To the Editor:
We read with interest the report of a novel deletion of part of the azoospermia factor c (AZFc [MIM 415000]) region of the human Y chromosome (Fernandes et al. 2004). This article reported that the deletion is found only in branch N of the Y-chromosome genealogical tree, occurs through one mutational pathway, is ∼2.2 Mb in size, and has no effect on spermatogenesis. We, too, recently reported this deletion, which Fernandes et al. termed the “g1/g3” deletion and which we termed the “b2/b3” deletion (Repping et al. 2004). Our findings, however, differed from those of Fernandes et al. in several important particulars: (1) our screening of 1,563 men demonstrated that this deletion is not confined to branch N and that it has at least four independent origins; (2) our analysis revealed two mutational pathways, rather than one, that can generate the deletion, and we confirmed the existence of the inverted AZFc organizations that are the intermediate steps in these pathways; (3) on the basis of the reference sequence of the Y chromosome, we concluded that the size of the deletion is 1.8 Mb, rather than ∼2.2 Mb; (4) using interphase FISH, we confirmed the amplicon organization that was postulated in the deletion and also identified three instances of duplication subsequent to the deletion; and (5) because of the possibility of a compensatory factor on Y chromosomes in branch N and because of the limited number of deletions outside this branch, we concluded that a possible effect of this deletion on risk of spermatogenic failure cannot be excluded (Repping et al. 2004).
Beyond these differences, however, the characterizations of this and other partial deletions of AZFc (Repping et al. 2003) highlight a more important question. At issue is the relative utility of sequence family variants (Saxena et al. 2000), compared with that of plus/minus STSs, for identification and differentiation of deletions involving AZFc. AZFc is composed entirely of amplicons—repeat units 115–678 kb in length that only differ by ∼1 nt per 3,000 bp. These rare differences are called “sequence family variants” (SFVs). We previously relied on SFVs to map and sequence the AZFc region of one man’s Y chromosome (Kuroda-Kawaguchi et al. 2001). The report by Fernandes et al. (2004) emphasized the use of SFVs in identification of the novel deletion, whereas our analysis relied on plus/minus STSs for identification of the deletion, followed, in most instances, by confirmation with FISH.
Two observations led us to ask whether SFVs, as opposed to plus/minus STSs, offer the simpler and more robust means of detecting and distinguishing deletions in AZFc. First, figures 1 and 4 in the report by Fernandes et al. (2004) indicated that negative results at the plus/minus STS sY1192 or 50f2/C combined with positive results at flanking STSs are sufficient to detect the deletion (table 1). Moreover, the b2/b3 deletion and other types of deletions involving AZFc can be distinguished by their plus/minus signatures, without the use of SFVs (table 1).
Table 1.
Plus/Minus STS Results Distinguishing Different Types of Deletions Involving AZFc[Note]
Second, table 2 in the report by Fernandes et al. (2004) showed that the SFV patterns of undeleted chromosomes vary considerably among different branches of the Y-chromosome genealogy and that the patterns also vary among individuals within branches. These observations suggested that the link between SFV patterns and particular types of deletions would likely not be consistent across the worldwide diversity of Y chromosomes.
The diversity of SFV patterns in undeleted chromosomes is not surprising, since AZFc is subject to large inversions, deletions, and duplications caused by ectopic homologous recombination between amplicons (Kuroda-Kawaguchi et al. 2001; Repping et al. 2003, 2004). Such events would rearrange the locations of particular variants and would blur the association between SFV patterns and particular types of deletions. The association would likely be further blurred by gene conversion, which frequently erases small sequence differences (i.e., SFVs) between amplicon copies on the Y chromosome (Rozen et al. 2003).
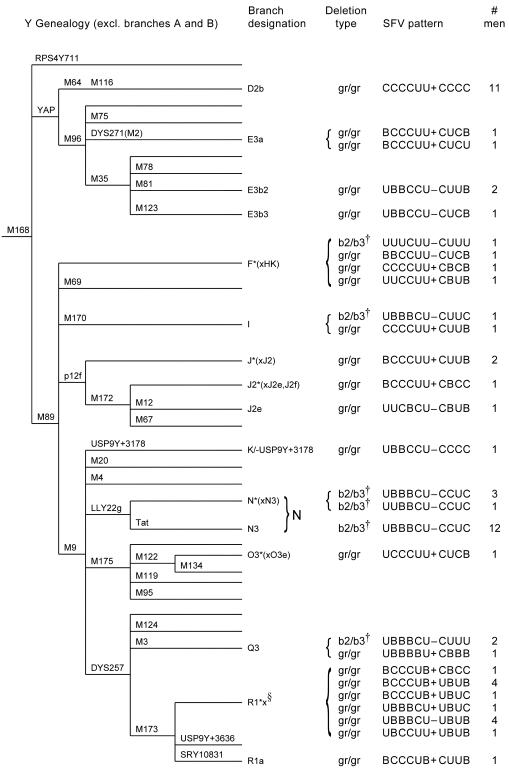
We experimentally investigated the consistency of SFV patterns in different types of deletions involving AZFc. First, using the SFVs employed by Fernandes et al. (2004), we typed 20 men reported elsewhere to have the b2/b3 deletion (Repping et al. 2004) (see GenBank Web site for SFV assays). These men represented branch N and three other branches of the Y-chromosome genealogy (fig. 1). Second, using the same SFVs, we typed 40 men reported elsewhere to have the gr/gr deletion, the other common partial AZFc deletion (Repping et al. 2003). These men represented 14 branches of the Y-chromosome genealogy (fig. 1).
Figure 1.
Genealogical analysis of SFV patterns associated with b2/b3 and gr/gr deletions. In the SFV patterns, “C” indicates the cut variant described by Fernandes et al. (2004), “U” indicates the uncut variant, “B” indicates both variants, and + and − indicate the presence or absence, respectively, of the Y-DAZ3 variant. The order of SFVs is as shown in table 2 in the work of Fernandes et al. (2004): DAZ-SNV I, DAZ-SNV II, sY586 (DAZ-SNV III), DAZ-SNV IV, sY587 (DAZ-SNV V), DAZ-SNV VI, AZFc SFV 18 (assayed by Y-DAZ3), TTY4-SNV I, BPY2-SNV, GOLY-SNV I, and AZFc SFV 20 (AZFc-P1-SNV I) (Saxena et al. 2000; Kuroda-Kawaguchi et al. 2001 [Web table E]; Fernandes et al. 2002, 2004). The genealogical tree of extant human Y chromosomes and the branch designations are from the studies by Underhill et al. (2000) and the Y-Chromosome Consortium (2002). §, R1*x is an abbreviation for R1*(xR1a,R1/-USP9Y+3636). †, Termed “g1/g3” by Fernandes et al. (2004).
The b2/b3 deletions outside branch N showed diverse SFV patterns, and the gr/gr deletions showed even greater diversity (fig. 1). This greater diversity was likely due to the larger number of independent gr/gr deletions studied. Two branches, F*(xHK) and R1*x, contained numerous deletions and a high diversity of SFV patterns (fig. 1). In these branches, multiple independent deletion events probably account for the high diversity. By contrast, two other branches, D2b and N, contained numerous deletions but uniform SFV patterns. This uniformity is explained by the fact that all chromosomes in these branches descended from deleted founders (Repping et al. 2003, 2004; Fernandes et al. 2004). Thus, the chromosomes in each of these branches represent a single deletion event.
Our data also showed that the SFV patterns of b2/b3 and gr/gr deletions are not distinct from each other. For example, the b2/b3 pattern UUUCUU−CUUU (branch F*[xHK]) is more similar to the gr/gr pattern UUCCUU+CBUB (branch F*[xHK], four differences [underlined]) than to the b2/b3 pattern UBBBCU−CCUC (branch N, six differences). In another example, the gr/gr pattern UBBBCU−UBUB (branch R1*x) is more similar to the b2/b3 pattern UBBBCU−CUUC (branch I, three differences) than to the gr/gr pattern BCCCUB+CBCC (branch R1*x, 10 differences).
In conclusion, the SFV patterns of b2/b3 and gr/gr deletions vary widely and are not clearly distinct. SFVs can offer insight only if one knows the common SFV organizations in the genealogical branches represented by the Y chromosomes being tested. However, SFV organizations across the Y-chromosome genealogical tree are largely unknown, and SFV patterns vary even among individuals in the same branch. Just as important is that a large number of two-step assays are needed for SFV typing and for determining the Y-chromosome branch. By contrast, six simple plus/minus STSs distinguish between the deletions involving AZFc (table 1). Thus, plus/minus STSs provide a straightforward means of identifying and distinguishing the deletions of part of AZFc, whereas, in most situations, SFVs do not.
Acknowledgments
We thank J. Lange and H. Skaletsky, for comments, and C. Disteche, B. Gilbert, K. Keppler, T. Kuroda-Kawaguchi, and T. Ogata, for samples. This work was supported by the National Institutes of Health, the Howard Hughes Medical Institute, and the Academic Medical Center.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for STSs 50f2/C [accession number Y07728], sY142 [accession number G38345], sY1191 [accession number G73809], sY1192 [accession number G67166], sY1197 [accession number G67168], sY1201 [accession number G67170], sY1206[accession number G67171], and sY1291 [accession number G72340] and for SFV assays DAZ-SNV I [accession number G73167], DAZ-SNV II [accession number G73166], sY586 [accession number G63907], DAZ-SNV IV [accession number G73168], sY587 [accession number G63908], DAZ-SNV VI [accession number G73169], Y-DAZ3 [accession number G73170], TTY4-SNV I [accession number BV012731], BPY2-SNV [accession number BV012732], GOLY-SNV I [accession number BV012733], and AZFc SFV 20 [AZFc-P1-SNV I] [accession number G73351])
- Nature Genetics,http://www.nature.com/ng/journal/v29/n3/extref/ng757-S6.doc (for AZFc SFVs 18 and 20 in Web table E in Kuroda-Kawaguchi et al. 2001)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for AZFc) [PubMed]
References
- Fernandes S, Huellen K, Goncalves J, Dukal H, Zeisler J,Rajpert De Meyts E, Skakkebaek NE, Habermann B, Krause W, Sousa M, Barros A, Vogt PH (2002) High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod 8:286–298 10.1093/molehr/8.3.286 [DOI] [PubMed] [Google Scholar]
- Fernandes S, Paracchini S, Meyer LH, Floridia G, Tyler-Smith C, Vogt PH (2004) A large AZFc deletion removes DAZ3/DAZ4 and nearby genes from men in Y haplogroup N. Am J Hum Genet 74:180–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC (2001) The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 29:279–286 10.1038/ng757 [DOI] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Brown L, van Daalen SKM, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JWA, Oates RD, Silber S, van der Veen F, Page DC, Rozen S (2003) Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet 35:247–251 10.1038/ng1250 [DOI] [PubMed] [Google Scholar]
- Repping S, van Daalen SKM, Korver CM, Brown LG, Marszalek JD, Gianotten J, Oates RD, Silber S, van der Veen F, Page DC, Rozen S (2004) A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8-Mb deletion in the azoospermia factor c region. Genomics 83:1046–1052 10.1016/j.ygeno.2003.12.018 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston JH, Wilson RK, Page DC (2003) Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423:873–876 10.1038/nature01723 [DOI] [PubMed] [Google Scholar]
- Saxena R, de Vries JWA, Repping S, Alagappan RK, Skaletsky H, Brown LG, Ma P, Chen E, Hoovers JMN, Page DC (2000) Four DAZ genes in two clusters found in the AZFc region of the human Y chromosome. Genomics 67:256–267 10.1006/geno.2000.6260 [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, et al (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837 10.1038/nature01722 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza L, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 10.1038/81685 [DOI] [PubMed] [Google Scholar]
- Y-Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 10.1101/gr.217602 [DOI] [PMC free article] [PubMed] [Google Scholar]