Abstract
Bardet-Biedl syndrome (BBS) is a genetically heterogeneous, pleiotropic human disorder characterized by obesity, retinopathy, polydactyly, renal and cardiac malformations, learning disabilities, and hypogenitalism. Eight BBS loci have been mapped, and seven genes have been identified. BBS3 was previously mapped to chromosome 3 by linkage analysis in a large Israeli Bedouin kindred. The rarity of other families mapping to the BBS3 locus has made it difficult to narrow the disease interval sufficiently to identify the gene by positional cloning. We hypothesized that the genomes of model organisms that contained the orthologues to known BBS genes would also likely contain a BBS3 orthologue. Therefore, comparative genomic analysis was performed to prioritize BBS candidate genes for mutation screening. Known BBS proteins were compared with the translated genomes of model organisms to identify a subset of organisms in which these proteins were conserved. By including multiple organisms that have relatively small genome sizes in the analysis, the number of candidate genes was reduced, and a few genes mapping to the BBS3 interval emerged as the best candidates for this disorder. One of these genes, ADP-ribosylation factor-like 6 (ARL6), contains a homozygous stop mutation that segregates completely with the disease in the Bedouin kindred originally used to map the BBS3 locus, identifying this gene as the BBS3 gene. These data illustrate the power of comparative genomic analysis for the study of human disease and identifies a novel BBS gene.
Introduction
Bardet-Biedl syndrome (BBS [MIM 209900]) is a pleiotropic autosomal recessive disorder characterized by obesity, pigmentary retinopathy, polydactyly, renal abnormalities, learning disabilities, and hypogenitalism (Bardet 1920; Biedl 1922; Green et al. 1989). The disorder is also associated with an increased susceptibility to diabetes mellitus, hypertension, and congenital heart disease (Harnett et al. 1988; Green et al. 1989; Elbedour et al. 1994). The disorder shows variable expressivity within and between families, a finding suggesting genetic complexity. BBS displays extensive genetic heterogeneity and a requirement for full penetrance of two mutations at one locus and a third mutation at a second locus has been suggested (Katsanis et al. 2001). To date, eight BBS loci have been mapped, and seven BBS genes have been identified. The first three BBS genes to be discovered (BBS6, BBS2, and BBS4) are not homologous to one another and were identified using positional cloning (Katsanis et al. 2000; Slavotinek et al. 2000; Mykytyn et al. 2001; Nishimura et al. 2001). Subsequently, bioinformatics comparisons of protein sequences have aided in the identification of additional BBS genes. For example, the positional cloning of the BBS1 gene was aided by the finding of limited sequence homology to BBS2 (Mykytyn et al. 2002). BBS7 and BBS8 were identified by database searches for proteins with partial homology to BBS2 and BBS4, respectively (Ansley et al. 2003; Badano et al. 2003). BBS5 was recently identified using a combination of comparative genomics and positional cloning (Li et al. 2004).
The BBS3 locus was initially mapped to chromosome 3 in a large, inbred Israeli Bedouin kindred in a study that showed the utility of using pooled DNA samples for genetic mapping of human disorders (Sheffield et al. 1994). Genotyping of the Bedouin DNA samples initially localized the disease interval to an ∼10-cM region. Few additional BBS3-linked families have been identified, and this has limited extensive refinement of the disease locus. The sequencing of the human genome (Lander et al. 2001; Venter et al. 2001), as well as the sequencing of other model organisms, has made it possible to perform cross-species comparisons of genomes (comparative genomics). In the present study, we reasoned that organisms containing orthologues to the known BBS genes were likely to also contain orthologues to as-yet-unidentified BBS genes. Comparisons of the human genome with the genomes of other model organisms, particularly lower eukaryotic organisms with relatively fewer genes, were used to prioritize BBS candidate genes for mutation screening and assisted in the identification of the BBS3 gene.
Material and Methods
Subjects
Signed informed-consent forms, approved by the Institutional Review Board at the University of Iowa and collaborating institutions, were obtained from all study participants on entry into the study. The diagnosis of BBS in the large Bedouin Arab family was based on the presence of at least two of the cardinal features of BBS (obesity, polydactyly, renal anomalies, retinopathy, hypogonadism, and learning disabilities). The diagnosis of BBS in individuals without a family history of the disorder was based on the presence of at least three of the cardinal features of BBS, most commonly obesity, polydactyly, and retinal degeneration. The clinical features of BBS in the Bedouin family have been reported elsewhere (Kwitek-Black et al. 1993; Sheffield et al. 1994; Carmi et al. 1995).
Bioinformatic Analysis
Identification and prioritization of BBS candidate genes in this study used a computational comparative genomics technique with similarities to previous methods (Avidor-Reiss et al. 2004; Li et al. 2004). This approach is briefly described, in general terms, in this section, followed by a description of its implementation. Specific details of the application of this method to the study of BBS can be found in the “Results” section.
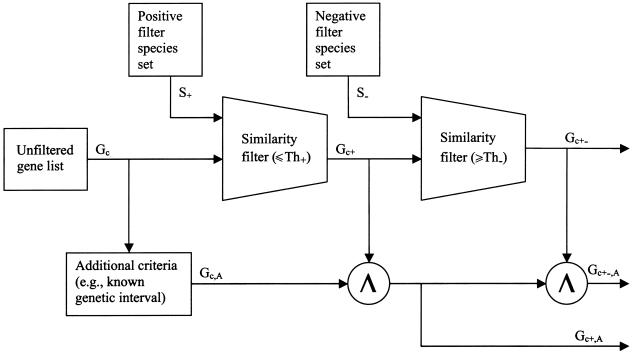
A basic principle of this approach is that specific biological features are manifest in an evolutionarily conserved structural and/or functional characteristic shared among a subset of species. In addition, a subset of species is also identifiable that explicitly lacks this same characteristic. Together, these two subsets of species serve as positive and negative filters, respectively. Employing computational sequence-similarity tools and available protein and genomic sequences, these filters are applied to a candidate set of genes to prioritize them for experimental analysis, such as mutation screening. Figure 1 depicts this process, which is outlined as follows:
-
1.
Initial determination of candidate gene set Gc.
-
2.
Selection of positive and negative sets of species: S+ and S-.
-
3.
Determination of thresholds for similarity filters: Th+ and Th-.
-
4.
Computational application of filters S+ and S- to candidate gene set Gc.
-
a.
Similarity analysis of Gc relative to S+, and retention of candidates exceeding Th+. We refer to this subset as “Gc+”.
-
b.
Similarity analysis of Gc+ relative to S-, and rejection of candidates falling below Th- We refer to this subset as “Gc+-”.
-
a.
-
5.
Application of additional criteria, A, for candidate gene ranking.
-
a. Utilization of known or suspected linkage interval.
-
i. Intersection of the subset of genes in the interval, Gc,A, with Gc+ and Gc+-. We refer to these subsets as “Gc+,A” and “Gc+-,A”.
-
i.
-
b.
Utilization of available expression information.
-
c.
Utilization of other annotations (e.g., Gene Ontology Consortium [GO] biological process, molecular function, cellular component, etc. [Ashburner et al. 2000]).
-
a.
Figure 1.
General outline of the candidate ranking process. A list of unfiltered genes (Gc) can be prioritized through a set of positive filter species (S+) on the basis of a similarity filter threshold (Th+), which yields a subset of genes (Gc+) found in all positive filter species (S+). These can be further screened with similarity filter threshold (Th-) in a set of negative filter species (S-) to yield a more restricive subset of genes (Gc+-). Further filtering continues by intersection (Λ) with additional criteria (A) to generate (Gc+,A). An even more refined set (Gc+-,A) can be obtained by intersecting Gc+,A with Gc+-.
The implementation of this process for identification of BBS3 candidate genes employed BLAST (Altschul et al. 1990) as the similarity-analysis tool. The thresholds (Th+ and Th-) were expressed as BLAST e-values. These values were determined based on similarity scores of the known BBS genes. The test for S+ was implemented as a “less than” comparison with Th+; the test for S− was a “greater than” comparison with Th-. The linkage interval for BBS3 was applied as additional criteria A. The selection of sets S+ and S- was based upon the presence or absence of ciliated structures, as further described in the Results section.
Data Sources
Genomic information was obtained from Ensembl (Caenorhabditis elegans, Danio rerio, Drosophila melanogaster, and Mus musculus), from the U.S. Department of Energy Joint Genome Institute (JGI) (Ciona intestinalis and Chlamydomonas reinhardtii), from The Institute for Genomic Research (TIGR) (Trypanosoma brucei and T. cruzi), from the Saccharomyces Genome Database (SGD) (Saccharomyces cerevisiae), from The Arabidopsis Information Resource (TAIR) (Arabidopsis thaliana), and from the Broad Institute (Aspergillus nidulans). Predicted Ensembl genes for humans (Release 22.34a) were retrieved for human genomewide analysis.
Genotyping
PCR amplification for the analysis of STRPs was performed using 40 ng genomic DNA in 8.4 μl reactions containing 1.25 μl 10× PCR buffer (100 mM Tris-HCl [pH 8.8], 500 mM KCl, 15 mM MgCl2, 0.01% gelatin [w/v]), 200 uM each of dATP, dCTP, dGTP and dTTP, 2.5 pmol of each primer, and 0.2 U of Taq polymerase. Samples were subjected to 35 cycles of 94°C for 30 s; 50, 52, 55, or 57°C (as required) for 30 s; and 72°C for 30 s. Amplification products were electrophoresed on 6% polyacrylamide gels containing 7.7 M urea at 60 W for ∼2 h. The bands were visualized by silver staining. Oligonucleotide primers for the STRPs were obtained as MapPairs (Research Genetics) or were custom designed and synthesized commercially.
DNA Sequencing and Mutation Screening
PCR products for sequencing were amplified in a 25-μl reaction volume and were visualized on a 1.2% agarose gel. The corresponding bands were excised and purified using the QIAquick gel extraction kit (Qiagen). 4.5 μl of purified PCR product was used as template for sequencing reactions using dye-terminator chemistry (Applied Biosystems). PCR product sequencing reactions were precipitated in the presence of glycogen and isopropanol. The reactions were analyzed on an ABI 3730 DNA Sequencer. All sequence variants were verified bidirectionally by direct DNA sequencing and/or by restriction enzyme digestion. Primer sequences used to screen the entire ARL6 gene are available in table A1 (online only).
In some cases, the coding sequence of candidate genes was screened by SSCP analysis. Amplicons for SSCP analysis were designed to be ∼200 bp in size. For SSCP, PCR products were electrophoresed on SSCP gels (7 ml 50% glycerol, 3.5 ml 5× TBE, 8.8 ml 37.5:1 acrylamide/bisacrylamide, and 50.7 ml ddH20) for 3–4 h in 0.5× TBE at room temperature, with the temperature controlled by a cooling fan. The gels were silver stained to visualize DNA bands. Abnormal variants were sequenced and compared with a control sample (CEPH sample 1331–01) to detect any changes from that of the normal sequence.
Results
Genetic Fine Mapping
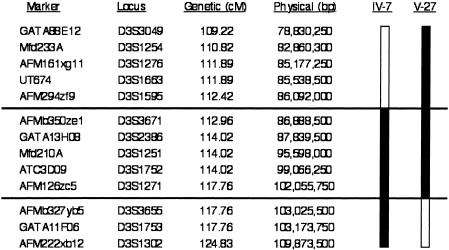
The BBS3 pedigree from a large Israeli Bedouin kindred has been previously published (Sheffield et al. 1994). DNA samples were obtained from the 13 affected individuals in this pedigree and from numerous other individuals in this Bedouin population, including 21 unaffected siblings and 12 parents (some of whom were also siblings of affected individuals). To narrow the disease interval, genotyping with STRPs was performed across the linked interval, focusing on two affected individuals who were not homozygous for all markers across the genetic interval initially reported (Sheffield et al. 1994). Mapping of the homozygous interval in these two individuals allowed us to refine the disease interval to ∼5.3 cM (fig. 2). This interval proved to be a region of lower-than-average recombination, in part because the 16.9-Mb region between the flanking markers (D3S1595 and D3S3655) crosses the centromere. Analysis of the human genome (UCSC Genome Browser) across the BBS3 interval revealed a minimum of 67 UniGene clusters. In an attempt to further narrow the BBS3 interval, we genotyped 54 small kindreds, including many that had a single affected individual, looking for families consistent with linkage to the BBS3 locus and/or for isolated affected individuals that were homozygous across this interval. Only one small pedigree was identified that was consistent with linkage on the basis of a single homozygous affected individual, indicating that BBS3 is a rare cause of BBS.
Figure 2.
Refinement of genetic localization of the BBS3 candidate interval. The genetic map was obtained from the Marshfield Medical Clinic Web site, and the physical map distances were obtained from the UCSC Genome Browser on the basis of the July 2003 data release. Critical recombination events are also illustrated. The patient identification terminology is the same as was published previously (Sheffield et al. 1994).
Comparative Genomic Analysis to Identify BBS Candidate Genes
The existence of several known BBS genes, previously identified by positional cloning, provided the opportunity to use bioinformatics methods to identify additional BBS genes. Since prior studies of BBS4 (Kim et al. 2004), BBS8 (Ansley et al. 2003), and the BBS4 mouse model (Mykytyn et al. 2004) implicated the involvement of BBS proteins in ciliary function, we included within the set of genomes to be analyzed, the genomes of well-studied eukaryotic ciliated organisms (S+), as well as some nonciliated organisms (S-) for which complete genome sequence was available.
The value of Th+ was determined on the basis of BLAST analysis of five known BBS proteins (BBS1, BBS2, BBS4, BBS7, and BBS8) against available genomic sequence from ciliated organisms, including nematode, zebrafish, fruit fly, laboratory mouse, sea squirt, biflagellated green algae, and flagellated parasites. Putative BBS orthologues were identified for many but not all organisms at a Th+ significance level of ⩽e-35. The genome of M. musculus and D. rerio contain BBS orthologues with highly significant expected values, sequence percent identity (>63%), and similarity (>75%) to five known BBS proteins. The genome of D. melanogaster contains only orthologues to BBS1, BBS4, and BBS8, whereas the remaining ciliated organisms (T. brucei, T. cruzi, C. reinhardtii, and C. intestinalis) showed significant expected values (⩽e-40), percent identity (>20%), and similarity (>40%) to BBS1, BBS2, BBS4, BBS7, and BBS8. We reasoned that the smaller genomes (compared with those of humans) of organisms such as T. brucei, T. cruzi, C. reinhardtii, C. intestinalis, and C. elegans, which contain orthologues to BBS genes, should be enriched for additional BBS genes. To further enhance for cilia-related genes, multiple ciliated organisms (T. brucei, T. cruzi, C. reinhardtii, and C. intestinalis) were chosen for comparison with the human genome, to define a set of BBS candidate genes. Similarly, the genomes of S. cerevisiae and A. thaliana, which do not contain putative orthologues of the known BBS genes (S- organisms), were selected to aid in exclusion of genes as potential BBS candidates. A Th- significance level of ⩾e-35 was selected on the basis of these comparisons.
The central step in the bioinformatics methods was the application of the positive and negative filter sets (S+ and S-; Th+ and Th-). We performed a BLAST analysis of the set of 21,184 candidate genes (Gc), as identified in Ensembl (Curwen et al. 2004) against the genomes of C. intestinalis, C. reinhardtii, T. brucei, and T. cruzi. These comparisons resulted in the identification of 1,588 genes (Gc+) in common between the known human genome (Gc) and these model organisms (S+), as shown in table 1. To remove genes that were less likely to be BBS genes, we compared this set of 1,588 genes against the S- genomes of S. cerevisiae and A. thaliana (two genomes that do not contain orthologues to the known BBS genes). This analysis resulted in a refined set of 114 BBS candidates (Gc+-) that were present in the S+ genomes of C. intestinalis, C. reinhardtii, T. brucei, and T. cruzi but were absent in the S- genomes of S. cerevisiae and A. thaliana. None of these 114 genes mapped to the BBS3 interval, as shown in table 1. However, two genes (Hs.388438 and Hs.446399) mapped to the BBS5 locus on chromosome 2q31. During the course of this study, the BBS5 gene was identified by others using a similar comparative genomic strategy (Li et al. 2004). It is of note that the BBS5 gene was one of the two BBS5 candidate genes (Hs.446399) identified in our study. The complete list of 1,588 genes in the Gc+ set and the 114 genes in the Gc+- set are available at the authors' Web site.
Table 1.
Ensembl Genes from Each Stage of the Comparative Genomics Approach[Note]
| All EnsemblGenes | S+ FilterOnly | S+ and S-Filters | |
| All chromosomes | 21,184 (Gc) | 1,588 (Gc+) | 114 (Gc+−) |
| BBS3 interval | 62 (Gc,A) | 4 (Gc+,A) | 0 (Gc+−,A) |
Note.— Each cell contains the intersection of the sets that define the row and the column.
In addition to the whole human genome analysis described above, we performed an intersection of the 1,588 Gc+ candidates with the 62 Ensembl genes contained within the human BBS3-linked interval. This analysis identified four BBS3 candidate genes (Gc+,A), as shown in table 1. We then prioritized the four Gc+,A genes for mutation analysis using two criteria: (1) a broad tissue pattern of expression similar to that of known BBS genes, on the basis of representation in dbEST, and (2) putative function, on the basis of GO data. On the basis of these criteria, candidate genes from the BBS3-linked interval, including a gene known as “ARL6,” were selected for mutation analysis. ARL6 (ADP-ribosylation factor [ARF]–like 6) is a member of a subgroup of the ARF family, proteins that regulate diverse cellular functions including regulation of intracellular traffic (Kahn and Gilman 1984; Price et al. 1988; Sewell and Kahn 1988; Pasqualato et al. 2002).
Mutation Analysis in the Bedouin Family
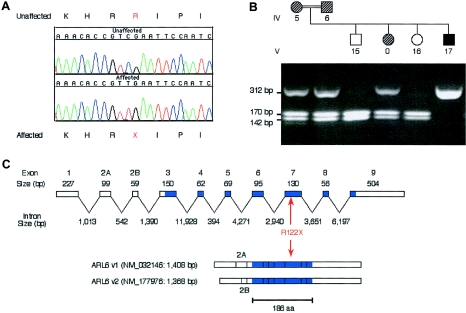
The coding sequence and splice sites from BBS3 candidate genes identified in the phylogenetic comparisons were initially sequenced in DNA amplified from two affected members of the BBS3-linked kindred. No nonsynonymous sequence variants were identified in the coding sequence of Hs.145172, Hs.16986 (FLJ11046), Hs.370688 (TOMM70A), Hs.23294 (MINA), Hs.369885, and Hs.223341. However, sequencing of ARL6 revealed a homozygous stop mutation in exon 7 (R122X) resulting in a predicted truncation of the protein from 186 amino acids to 121 amino acids. This mutation segregates completely with BBS in the Bedouin kindred in that all 13 affected individuals were homozygous for the mutation and each obligate carrier (12 parents of affected individuals) was heterozygous for the mutation. Twenty-one unaffected siblings of BBS patients either were heterozygous for the mutation or were homozygous for the normal allele (fig. 3). To determine whether R122X is a rare polymorphism, we genotyped 100 Arab control individuals (200 chromosomes) from the Middle East and 90 additional control individuals (180 chromosomes) from diverse ethnicities. No R122X alleles were detected in these 380 chromosomes. To exclude the possibility that a short, functional ARL6 isoform exists that excludes exon 7 (the exon containing the R122X mutation) as a functional component, we analyzed the 47 available ARL6 ESTs in dbEST. All ARL6 isoforms identified in this analysis include the complete exon 7 sequence.
Figure 3.
BBS3 mutation (R122X) detected in a large Bedouin kindred. A, Sequence from an affected individual from the Bedouin family and a control sample, showing the homozygous C→T change that results in premature termination at codon 122. B, An example of the TaqI restriction enzyme digest that was used to confirm the R122X mutation. The mutation results in the abolition of a TaqI site within exon 7. Following TaqI digestion of a PCR fragment containing exon 7, the wild-type allele is observed as two bands (142 bp and 170 bp), whereas the uncut mutant allele produces a 312-bp fragment. For the pedigree, the hatched symbols represent BBS carriers, as determined by genetic analysis; the filled symbol denotes an individual with BBS; and the open symbols are unaffected individuals. The patient-identification terminology is the same as was published previously (Sheffield et al. 1994), with the exception of the 0, which denotes a sample that was not previously available. C, The genomic structure of the ARL6 gene is shown, with the blue shading representing the translated region. The two ARL6 isoforms that are produced by alternative splicing are shown below. The location of the R122X mutation within the ARL6 gene is indicated in red.
In addition to the mutation analysis performed in the BBS3 Bedouin kindred, we performed mutation screening of 90 unrelated BBS patients, including a single patient that was shown to be homozygous across the BBS3 genetic interval. No nonsynonymous coding sequences were identified, consistent with previous genotyping data that indicate that BBS3 is a rare cause of the disorder.
Discussion
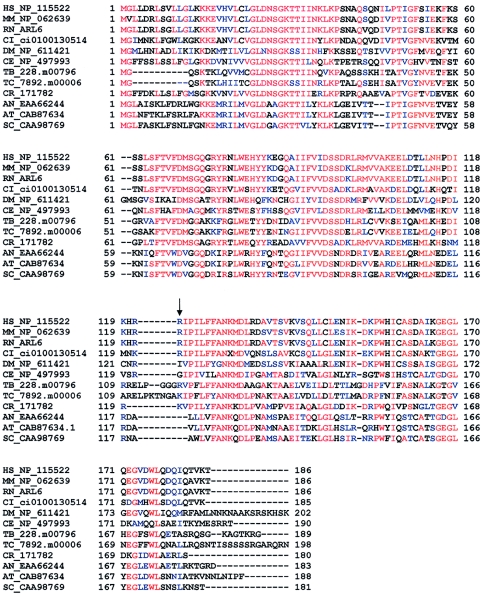
The identification of the BBS3 gene presented challenges, because of the extensive genetic heterogeneity of BBS, the paucity of families mapping to the BBS3 locus, and the diversity of BBS genes. Analysis of the BBS3 genetic interval in an attempt to find candidate genes, on the basis of homology to the known BBS genes, failed to produce attractive candidate genes. Genetic mapping narrowed the disease interval to a region containing at least 62 genes. To identify and prioritize the best BBS candidate genes, we used a comparative genomics approach to select genes shared by organisms harboring orthologues to known BBS genes. Sequencing of DNA from BBS patients in the large Bedouin kindred that was initially used to map the BBS3 locus, revealed that each affected individual harbored a homozygous stop mutation (R122X) in one of the genes identified in this manner, ARL6. R122X results in the predicted truncation of the protein from 186 to 121 residues, including many highly conserved amino acids (fig. 4). These data convincingly demonstrate that ARL6 is the BBS3 gene. The identification of mutations only in the large previously linked BBS3 Bedouin family is consistent with previous reports of the rarity of BBS3 as a cause of this syndrome (Bruford et al. 1997). The reported tissue-expression pattern of ARL6 (Jacobs et al. 1999) and analysis of ESTs from dbEST indicate that ARL6 is widely expressed in human tissues including brain, eye, heart, and kidney, further supporting this gene as a cause of BBS.
Figure 4.
Multiple alignment of ARL6 (HS_NP_115522) and the corresponding best BLAST hit in 11 other model organisms. The mutation (R122X) is denoted by an arrow. Each sequence is denoted by the first letter of the genus-species name, followed by a GenBank accession number (whenever possible) or a unique identifier (e.g., “DM_NP_611421” refers to the protein represented by NP_611421 in the genome of Drosophila melanogaster). Consensus residues are shown in red; conserved residues are shown in blue. Numbers flanking sequences correspond to the position of the residue within each sequence (excluding gaps). Abbreviations are as follows: HS, Homo sapiens; MM, Mus musculus; RN, Rattus norvegicus; CI, Ciona intestinalis; DM, Drosophila melanogaster; CE, Caenorhabditis elegans; TB, Trypanosoma brucei; TC, Trypanosoma cruzi; CR, Chlamydomonas reinhardtii; AN, Aspergillus nidulans; AT, Arabidopsis thaliana; and SC, Saccharomyces cerevisiae.
The computational comparative genomics approach used in our study is similar to the approach used recently to identify the BBS5 gene (Li et al. 2004). In that study, genes that were conserved in the genome from the model organism C. reinhardii and not in A. thaliana and that mapped to the BBS5 locus were selected as BBS candidate genes. Of note, the BBS5 gene was found within the comparative genomics set of 114 Gc+ genes in our study. When this set of 114 genes was intersected with the set of 62 Ensembl genes in the linked BBS3 interval, no genes were identified. It is possible that different choices for S- and for Th- would have resulted in a nonempty set of candidates in the set Gc+-,A. However, one of the strengths of this method is the ability to employ additional criteria, such as a known linked interval. As can be seen in table 1, both dimensions of filtering allow a distillation of candidates to a number, which then may be individually examined for functional annotation and/or direct mutation screening. Since other BBS genes likely exist, the comparative genomics approach should prove useful in identifying additional genes causing rare cases of BBS.
Little is known about the pathophysiology of BBS and the specific functions of BBS proteins. BBS6 is caused by mutations in the MKKS gene (Katsanis et al. 2000; Slavotinek et al. 2000), mutations that also cause McKusick-Kaufman syndrome (hydrometrocolpos, post-axial polydactyly, and congenital heart defects) (Robinow and Shaw 1979; Stone et al. 2000). MKKS has sequence homology to the α subunit of the Thermoplasma acidophilum thermosome (Stone et al. 2000), a prokaryotic chaperonin complex with similarity to a eukaryotic chaperonin called “tailless complex polypeptide ring complex” (TRiC) (Frydman et al. 1992). However, the other known BBS proteins have no significant similarity to chaperonins or other proteins of known function. BBS4 and BBS8 contain tetratricopeptide repeat (TPR) domains, indicating that they may interact with other proteins. Recently, it was hypothesized that BBS8 and other BBS proteins are involved in cilia function, on the basis of localization of BBS8 to the basal body of ciliated cells (Ansley et al. 2003). In addition, BBS4 was recently shown to localize to the centriolar satellites of centrosomes and basal bodies of primary cilia where it interacts with components of the dynein transport machinery (Kim et al. 2004). We recently demonstrated that BBS4 knockout mice have features of the human disorder and that absence of this protein leads to failure of spermatozoa flagellar formation but not to the failure of the formation of cilia in general (Mykytyn et al. 2004). Furthermore, we demonstrated that absence of this protein did not disrupt initial formation of photoreceptor outer segments, including the connecting cilia; rather, photoreceptors underwent cell death due to apoptosis. Collectively, these results support the hypothesis that BBS proteins are involved in ciliary function and/or intracellular transport.
The comparative genomic analysis of the known BBS genes performed to identify the BBS3 gene provides some insight into the function of BBS genes. A striking finding is that, in general, ciliated organisms have orthologous sequences to BBS proteins, whereas nonciliated organisms do not (Mykytyn et al. 2004). For example, even lower, single-cell ciliated organisms—such as T. brucei, T. cruzi, and C. reinhardtii—have homologous sequences to the known BBS proteins. The ciliated organism C. intestinalis also has homologous sequences to BBS proteins. In contrast, organisms without cilia (e.g., S. cerevisiae, S. pombe, A. nidulans, and A. thaliana) do not have sequences that are homologous to BBS proteins. These data support a role for BBS genes in cilia function or some other specialized function common to ciliated organisms. This hypothesis is supported by two previous studies, in which comparative genomics were used to identify genes found in ciliated organisms (Avidor-Reiss et al. 2004; Li et al. 2004).
The finding that some organisms do not have orthologues to BBS genes indicates that the BBS genes do not play a role in a process required of all eukaryotic cells—or, at a minimum, that BBS genes add specificity to a general cellular process found in all eukaryotic organisms. This finding is consistent with the BBS genes playing a role in cilial function and/or a role in intracellular transport.
Although the precise function of ARL6 is not known (Ingley et al. 1999; Jacobs et al. 1999; Pasqualato et al. 2002), the identification of this gene as the BBS3 gene supports a general role for BBS genes in intracellular transport. The ADP-ribosylation factor group of proteins is part of the Ras superfamily, which is subdivided into ARF and ARF-like (ARL) subgroups (Pasqualato et al. 2002). ARF proteins were initially identified because of their ability to stimulate ADP-ribosyltransferase activity of cholera toxin (Kahn and Gilman 1984; Price et al. 1988; Sewell and Kahn 1988; Pasqualato et al. 2002). The ARL proteins were identified on the basis of their similarity to ARF proteins. These proteins are thought to have diverse roles, including regulation of intracellular vesicle and membrane trafficking and microtubule assembly (Bhamidipati et al. 2000; Pasqualato et al. 2002). Further elucidation of the precise function of ARL6 and the other BBS proteins, as well as their interactions, should provide important clues to the pathophysiology of diverse phenotypes, including obesity, diabetes, and retinal degeneration.
Acknowledgments
We are grateful to the patients and their families for participating in this study. We thank G. Beck and J. Beck for technical assistance and D. Aguiar-Crouch for administrative assistance. This work was supported by the following grants and organizations: National Institutes of Health grants P50-HL-55006 (V.C.S.) and R01-EY-11298 (V.C.S. and E.M.S.); the Carver Endowment for Molecular Ophthalmology (E.M.S. and V.C.S.); and Research to Prevent Blindness, New York, NY (Department of Ophthalmology, University of Iowa). V.C.S. and E.M.S. are investigators of the Howard Hughes Medical Institute.
Appendix A
Table A1.
ARL6 Primer Sequences
|
Primer |
|||
| Locus Name | Forward | Reverse | Size(bp) |
| Hs.196918-01 | GCTTCAGGACCGGAAGAAGCT | ACGGGGACAGAGGGTGGCGCT | 318 |
| Hs.196918-02 | GTTTAAAGGCTCTATGATAGT | ATCTACTATAAAGCACTGAGT | 223 |
| Hs.196918-03 | ATTTCTACCCATAATCATATC | GTTGCATAAGCAACCTGTCAT | 198 |
| Hs.196918-04 | CTACCAATATTTTCCATAACT | ATATAACATAATTCTGCACAT | 286 |
| Hs.196918-05 | GAAACTGAAAATCTGGATACT | AGGAAGAATGACCAGGTAGAC | 223 |
| Hs.196918-06 | TGCTTTAGTTTATAATGTAGT | AAAGCCAAACAACCTAGCCAT | 207 |
| Hs.196918-07 | TTAGGTTAGGAAATGCCCATG | AATGAAAGATAATTACATAGT | 281 |
| Hs.196918-08 | TGTTTCTGTGTGTGTGATATG | TGAAGCTTGTATATTTTTCAG | 312 |
| Hs.196918-09 | GTAATCCTTTATTCTTATAGT | CAAAGGCTGAAGACACACAGT | 159 |
| Hs.196918.10 | ACATGTTGTATAGATTTGACT | CTTAAAAAGGTCAGAGTCCAT | 253 |
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Authors' Web site, http://genome.uiowa.edu/data/BBS/BBS3/index.html (for BBS candidate gene sets)
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Broad Institute, http://www.broad.mit.edu/annotation/fungi/aspergillus/
- Ensembl Genome Browser, http://www.ensembl.org/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/
- JGI, http://www.jgi.doe.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nih.gov/entrez/query.fcgi?db=OMIM
- SGD, http://www.yeastgenome.org/
- TAIR, http://www.arabidopsis.org/
- TIGR, http://www.tigr.org/
- UCSC Genome Browser, http://genome-test.cse.ucsc.edu/cgi-bin/hgGateway
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, Mah AK, Johnsen RC, Cavender JC, Lewis RA, Leroux MR, Beales PL, Katsanis N (2003) Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425:628–633 10.1038/nature02030 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS (2004) Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117:527–539 10.1016/S0092-8674(04)00412-X [DOI] [PubMed] [Google Scholar]
- Badano JL, Ansley SJ, Leitch CC, Lewis RA, Lupski JR, Katsanis N (2003) Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am J Hum Genet 72:650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardet G (1920) Sur un syndrome d’obesite infantile avec polydactylie et retinite pigmentaire (contribution a l’etude des formes cliniques de l’obesite hypophysaire). Thesis, University of Paris, Paris [Google Scholar]
- Bhamidipati A, Lewis SA, Cowan NJ (2000) ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J Cell Biol 149:1087–1096 10.1083/jcb.149.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedl A (1922) Ein Geschwisterpaar mit adipose-genitaler Dystrophie. Dtsch Med Wschr 48:1630 [Google Scholar]
- Bruford EA, Riise R, Teague PW, Porter K, Thomson KL, Moore AT, Jay M, Warburg M, Schinzel A, Tommerup N, Tornqvist K, Rosenberg T, Patton M, Mansfield DC, Wright AF (1997) Linkage mapping in 29 Bardet-Biedl syndrome families confirms loci in chromosomal regions 11q13, 15q22.3-q23, and 16q21. Genomics 41:93–99 10.1006/geno.1997.4613 [DOI] [PubMed] [Google Scholar]
- Carmi R, Elbedour K, Stone EM, Sheffield VC (1995) Phenotypic differences among patients with Bardet-Biedl syndrome linked to three different chromosome loci. Am J Med Genet 59:199–203 [DOI] [PubMed] [Google Scholar]
- Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SM, Clamp M (2004) The Ensembl automatic gene annotation system. Genome Res 14:942–950 10.1101/gr.1858004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbedour K, Zucker N, Zalzstein E, Barki Y, Carmi R (1994) Cardiac abnormalities in the Bardet-Biedl syndrome: echocardiographic studies of 22 patients. Am J Med Genet 52:164–169 [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl FU (1992) Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. Embo J 11:4767–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JS, Parfrey PS, Harnett JD, Farid NR, Cramer BC, Johnson G, Heath O, McManamon PJ, O’Leary E, Pryse-Phillips W (1989) The cardinal manifestations of Bardet-Biedl syndrome, a form of Laurence-Moon-Biedl syndrome. N Engl J Med 321:1002–1009 [DOI] [PubMed] [Google Scholar]
- Harnett JD, Green JS, Cramer BC, Johnson G, Chafe L, McManamon P, Farid NR, Pryse-Phillips W, Parfrey PS (1988) The spectrum of renal disease in Laurence-Moon-Biedl syndrome. N Engl J Med 319:615–618 [DOI] [PubMed] [Google Scholar]
- Ingley E, Williams JH, Walker CE, Tsai S, Colley S, Sayer MS, Tilbrook PA, Sarna M, Beaumont JG, Klinken SP (1999) A novel ADP-ribosylation like factor (ARL-6), interacts with the protein-conducting channel SEC61beta subunit. FEBS Lett 459:69–74 10.1016/S0014-5793(99)01188-6 [DOI] [PubMed] [Google Scholar]
- Jacobs S, Schilf C, Fliegert F, Koling S, Weber Y, Schurmann A, Joost HG (1999) ADP-ribosylation factor (ARF)-like 4, 6, and 7 represent a subgroup of the ARF family characterization by rapid nucleotide exchange and a nuclear localization signal. FEBS Lett 456:384–388 10.1016/S0014-5793(99)00759-0 [DOI] [PubMed] [Google Scholar]
- Kahn RA, Gilman AG (1984) Purification of a protein cofactor required for ADP-ribosylation of the stimulatory regulatory component of adenylate cyclase by cholera toxin. J Biol Chem 259:6228–6234 [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR (2001) Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 293:2256–2259 10.1126/science.1063525 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, Ansley SJ, Davidson WS, Lupski JR (2000) Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet 26:67–70 10.1038/79201 [DOI] [PubMed] [Google Scholar]
- Kim JC, Badano JL, Sibold S, Esmail MA, Hill J, Hoskins BE, Leitch CC, Venner K, Ansley SJ, Ross AJ, Leroux MR, Katsanis N, Beales PL (2004) The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat Genet 36:462–470 10.1038/ng1352 [DOI] [PubMed] [Google Scholar]
- Kwitek-Black AE, Carmi R, Duyk GM, Buetow KH, Elbedour K, Parvari R, Yandava CN, Stone EM, Sheffield VC (1993) Linkage of Bardet-Biedl syndrome to chromosome 16q and evidence for non-allelic genetic heterogeneity. Nat Genet 5:392–396 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK (2004) Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117:541–552 10.1016/S0092-8674(04)00450-7 [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothschild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, Sheffield VC (2001) Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet 28:188–191 10.1038/88925 [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Mullins RF, Andrews M, Chiang AP, Swiderski RE, Yang B, Braun T, Casavant T, Stone EM, Sheffield VC (2004) Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc Natl Acad Sci USA 101:8664–8669 10.1073/pnas.0402354101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, Fulton AB, Carmi R, Luleci G, Chandrasekharappa SC, Collins FS, Jacobson SG, Heckenlively JR, Weleber RG, Stone EM, Sheffield VC (2002) Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet 31:435–438 [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Searby CC, Carmi R, Elbedour K, Van Maldergem L, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, Haider NB, Kwitek-Black AE, Ying L, Duhl DM, Gorman SW, Heon E, Iannaccone A, Bonneau D, Biesecker LG, Jacobson SG, Stone EM, Sheffield VC (2001) Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Hum Mol Genet 10:865–874 10.1093/hmg/10.8.865 [DOI] [PubMed] [Google Scholar]
- Pasqualato S, Renault L, Cherfils J (2002) Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for “front-back” communication. EMBO Rep 3:1035–1041 10.1093/embo-reports/kvf221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR, Nightingale M, Tsai SC, Williamson KC, Adamik R, Chen HC, Moss J, Vaughan M (1988) Guanine nucleotide-binding proteins that enhance choleragen ADP-ribosyltransferase activity: nucleotide and deduced amino acid sequence of an ADP-ribosylation factor cDNA. Proc Natl Acad Sci USA 85:5488–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow M, Shaw A (1979) The McKusick-Kaufman syndrome: recessively inherited vaginal atresia, hydrometrocolpos, uterovaginal duplications, anorectal anomalies, postaxial polydactyly, and congenital heart disease. J Pediatr 94:776–8 [DOI] [PubMed] [Google Scholar]
- Sewell JL, Kahn RA (1988) Sequences of the bovine and yeast ADP-ribosylation factor and comparison to other GTP-binding proteins. Proc Natl Acad Sci USA 85:4620–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield VC, Carmi R, Kwitek-Black A, Rokhlina T, Nishimura D, Duyk GM, Elbedour K, Sunden SL, Stone EM (1994) Identification of a Bardet-Biedl syndrome locus on chromosome 3 and evaluation of an efficient approach to homozygosity mapping. Hum Mol Genet 3:1331–1335 [DOI] [PubMed] [Google Scholar]
- Slavotinek AM, Stone EM, Mykytyn K, Heckenlively JR, Green JS, Heon E, Musarella MA, Parfrey PS, Sheffield VC, Biesecker LG (2000) Mutations in MKKS cause Bardet-Biedl syndrome. Nat Genet 26:15–16 10.1038/79116 [DOI] [PubMed] [Google Scholar]
- Stone DL, Slavotinek A, Bouffard GG, Banerjee-Basu S, Baxevanis AD, Barr M, Biesecker LG (2000) Mutation of a gene encoding a putative chaperonin causes McKusick-Kaufman syndrome. Nat Genet 25:79–82 10.1038/75637 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]