
| Chromosomal Regionand Family Subset(s) | LOD* | 1-LOD Unit DownSupport Interval(cM) |
| 3q13: | ||
| All families | 3.50 | 134–152 |
| Families with ACS | 3.16 | 134–152 |
| Families with NDIA | 2.42 | 112–154 |
| 5q31: | ||
| All families | 1.4 | 138–205 |
| 1q25: | ||
| Families with ACS | 2.17 | 194–217 |
| 7p14: | ||
| Families with ADL | 1.61 | 42–71 |
| 19p13: | ||
| Families with NDIA | 1.72 | 22–35 |
The subset of patients with ACS gave two regions with MLOD scores >1.5 (chromosomes 1q25 and 3q13) and three regions with LOD* scores >1.0 (chromosomes 1q25, 3q13, and 19p13), with the regions on chromosomes 1 and 3 meeting the criteria for follow-up. The region on chromosome 1q25 achieved an MLOD result of 2.44 at D1S518, with a LOD* result of 2.17 nearby at 202.4 cM (fig. 2C). It is interesting that the Merlin pairs-score LOD score, although maximizing at the same location, was somewhat higher—2.85, with an empirical P value of .006. The locus-counting method gave a genomewide significant LOD score of 2.70 (P<.05), under the null hypothesis of no linkage. The region on chromosome 3 provided maximum LOD scores at the same location as in the overall group, with a maximum LOD score of 3.16 and an empirical P value of .002. Both of these regions met the criteria for genomewide significance in the subset of patients with ACS.
The subset of patients with ADL gave two regions with MLOD scores >1.5 (chromosomes 1q23 and 7p14) and five regions with LOD* scores >1.0 (chromosomes 1p36, 6p12, 7p14, 17q25, and 18q21). The chromosome 7p14 MLOD score was 1.87 at D7S2846, and the maximum LOD* score was 1.61 at 55.3 cM (empirical P value = .061). The LOD score cutoff for genomewide significance was 2.80.
The subset of patients with NDIA provided six regions with MLOD scores >1.5 (chromosomes 3q13, 10p15, 16p13, 17q25, 19p13, and 20q13) and seven regions with LOD* scores >1.0 (chromosomes 1p22, 3q13, 7p21, 14q11, 18q11, 19p13, and 20q13). Again, the results on chromosome 3 were consistent with the results in the overall group, with an MLOD score of 1.59 at D3S2460 and a LOD* score of 2.42 at 140 cM (empirical P value = .025). The region on chromosome 19 gave an MLOD score of 1.98 at D19S252, with a LOD* score of 1.72, also at D19S252 (44 cM). However, the Merlin pairs-score LOD score was only 0.53, with an empirical P value of .364. Although chromosome 20 provided LOD scores above the cut points for each analysis, the locations were >10 cM apart: MLOD=1.50 at D20S102 (90.4 cM) and LOD* = 1.08 at 102 cM. Again, none of the regions in the subset of patients with NDIA met the criteria for genomewide significance (P<.05), with a LOD score cutoff of 2.80.
The results on chromosome 3 are particularly notable because the evidence for linkage was strong and consistent (multipoint LOD=3.50 at 140 cM; empirical P value <.001) in the overall sample, as well as in phenotypic subsets (fig. 2A). This region meets the criteria for genomewide significance (P<.05) in the set of all families, as well as in the subset of patients with ACS. Furthermore, the LOD score from the multipoint analysis exceeded the LOD score from the two-point analysis, which suggests that the markers surrounding D3S2460 increase the evidence for linkage. The peaks were broad, with LOD scores consistently >1.0 from D3S2406 to D3S2322 (112–146 cM) for all families, as well as for the families with ACS and NDIA. The families with ADL provided no evidence for linkage in this region on chromosome 3. However, the LOD score in all families was higher than the LOD score in any of the phenotypic subsets, which suggests that the evidence for linkage resides in the collection of all GENECARD families.
The evidence for linkage on chromosome 5 (fig. 2B) was also stronger in the entire GENECARD sample set than in any of the subsets. However, the evidence appears to derive predominantly from the families with NDIA. The region was large, from D5S505 to D5S1471, covering nearly 50 cM, with flat LOD scores across this region. Furthermore, the microsatellite marker map was less dense in this region, with an intermarker distance of >20 cM between D5S1471 and D5S1354, which suggests that adding more markers will better define the linkage region.
The results for chromosome 1 (fig. 2C) illustrate that phenotypic stratification may increase evidence for linkage in genetically homogeneous subgroups in genomic regions with modest evidence for linkage in the entire sample set. Analyses of this chromosome yielded LOD scores above the cut points for several strata; however, the location estimates were very different. The results for the entire GENECARD sample were modest, with the maximum LOD score (LOD* = 0.95) at 197 cM. When stratified by ACS, this modest maximum in the overall group became a well-defined LOD* peak, with a maximum of 2.17. It is interesting that the Merlin pairs-score LOD score was somewhat larger (2.85) and met the criteria for genomewide significance (P<.05) in the families with ACS. This difference suggests considerable variability in linkage information, perhaps related to family size. Analysis of this region on chromosome 1 has revealed a gene for familial combined hyperlipidemia (FCH), upstream transcription factor USF1. To evaluate whether this peak was a result of a subset of families with FCH, we evaluated lipid values in the GENECARD families and identified 12 families with lipid values characteristic of FCH. These 12 families did not account for the linkage evidence on chromosome 1, either as a group or individually. A LOD* score of 1.20 at 2.7 cM in the group with ADL and a LOD* score of 1.11 at 62 cM in the group with NDIA suggest the possibility of three distinct regions of linkage on chromosome 1.
Discussion
We have completed the first phase of the genetic analysis of CAD in 420 of the GENECARD families. The most significant result was a nonparametric multipoint LOD score of 3.5 on chromosome 3q13, which meets the criteria for genomewide significance. Regions of chromosome 3q have been implicated in three other genome screens for CAD (Francke et al. 2001; Broeckel et al. 2002; Harrap et al. 2002). However, in each of these genome screens, the evidence of linkage to CAD is >60 cM distal to the peak in the GENECARD analysis, as estimated by a meta-analysis (Chiodini and Lewis 2003). It is interesting that this peak is also located in a region that was identified in a genome screen of metabolic syndrome (Kissebah et al. 2000). Quantitative-trait linkage analysis of plasma lipids has shown evidence of linkage with triglyceride-HDL ratio, which provides a LOD score of 1.8 in our peak region (Shearman et al. 2000). There is also evidence for linkage to HDL itself (Imperatore et al. 2000; Coon et al. 2001) and fractionated low-density lipoprotein (LDL) particles (Rainwater et al. 1999) in this region. The location of these QTL linkage results in the proximity of the GENECARD peak is particularly interesting, since there is very little evidence for linkage in the subset of patients with ADL. Together these results provide consistent evidence that one or more genes for CAD reside on chromosome 3q.
In addition to the chromosome 3 region, several other regions of linkage include interesting candidate genes. Despite the small numbers of families with apparent FCH in the GENECARD sample, it would be useful to examine the USF1 gene in patients with CAD (Pajukanta et al. 2004). Also in the 1q region are the C-reactive protein (CRP) precursor locus and the P-selectin and E-selectin loci, all excellent candidates for early-onset CAD, especially in the context of ACS. There is evidence for a locus on chromosome 1q from QTL linkage analysis of lipid phenotypes (Shearman et al. 2000; Broeckel et al. 2002) and from genome screens of diabetes (Wiltshire et al. 2001; Hsueh et al. 2003; Xiang et al. 2004) at approximately the same location as the GENECARD 1q peak. However, the evidence in the accumulated CAD scans performed to date is not nearly as compelling for chromosome 1q as it is for chromosome 3q.
To date, there have been five published genome screens for CAD (Pajukanta et al. 2000; Francke et al. 2001; Broeckel et al. 2002; Harrap et al. 2002; Wang et al. 2004) that have highlighted six chromosomal regions in addition to 3q. The GENECARD families did not provide consistent evidence of linkage at 2q21-22 or Xq23-26 (Pajukanta et al. 2000), 2q36 (Harrap et al. 2002), or 14q (Broeckel et al. 2002). For the region on 16p13 identified by Francke et al. (2001), MLOD scores of 1.16, 1.23, and 1.65 (marker D16S764) were obtained in all GENECARD families, families with ACS, and families with NDIA, respectively. However, these LOD scores achieved neither empirical P values <.05 nor genomewide significance. In addition, the multipoint analysis did not provide similar evidence for linkage (LOD* = 0.20) at this locus. A recent study by Wang et al. (2004) identified a strong signal at chromosome 1p34-36 in a set of families with early-onset MI, in approximately the same region on chromosome 1 as the peak in the GENECARD subset of patients with ADL. As a group, these linkage studies do not provide consistent evidence for linkage in any single region, although consistencies are seen across pairs of studies. Possible reasons for the differences in linkage evidence for these regions include differences in the genetic maps and marker density, as well as phenotypic or population differences, such as genetic heterogeneity, which may lead to variable evidence for linkage across the five studies. Additional fine mapping in these regions will be required to clarify the linkage evidence in the GENECARD families. Given the results from the stratified analysis in the GENECARD study, as well as the differences among the various genome screens, it is likely that close attention to the clinical and metabolic characteristics of CAD will be required.
Through our subgroups of patients with CAD, we have attempted to address some of the heterogeneity believed to be behind the difficulties in replication of genetic studies of CAD. Although we hope that this strategy will allow genetic effects to be more easily detected in a homogeneous subgroup, we also run the risk of reducing statistical power in our sample, as well as increasing the variability in the results, as we observed when the MLOD, LOD*, and Merlin pairs-score statistics were compared. The addition of another set of 400 families, along with collaboration among the different groups to identify phenotypic characteristics related to linkage evidence, provides hope that we can establish regions of consistency and detect smaller genetic effects. Furthermore, understanding how phenotypic heterogeneity relates to evidence for linkage will provide more homogeneous subsets in which to perform additional genetic studies. More analyses, including ordered subset analysis (Hauser et al. 2004) and quantitative-trait linkage analysis, will be required to understand the possible relationship between evidence for linkage and metabolic risk factors such as lipid levels and insulin resistance. The relationships will need to be examined not only within the GENECARD families but also across the different family sets assembled for analysis of CAD (Pajukanta et al. 2000; Francke et al. 2001; Broeckel et al. 2002; Harrap et al. 2002; Wang et al. 2004), hyperlipidemias (Naoumova et al. 2003; Pajukanta et al. 2004), metabolic syndrome (Kissebah et al. 2000; Tang et al. 2003), and diabetes (Ghosh et al. 2000; Watanabe et al. 2000; Du et al. 2001; Wiltshire et al. 2001; Hsueh et al. 2003; Xiang et al. 2004), conditions that may share genetic factors.
Review of the GENECARD family histories reveals a significant burden of disease. The individuals sampled represent the CAD survivors, and thus we are not able to include individuals in whom CAD presents as sudden death. This is a limitation in any of the CAD or MI genome screens. One way to examine the full spectrum of CAD is to study the offspring of the GENECARD probands and siblings prior to the onset of disease. These offspring are at very high risk of CAD but have not had clinical symptoms. By studying the offspring prospectively, we can evaluate the risk of sudden death. In addition to facilitating the study of the genetic basis for CAD, this approach will also allow us to examine the metabolic risk factors prior to treatment that appear to contribute to CAD in these families (Jomini et al. 2002; Hauser et al. 2003).
This work represents one of the largest systematic analyses of the genetics of CAD and provides a solid basis for future candidate-gene analysis. The region 3q13 provides strong evidence for the existence of genes for CAD in the GENECARD families. Interesting results were also obtained for chromosome 5q31 and 1q25, although work remains to be done to establish the genetic effects in these regions. The genes ultimately identified in these regions may drive or contribute to major pathophysiological processes that lead to CAD—processes that are either already established or hitherto unrecognized. A greater understanding of the genetic basis of the disease could potentially lead to the development of novel preventive or therapeutic strategies for CAD. Fine mapping, candidate-gene analyses, and analyses of phenotypic heterogeneity are under way and represent the next steps toward the realization of that potential.
Acknowledgments
We gratefully acknowledge the participation and enthusiasm of the families who gave their DNA samples for this work, and we thank the medical caregivers who helped us collect the primary data sources (Hauser et al. 2003). We are grateful for the assistance of staff members at each of the clinical collection sites, GlaxoSmithKline, and the Center for Human Genetics at Duke University. This work was supported by contracts to the participating institutions from GlaxoSmithKline and by grants from the National Institutes of Health (HL073389 and MH059528 [to E.R.H.]).
Electronic-Database Information
The URL for data presented herein is as follows:
- Duke Center for Human Genetics, http://www.chg.duke.edu/research/support.html
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin: rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- American Heart Association (2004) Heart and stroke statistical update. American Heart Association, Dallas [Google Scholar]
- Bass MP, Martin ER, Hauser ER (2004) Pedigree generation for analysis of genetic linkage and association. In: Altman RB, Dunker AK, Hunter L, Jung TA, Klein TE (eds) Pacific symposium on biocomputing 2004. World Scientific, Hackensack, NJ [DOI] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ (1997) Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow JL (2000) Genetics of lipoprotein abnormalities associated with coronary artery disease susceptibility. Annu Rev Genet 34:233–254 10.1146/annurev.genet.34.1.233 [DOI] [PubMed] [Google Scholar]
- Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, Comuzzie AG, Blangero J, Nurnberg P, Reis A, Riegger GA, Jacob HJ, Schunkert H (2002) A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet 30:210–214 10.1038/ng827 [DOI] [PubMed] [Google Scholar]
- Broman KW (2001) Estimation of allele frequencies with data on sibships. Genet Epidemiol 20:307–315 10.1002/gepi.2.abs [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individuals and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodini BD, Lewis CM (2003) Meta-analysis of 4 coronary heart disease genome-wide linkage studies confirms a susceptibility locus on chromosome 3q. Arterioscler Thromb Vasc Biol 23:1863–1868 10.1161/01.ATV.0000093281.10213.DB [DOI] [PubMed] [Google Scholar]
- Coon H, Leppert MF, Eckfeldt JH, Oberman A, Myers RH, Peacock JM, Province MA, Hopkins PN, Heiss G (2001) Genome-wide linkage analysis of lipids in the Hypertension Genetic Epidemiology Network (HyperGEN) blood pressure study. Arterioscler Thromb Vasc Biol 21:1969–1976 [DOI] [PubMed] [Google Scholar]
- Du W, Sun H, Wang H, Qiang B, Shen Y, Yao Z, Gu J, Xiong M, Huang W, Chen Z, Zuo J, Hua X, Gao W, Sun Q, Fang F (2001) Confirmation of susceptibility gene loci on chromosome 1 in northern China Han families with type 2 diabetes. Chin Med J (Engl ) 114:876–878 [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M (2000) Improved inference of relationship for pairs of individuals. Am J Hum Genet 67:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke S, Manraj M, Lacquemant C, Lecoeur C, Lepretre F, Passa P, Hebe A, Corset L, Yan SL, Lahmidi S, Jankee S, Gunness TK, Ramjuttun US, Balgobin V, Dina C, Froguel P (2001) A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet 10:2751–2765 10.1093/hmg/10.24.2751 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, et al (2000) The Finland-United States investigation of non–insulin-dependent diabetes mellitus genetics (FUSION) study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet 67:1174–1185 [PMC free article] [PubMed] [Google Scholar]
- Grundy SM (1995) Atherogenic dyslipidemia: lipoprotein abnormalities and implications for therapy. Am J Cardiol 75:45B–52B 10.1016/0002-9149(95)80011-G [DOI] [PubMed] [Google Scholar]
- Harrap SB, Stebbing M, Hopper JL, Hoang HN, Giles GG (2000) Familial patterns of covariation for cardiovascular risk factors in adults: the Victorian Family Heart Study. Am J Epidemiol 152:704–715 10.1093/aje/152.8.704 [DOI] [PubMed] [Google Scholar]
- Harrap SB, Zammit KS, Wong ZY, Williams FM, Bahlo M, Tonkin AM, Anderson ST (2002) Genome-wide linkage analysis of the acute coronary syndrome suggests a locus on chromosome 2. Arterioscler Thromb Vasc Biol 22:874–878 10.1161/01.ATV.0000016258.40568.F1 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Boehnke M, Guo SW, Risch N (1996) Affected-sib-pair interval mapping and exclusion for complex genetic traits: sampling considerations. Genet Epidemiol 13:117–137 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Mooser V, Crossman DC, Haines JL, Jones CH, Winkelmann BR, Schmidt S, Scott WK, Roses AD, Pericak-Vance MA, Granger CB, Kraus WE (2003) Design of the genetics of early onset cardiovascular disease (GENECARD) study. Am Heart J 145:602–613 10.1067/mhj.2003.13 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M (2004) Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol 27:53–63 10.1002/gepi.20000 [DOI] [PubMed] [Google Scholar]
- Holmans P, Craddock N (1997) Efficient strategies for genome scanning using maximum-likelihood affected-sib-pair analysis. Am J Hum Genet 60:657–666 [PMC free article] [PubMed] [Google Scholar]
- Hsueh WC, St Jean PL, Mitchell BD, Pollin TI, Knowler WC, Ehm MG, Bell CJ, Sakul H, Wagner MJ, Burns DK, Shuldiner AR (2003) Genome-wide and fine-mapping linkage studies of type 2 diabetes and glucose traits in the Old Order Amish: evidence for a new diabetes locus on chromosome 14q11 and confirmation of a locus on chromosome 1q21-q24. Diabetes 52:550–557 [DOI] [PubMed] [Google Scholar]
- Imperatore, G, Knowler WC, Pettitt DJ, Kobes S, Fuller JH, Bennett PH, Hanson RL (2000) A locus influencing total serum cholesterol on chromosome 19p: results from an autosomal genomic scan of serum lipid concentrations in Pima Indians. Arterioscler Thromb Vasc Biol 20:2651–2656 [DOI] [PubMed] [Google Scholar]
- Jomini V, Oppliger-Pasquali S, Wietlisbach V, Rodondi N, Jotterand V, Paccaud F, Darioli R, Nicod P, Mooser V (2002) Contribution of major cardiovascular risk factors to familial premature coronary artery disease: the GENECARD project. J Am Coll Cardiol 40:676–684 10.1016/S0735-1097(02)02017-X [DOI] [PubMed] [Google Scholar]
- Katzmarzyk PT, Malina RM, Perusse L, Rice T, Province MA, Rao DC, Bouchard C (2000) Familial resemblance in fatness and fat distribution. Am J Human Biol 12:395–404 [DOI] [PubMed] [Google Scholar]
- Kissebah AH, Sonnenberg GE, Myklebust J, Goldstein M, Broman K, James RG, Marks JA, Krakower GR, Jacob HJ, Weber J, Martin L, Blangero J, Comuzzie AG (2000) Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci USA 97:14478–14483 10.1073/pnas.97.26.14478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lusis AJ (2000) Atherosclerosis. Nature 407:233–241 10.1038/35025203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis AJ, Rotter JI, Sparkes RS (eds) (1992) Molecular genetics of coronary artery disease: candidate genes and processes in atherosclerosis. Karger, Los Angeles [Google Scholar]
- Marenberg, ME, Risch N, Berkman LF, Floderus B, De Faire U (1994) Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med 330:1041–1046 10.1056/NEJM199404143301503 [DOI] [PubMed] [Google Scholar]
- Naoumova RP, Bonney SA, Eichenbaum-Voline S, Patel HN, Jones B, Jones EL, Amey J, Colilla S, Neuwirth CK, Allotey R, Seed M, Betteridge DJ, Galton DJ, Cox NJ, Bell GI, Scott J, Shoulders CC (2003) Confirmed locus on chromosome 11p and candidate loci on 6q and 8p for the triglyceride and cholesterol traits of combined hyperlipidemia. Arterioscler Thromb Vasc Biol 23:2070–2077 10.1161/01.ATV.0000095975.35247.9F [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1995) The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet 11:402–408 [DOI] [PubMed] [Google Scholar]
- ——— (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1991) Analysis of human genetic linkage. Rev ed. Johns Hopkins University Press, Baltimore [Google Scholar]
- Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Sato H, Hori M, Nakamura Y, Tanaka T (2002) Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat Genet 32:650–654 10.1038/ng1047 [DOI] [PubMed] [Google Scholar]
- Pajukanta P, Cargill M, Viitanen L, Nuotio I, Kareinen A, Perola M, Terwilliger JD, Kempas E, Daly M, Lilja H, Rioux JD, Brettin T, Viikari JS, Ronnemaa T, Laakso M, Lander ES, Peltonen L (2000) Two loci on chromosomes 2 and X for premature coronary heart disease identified in early- and late-settlement populations of Finland. Am J Hum Genet 67:1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, Gentile M, Duan XJ, Soro-Paavonen A, Naukkarinen J, Saarela J, Laakso M, Ehnholm C, Taskinen MR, Peltonen L (2004) Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1). Nat Genet 36:371–376 10.1038/ng1320 [DOI] [PubMed] [Google Scholar]
- Perusse L, Rice T, Despres JP, Bergeron J, Province MA, Gagnon J, Leon AS, Rao DC, Skinner JS, Wilmore JH, Bouchard C (1997) Familial resemblance of plasma lipids, lipoproteins and postheparin lipoprotein and hepatic lipases in the HERITAGE Family Study. Arterioscler Thromb Vasc Biol 17:3263–3269 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P (2000) Association mapping in structured populations. Am J Hum Genet 67:170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainwater DL, Almasy L, Blangero J, Cole SA, VandeBerg JL, MacCluer JW, Hixson JE (1999) A genome search identifes major quantitative trait loci on human chromosomes 3 and 4 that influence cholesterol concentrations in small LDL particles. Arterioscler Thromb Vasc Biol 19:777–783 [DOI] [PubMed] [Google Scholar]
- Rice T, Despres JP, Daw EW, Gagnon J, Borecki IB, Perusse L, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C (1997) Familial resemblance for abdominal visceral fat: the HERITAGE family study. Int J Obes Relat Metab Disord 21:1024–1031 10.1038/sj.ijo.0800511 [DOI] [PubMed] [Google Scholar]
- Rissanen AM (1979) Familial occurrence of coronary artery disease: effect of age at diagnosis. Am J Cardiol 44:60–66 10.1016/0002-9149(79)90251-0 [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) Arlequin, v. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva [Google Scholar]
- Shea S, Ottman R, Gabrieli C, Stein Z, Nichols A (1984) Family history as an independent risk factor for coronary artery disease. J Am Coll Cardiol 4:793–801 [DOI] [PubMed] [Google Scholar]
- Shearman AM, Ordovas JM, Cupples LA, Schaefer EJ, Harmon MD, Shao Y, Keen JD, Destefano AL, Joost O, Wilson PW, Housman DE, Myers RH (2000) Evidence for a gene influencing the TG/HDL-C ratio on chromosome 7q32.3- qter: a genome-wide scan in the Framingham study. Hum Mol Genet 9:1315–1320 10.1093/hmg/9.9.1315 [DOI] [PubMed] [Google Scholar]
- Tang W, Miller MB, Rich SS, North KE, Pankow JS, Borecki IB, Myers RH, Hopkins PN, Leppert M, Arnett DK (2003) Linkage analysis of a composite factor for the multiple metabolic syndrome: the National Heart, Lung, and Blood Institute Family Heart Study. Diabetes 52:2840–2847 [DOI] [PubMed] [Google Scholar]
- Vance JM, Ben Othmane K (1998) Methods of genotyping. In: Haines JL, Pericak-Vance MA (eds) Approaches to gene mapping in complex human diseases. Wiley-Liss, New York [Google Scholar]
- Vance JM, Slotterbeck B, Yamaoka L, Haynes C, Roses AD, Pericak-Vance MA (1996) A fluorescent genotyping system for multiple users, flexibility and high output. Am J Hum Genet 59:A239 [Google Scholar]
- Wang Q, Rao S, Shen GQ, Li L, Moliterno DJ, Newby LK, Rogers WJ, Cannata R, Zirzow E, Elston RC, Topol EJ (2004) Premature myocardial infarction novel susceptibility locus on chromosome 1P34–36 identified by genomewide linkage analysis. Am J Hum Genet 74:262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe RM, Ghosh S, Langefeld CD, Valle TT, Hauser ER, Magnuson VL, Mohlke KL, et al (2000) The Finland-United States investigation of non–insulin-dependent diabetes mellitus genetics (FUSION) study. II. An autosomal genome scan for diabetes-related quantitative-trait loci. Am J Hum Genet 67:1186–1200 [PMC free article] [PubMed] [Google Scholar]
- Wiltshire S, Cardon LR, McCarthy MI (2002) Evaluating the results of genomewide linkage scans of complex traits by locus counting. Am J Hum Genet 71:1175–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire S, Hattersley AT, Hitman GA, Walker M, Levy JC, Sampson M, O’Rahilly S, et al (2001) A genomewide scan for loci predisposing to type 2 diabetes in a U.K. population (the Diabetes UK Warren 2 Repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q. Am J Hum Genet 69:553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Wang Y, Zheng T, Jia W, Li J, Chen L, Shen K, Wu S, Lin X, Zhang G, Wang C, Wang S, Lu H, Fang Q, Shi Y, Zhang R, Xu J, Weng Q (2004) Genome-wide search for type 2 diabetes/impaired glucose homeostasis susceptibility genes in the Chinese: significant linkage to chromosome 6q21-q23 and chromosome 1q21-q24. Diabetes 53:228–234 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Izawa H, Ichihara S, Takatsu F, Ishihara H, Hirayama H, Sone T, Tanaka M, Yokota M (2002) Prediction of the risk of myocardial infarction from polymorphisms in candidate genes. N Engl J Med 347:1916–1923 10.1056/NEJMoa021445 [DOI] [PubMed] [Google Scholar]