Abstract
The 8–10 million European Roma/Gypsies are a founder population of common origins that has subsequently split into multiple socially divergent and geographically dispersed Gypsy groups. Unlike other founder populations, whose genealogy has been extensively documented, the demographic history of the Gypsies is not fully understood and, given the lack of written records, has to be inferred from current genetic data. In this study, we have used five disease loci harboring private Gypsy mutations to examine some missing historical parameters and current structure. We analyzed the frequency distribution of the five mutations in 832–1,363 unrelated controls, representing 14 Gypsy populations, and the diversification of chromosomal haplotypes in 501 members of affected families. Sharing of mutations and high carrier rates supported a strong founder effect, and the identity of the congenital myasthenia 1267delG mutation in Gypsy and Indian/Pakistani chromosomes provided the best evidence yet of the Indian origins of the Gypsies. However, dramatic differences in mutation frequencies and haplotype divergence and very limited haplotype sharing pointed to strong internal differentiation and characterized the Gypsies as a founder population comprising multiple subisolates. Using disease haplotype coalescence times at the different loci, we estimated that the entire Gypsy population was founded ∼32–40 generations ago, with secondary and tertiary founder events occurring ∼16–25 generations ago. The existence of multiple subisolates, with endogamy maintained to the present day, suggests a general approach to complex disorders in which initial gene mapping could be performed in large families from a single Gypsy group, whereas fine mapping would rely on the informed sampling of the divergent subisolates and searching for the shared genomic region that displays the strongest linkage disequilibrium with the disease.
Introduction
Founder populations have been an invaluable resource for understanding the molecular basis of Mendelian disorders (Motulsky 1995; Risch et al. 1995; de la Chapelle and Wright 1998; Sheffield et al. 1998; Peltonen et al. 1999; Ostrer 2001; Arcos-Burgos and Muenke 2002), and their (still-disputed) potential to contribute to research into genetically complex disorders is closely related to demographic history and population structure. Linkage disequilibrium (LD) approaches to gene mapping are sensitive to time since founding, number of founders, extent of genetic isolation, substructure, and frequency and age of the genetic variant (de la Chapelle and Wright, 1998; Kruglyak 1999; Wright et al. 1999; Peltonen et al. 2000; Shifman and Darvasi 2001; Heutink and Oostra 2002).
Recent studies have identified novel single-gene disorders and private mutations among the Roma (Gypsies), drawing attention to this previously ignored founder population (Kalaydjieva et al. 1996, 2000; Piccolo et al. 1996; Abicht et al. 1999; Rogers et al. 2000; Varon et al. 2003). Unlike other founder populations, whose history and genealogy have been extensively documented (Gulcher and Stefansson 1998; Agarwala et al. 1999; Scriver 2001; Norio 2003), the Gypsy population is characterized by a lack of reliable records, a nomadic tradition, and dispersal as an underprivileged ethnic minority in numerous countries, which has resulted in the need to infer their demographic history and genealogy from current genetic data.
Cultural anthropology, linguistics, and limited historical records from the surrounding majority populations describe the Gypsies as a population of Indian origins, with their exodus from India dated to approximately the 5th–10th century a.d., their arrival in Byzantium dated to the 11th or 12th century, and their dispersal throughout Europe documented by the end of the 15th century (Fraser 1992; Marushiakova and Popov 1997). Upon arrival in Europe, a large fraction of the initial migrant population (referred to as “Balkan Gypsies”) settled permanently in the Balkans south of the Danube. Others, known as “Vlax Roma,” moved north into Wallachia (present-day Romania), while the remainder continued the journey to all parts of the continent (Fraser 1992). Superimposed on these early migrations are several recent migration waves—out of Romania at the end of the 19th century and out of the Balkans in the second half of the 20th century—as well as multiple movements of small nomadic groups (Fraser 1992). The social organization of the Gypsies, similar to the endogamous professional jatis of India, includes numerous Gypsy groups with ethnonyms reflecting traditional trades. Group identity is based on traditions, customs, language, trades, history of migrations, and religion (Petulengro 1915–1916; Fraser 1992; Liegeois 1994; Marushiakova and Popov 1997).
Geographically dispersed and socially and linguistically divergent Gypsy groups have been shown to share unique Mendelian disorders and founder mutations (Kalaydjieva et al. 1996, 2000; Piccolo et al. 1996; Abicht et al. 1999; Rogers et al. 2000; Varon et al. 2003), as well as ancestral Y chromosome and mtDNA lineages (Gresham et al. 2001; Kalaydjieva et al. 2001a). Analysis of the distribution and diversity of these lineages has resulted in characterization of the current genetic profile of the Gypsies as the product of profound population bottlenecks, random genetic drift, and differential admixture, correlating best with the historical migrations within Europe (Gresham et al. 2001; Kalaydjieva and Morar 2003).
In this study, we have obtained new information on five loci harboring private disease-causing mutations that are thus likely to reflect the history of the Gypsy population rather than recent admixture. The combined data were used to infer some of the missing parameters relevant to the comprehensive characterization of the population history of the Gypsies. The frequency and distribution of the selected mutations and the diversification of the surrounding chromosomal haplotypes were used to examine the times of founding, the extent of genetic differentiation, and the implications of population history for the mapping of disease genes.
Subjects and Methods
Subjects
The study included a total of 1,870 subjects. The Gypsy sample consisted of 1,363 unrelated healthy control individuals and 501 members of affected families (241 affected subjects and 260 unaffected relatives). The remaining six individuals were patients with congenital myasthenia syndrome (CMS [MIM 254210]) who were of Indian or Pakistani origin and were homozygous for the 1267delG mutation.
The 1,363 Gypsy control individuals were tested for the presence of the five disease mutations (table 1), to estimate carrier frequencies in different Gypsy groups. The number of subjects tested for the individual mutations ranged from 832, for congenital cataracts facial dysmorphism neuropathy (CCFDN [MIM 604168]), to 1,363, for limb-girdle muscular dystrophy 2C (LGMD2C [MIM 253700]). The samples from Bulgaria were collected as part of a pilot community-based carrier-testing program, with pre- and postdiagnostic information and counseling provided to all participating individuals.
Table 1.
Sample Sizes and Estimated Carrier Rates (%) in Gypsy Groups Screened for Five Founder Mutations
|
No. of Individuals Tested (N) and Carrier Rate (CR) for Mutation |
||||||||||
| 1267delG(CMS) |
R148X(HMSNL) |
P28T(GKD)a |
C283Y(LGMD2C) |
IVS6+389C→T(CCFDN)b |
||||||
| Migrational Category and Gypsy Group Identity | N | CR(%) | N | CR(%) | N | CR(%) | N | CR(%) | N | CR(%) |
| Balkan: | ||||||||||
| Darakchii | 54 | 5.56 | 54 | .00 | 54 | 1.85 | 54 | 5.56 | 54 | .00 |
| Kalaidjii North | 96 | 5.21 | 91 | 4.40 | 95 | 1.05 | 95 | .00 | 83 | .00 |
| Blacksmiths | 63 | 6.35 | 63 | .00 | 60 | .00 | 63 | .00 | 63 | .00 |
| Musicians | 62 | .00 | 62 | 3.23 | 62 | .00 | 61 | 1.64 | 57 | 1.75 |
| Feredjelli | 43 | .00 | 43 | .00 | 39 | .00 | 101 | .00 | 42 | 2.38 |
| Turgovzi | 65 | .00 | 56 | 1.79 | 56 | .00 | 224 | 6.25 | 57 | .00 |
| Xoroxane | 36 | 5.56 | 36 | 2.78 | 35 | 2.86 | 38 | .00 | 31 | .00 |
| Subtotal | 419 | 3.34 | 405 | 1.98 | 401 | .75 | 636 | 2.83 | 387 | .52 |
| Vlax: | ||||||||||
| Rudari | 138 | 1.45 | 139 | 11.51 | 139 | 2.88 | 113 | .00 | 115 | 6.96 |
| Kalderash | 41 | .00 | 41 | 9.76 | 84 | 2.38 | 41 | .00 | 41 | 2.44 |
| Lom | 124 | 6.45 | 124 | 16.13 | 124 | 6.45 | 123 | .00 | 124 | .00 |
| Kalaidjii South | 63 | 7.94 | 63 | .00 | 63 | .00 | 63 | .00 | 61 | .00 |
| Subtotal | 366 | 4.10 | 367 | 10.90 | 410 | 3.41 | 340 | .00 | 341 | 2.64 |
| Western European: | ||||||||||
| Hungarian | 283 | 4.95 | 293 | .34 | 310 | .00 | 293 | 1.37 | NT | NT |
| Lithuanian | 20 | .00 | 20 | 5.00 | 19 | .00 | 20 | .00 | 20 | .00 |
| Spanish | 87 | 1.15 | 80 | 2.50 | 161 | 1.86 | 74 | 1.35 | 84 | 1.19 |
| Subtotal | 390 | 3.85 | 393 | 1.02 | 490 | .61 | 387 | 1.29 | 104 | .96 |
| Total | 1,175 | 3.74 | 1,165 | 4.46 | 1,301 | 1.54 | 1,363 | 1.69 | 832 | 1.44 |
GKD = galactokinase deficiency.
NT = not tested.
The 501 subjects from affected Gypsy families were participants in earlier studies aimed at the positional cloning of disease genes and the identification of pathogenic mutations. These samples, together with those from the six Indian/Pakistani subjects, were used for the analysis of chromosomal haplotype surrounding the hereditary motor and sensory neuropathy–Lom (HMSNL [MIM 601455]), CCFDN, CMS, and LGMD2C mutations (table 2). Disease haplotypes in the region of the GALK1 gene have been reported elsewhere (Kalaydjieva et al. 1999; Hunter et al. 2002).
Table 2.
Origins and Sample Sizes of Disease and Control Chromosomes Included in the Haplotype Analysis of the Four Gene Regions
|
No. of Disease Chromosomes |
|||||
| Migrational Category,Current Residence, andGypsy Group Identity | CHRNE(CMS) | NDRG1(HMSNL) | SGCG(LGMD2C) | CTDP1(CCFDN) | No. of NormalChromosomes |
| Balkan: | |||||
| Bulgaria: | |||||
| Darakchii | 3 | 0 | 0 | 0 | 0 |
| Kalaidjii North | 4 | 0 | 0 | 0 | 0 |
| Blacksmiths | 4 | 4 | 0 | 0 | 0 |
| Turgovzi | 0 | 0 | 48 | 0 | 0 |
| Xoroxane | 15 | 15 | 0 | 0 | 0 |
| Dassikane | 16 | 6 | 0 | 0 | 0 |
| Other Balkan countries: | |||||
| Unspecified | 22 | 0 | 0 | 0 | 0 |
| Subtotal | 64 | 25a | 48 | 0 | 56 |
| Vlax: | |||||
| Bulgaria: | |||||
| Rudari | 0 | 35 | 0 | 74 | 0 |
| Kalderash | 0 | 32 | 0 | 4 | 0 |
| Lom | 12 | 18 | 0 | 0 | 0 |
| Kalaidjii South | 9 | 0 | 0 | 0 | 0 |
| Other Balkan countries: | |||||
| Rudari | 0 | 0 | 0 | 42 | 0 |
| Unspecified | 2 | 16 | 0 | 0 | 0 |
| Subtotal | 23a | 101a | 0 | 120 | 163 |
| Western European | 47a | 14 | 30 | 9 | 41 |
| Other: | |||||
| India/Pakistan | 12 | 0 | 0 | 0 | 0 |
| Total | 146 | 140 | 78 | 129b | 260 |
Includes a nontransmitted chromosome from an affected family member of the proband.
Three recent recombinations in families with CCFDN were counted separately, giving an odd number of chromosomes.
Background LD in the HMSNL and CCFDN gene regions was examined using the nontransmitted chromosomes of the affected families from the Rudari, Kalderash, and Lom Gypsy groups. Additional genotyping data for the same regions were obtained by analyzing unaffected families from the Turgovzi group.
Informed consent has been obtained from all participants. The study complies with the ethical guidelines of the institutions involved.
Ethnic Composition of the Sample
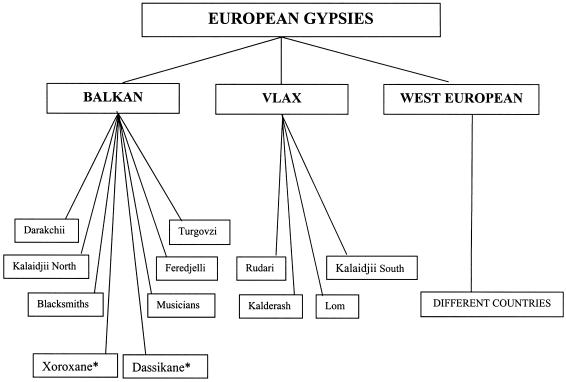
We have collected information aimed toward classifying the different Gypsy populations in accordance with historical and cultural anthropological criteria. Subjects from Bulgaria were assigned to Gypsy groups (tables 1 and 2 and fig. 1) on the basis of self-reported identity and a linguistic interview. Through use of existing cultural anthropology data (Marushiakova and Popov 1997), the groups were classified into larger migrational/linguistic categories: Balkan and Vlax. In two cases, self-reported identity did not specify an individual Gypsy group but referred to larger cultural-anthropological divisions within the Balkan category: the Xoroxane (Muslim) and Dassikane (Christian). The Rudari from other Balkan countries were classified on the basis of linguistic data. The remaining subjects were broadly assigned to migrational categories: Vlax and Balkan.
Figure 1.
Assignment of participating subjects to migrational/linguistic categories and individual Gypsy groups within the categories. The Xoroxane and Dassikane (indicated with an asterisk [*]) do not specify individual Gypsy groups but represent broader cultural anthropological divisions within the Balkan category, in which group identity has been lost. The western European migrational category comprises subjects from Hungary, Slovenia, the Czech Republic, Lithuania, Germany, France, Italy, Spain, and Portugal.
Individuals from Hungary, Slovenia, the Czech Republic, Lithuania, Germany, France, Italy, Spain, and Portugal, for whom information on Gypsy group identity was unavailable, partial, or contradictory, were classified together as “western European.” The available information on Indian/Pakistani subjects indicated diverse areas of origin (Uttar Pradesh, Andhra Pradesh, Southern India, and north of Islamabad in Pakistan), caste (Rajpoot, Kamma), and language groups (Urdu, Telugu).
Diseases and Founder Mutations
HMSNL, caused by a premature termination codon (R148X) in NDRG1 on 8q24, is a severe early-onset peripheral neuropathy, diagnosed in individuals from diverse Gypsy populations across Europe (Kalaydjieva et al. 1996, 1998, 2000).
CCFDN, caused by the splicing IVS6+389C→T mutation in CTDP1 on 18qter, is a developmental disorder confined to a single Gypsy group of the Vlax migrational category (Angelicheva et al. 1999; Tournev et al. 1999; Varon et al. 2003).
CMS occurs in diverse Gypsy groups (Abicht et al. 1999). The founder mutation, 1267delG in CHRNE on 17p13, was identified originally in patients from India/Pakistan (Croxen et al. 1999).
LGMD2C, caused by C283Y in SGCG on 13q12, is a Duchenne-like disorder in Gypsy patients from western Europe and in specific Balkan Gypsy groups (Piccolo et al. 1996; Merlini et al. 2000).
Galactokinase deficiency (MIM 230200), caused by P28T in GALK1 on 17q24, is an inborn error of galactose metabolism with development of infantile cataracts and is found across Europe; it is most common among the Vlax Gypsies (Kalaydjieva et al. 1999; Hunter et al. 2002).
Detailed accounts of the clinical phenotypes and of the genetic characterization of the disease loci have been provided in the references cited above.
Genotyping Analyses
Blood samples obtained during fieldwork in Bulgaria were collected in AS1 buffer (QIAGEN) or on FTA cards (Life Technologies). DNA was extracted following the manufacturers’ instructions. DNA samples from other countries were extracted from white blood cells through use of standard protocols.
PCR-based RFLP assays were used to detect the five mutations (table A [online only]). The digested products were resolved by agarose gel electrophoresis and visualized with ethidium bromide staining.
Table A.
Detection of the Five Founder Disease-Causing Mutations by PCR-Based RFLP Analysis
|
Digest Product size(s)(bp) |
||||||||
| Disease | Gene | Mutation | Primers (5′→ 3′) | PCR Product Size(bp) | Restriction Endonuclease | Normal Allele | Disease Allele | Reference |
| CMS | CHRNE | 1267delG | CAC GGA GCG AGC TCG TGT TTG A and CTG GAG ATG GGT GGG AAA TTG | 550/549 | EcoNI (XagI) | 355+195 | 549 | Abicht et al. 1999 |
| HMSNL | NDRG1 | R148X | AAC TGT GGA GAA TAC GGG and CTG TGC AGG CAG TTA CGG CAG C | 316 | TaqI | 190+126 | 316 | Kalaydjieva et al. 2001c |
| Galaktokinase deficiency | GALK1 | P28T | CGT CAT GGC TGC TTT GAG AC and TAG AGG GCG CCC TTC TTG AG | 272 | AvaI | 149+86+37 | 235+37 | Kalaydjieva et al. 1999 |
| LGMD2C | SGCG | C283Y | CCT GTC TGT GGC CGG TGT GA and GCG TTT ACT TCC CAT CCA CGC TGC | 168 | RsaI | 168 | 141+27 | Piccolo et al. 1996 |
| CCFDN | CTDP1 | IVS6+389C→T | CTT CCA GAG TTC ACG CCA TT and ACA CAA AAG CCC AGC TCA AG | 461 | NlaIII | 221+240 | 461 | Varon et al. 2003 |
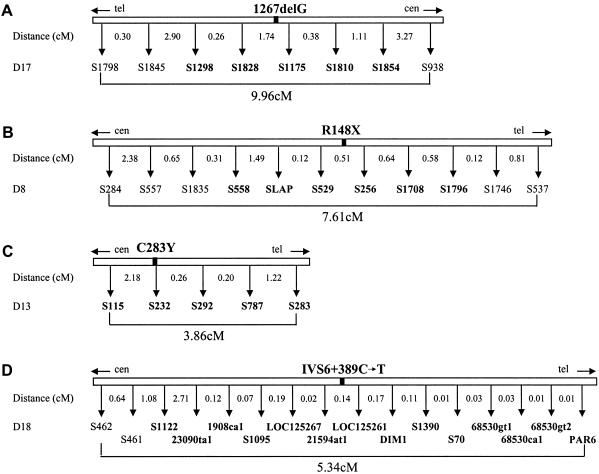
Haplotype analysis was performed using 8 markers over 9.96 cM for the CMS region, 11 markers over 7.61 cM for HMSNL, 5 markers over 3.86 cM for LGMD2C, and 16 markers over 5.34 cM for CCFDN (fig. 2). Most markers were microsatellite repeats, except LOC125267, LOC125261, DIM1, and PAR6 in the CCFDN region, which were insertion/deletions. Details on most markers are available in the public databases. Polymorphisms identified during the positional cloning of the HMSNL and CCFDN genes have been reported elsewhere (Chandler et al. 2000; Varon et al. 2003). For the genotyping, we used multiplex PCR with 5′ fluorescent labeling of one primer/pair, electrophoretic separation on an ABI 377 DNA Analyzer, and fragment analysis with Genotyper version 2.5.
Figure 2.
Genetic maps used in the haplotype analysis of four of the founder mutations. Markers are shown as equidistant from one another. The position of the mutation is indicated with a black square. Markers designated with an “S” number are known microsatellites found in public databases. Microsatellites identified during positional cloning studies include SLAP (NDRG1 region), 23090ta1, 1908ca1, 21594at1, 68530gt1, 68530ca1, and 68530gt2 (CTDP1 region). LOC125267, LOC125261, DIM1, and PAR6 in this region represent insertion/deletion polymorphisms. Markers in boldface type were included in the comparative analysis of regions spanning ∼3 cM around each disease mutation. A, CHRNE on 17p13.2. B, NDRG1 on 8q24.22. C, SGCG on 13q12.12. D, CTDP1on 18qter.
Data Processing
Allele frequencies for the five founder mutations were estimated using the carrier rates data. Pairwise differences in carrier frequencies were assessed for statistical significance through use of Fisher’s exact test. The genetic affinities of Gypsy groups were examined using the gene frequency data and Nei’s standard genetic distance (Nei 1987). The software package DISPAN (Ota 1993) was used to generate a population tree on the basis of the neighbor-joining method (Saitou and Nei 1987).
Haplotypes were constructed manually from the family genotyping data. Marker order and distances (fig. 2) followed the deCODE map (Kong et al. 2002; National Center for Biotechnology Information Genome View Web site). For markers not included in the public genetic maps, genetic distances were extrapolated from the physical maps. The core conserved haplotypes included markers close to the disease mutation, and variation in the peripheral markers was used to infer haplotype genealogy. Haplotypes sharing the same historical recombination closest to the disease locus were assigned to the same haplotype group, designated with a letter. Within each haplotype group, further diversification at more peripheral loci generated derivative haplotypes, which were given unique numbers. On the basis of these relationships, a haplotype network was constructed manually, in which the most likely founding haplotype was inferred from allele states at loci close to the disease mutation, the frequency and population distribution of the haplotype, and the diversity of its derivative haplotypes.
Arlequin version 2 (Schneider et al. 2000) was used to estimate haplotype diversity and FST values for each locus and each data set, as well as to examine population differentiation through use of exact tests based on haplotype frequencies. For direct comparisons of diversity, we analyzed haplotypes over comparable regions of ∼3 cM around the disease loci.
Haplotype coalescence times in generations (G) were estimated using a Bayesian approach and the DMLE+ software package (Rannala and Slatkin 1998; Reeve and Rannala 2002), which takes into account the marker information from the entire haplotype (independent of the inferred haplotype genealogy and of the network discussed in the previous section). We have used a generation time of 25 years and a population growth rate (g) of 1.32, as estimated previously (Hunter et al. 2002). Mutation carrier rates determined in the present study were used in conjunction with population size to estimate the fraction of the population sampled for each coalescence time calculation. The haplotypes on nontransmitted chromosomes, characterized in the family analyses, were used as control data. Each run was performed at least twice, with a minimum of 1,000,000 burn-ins preceding a minimum of 1,000,000 “real” iterations.
LD around the disease mutations was assessed using pexcess=(paffected-pnormal)/(1-pnormal), where paffected is the frequency of the disease-associated allele and pnormal is the frequency of that allele in the general population (Hastbacka et al. 1992, 1994). To evaluate background LD, we calculated D′ through use of GOLD software (Abecasis and Cookson 2000) and calculated empirical P values through use of the Markov-chain method of Guo and Thompson (1992) and Arlequin version 2 (Schneider et al. 2000). The step-down Holm Sidak procedure, described by Lautenberger et al. (2000), was used to correct for multiple marker-pair comparisons.
Results
Mutation Frequencies
We identified a total of 151 individuals heterozygous for one of the five disease-causing mutations. The cumulative carrier rate (averaged across the population) was 12.9%. Carrier rates for individual mutations ranged from ∼1.4% to 4.5% (table 1).
The distribution of mutations differed between migrational categories (table 1). The HMSNL, GALK, and CCFDN mutations were present in all categories but were more common among the Vlax Gypsies, whereas LGMD2C was fully confined to the Balkan and western European Gypsies. In contrast, the CMS mutation occurred at approximately equal frequencies in all migrational categories. Pairwise comparisons between the Vlax and Balkan migrational categories in Bulgaria revealed significant differences in the frequencies of the HMSNL (P<3×10-7), CCFDN (P<.029), galactokinase deficiency (P<.012), and LGMD2C (P<.0006) mutations but no difference for the CMS mutation. When all loci were taken into account, the difference between the Balkan and Vlax migrational categories was highly significant (P<.00014).
Mutation frequencies also differed markedly between individual Gypsy groups within each migrational category, as exemplified by CCFDN and LGMD2C, with carrier rates of ∼5%–7% in specific groups (the Rudari for CCFDN and Darakchii and Turgovzi for LGMD2C) and rates that were much lower or zero in other groups of the same migrational category.
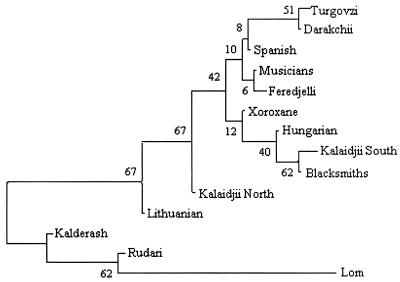
In the neighbor-joining tree based on all disease gene frequency data (fig. 3), the Rudari, Lom, and Kalderash groups from the Vlax migrational category were clustered separately from the Balkan groups. No pattern correlating with the Balkan and western European categories was apparent.
Figure 3.
Neighbor-joining tree of Gypsy groups, based on the gene frequency data obtained from the population screening of the five disease mutations and Nei’s standard genetic distance (Nei 1987). The tree was generated using the DISPAN package (Ota 1993), on the basis of the neighbor-joining method (Saitou and Nei 1987). The numbers indicate the robustness of the branches in the tree, assessed with a bootstrap approach.
Disease Haplotypes
Closely related haplotypes at the CMS, HMSNL, CCFDN, and LGMD2C loci were observed in all Gypsy chromosomes, supporting the common origins of the mutations (tables B–E [online only]). In the CMS region, both Indian/Pakistani and Gypsy chromosomes displayed allele 4 at marker D17S1175, 9.76 kb from the disease mutation, and 121/134 Gypsy and 10/12 Indian/Pakistani chromosomes also shared an identical 1.49-cM three-marker haplotype (table B [online only]). We conclude that the CMS 1267delG mutation in the two populations is derived from a common ancestor. The diverse geographic, linguistic, caste, and religious affiliations of the Indian/Pakistani patients suggest both an old age and a wide geographic dispersal of the mutation in the Indian subcontinent.
Table B.
Frequency of Unique CMS Disease Haplotypes in Individuals from India/Pakistan and in European Gypsies
|
Allele at Marker |
Frequency of Haplotype in |
||||||||||||
| Haplotype | D17S1798 | D17S1845 | D17S1298 | D17S1828 | D17S1175 | D17S1810 | D17S1854 | D17S938 | OverallSample | Balkan Migrational Category | Vlax Migrational Category | West European Migrational Category | Indian/Pakistani Population |
| A1 | 6 | 3 | 3 | 5 | 4 | 9 | 7 | 9 | 11 | 3 | 8 | ||
| A2 | 6 | 3 | 3 | 5 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| A3 | 5 | 3 | 3 | 5 | 4 | 9 | 7 | 2 | 1 | 1 | |||
| A4 | 6 | 7 | 3 | 5 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| A5 | 5 | 12 | 3 | 5 | 4 | 9 | 7 | 2 | 3 | 1 | 2 | ||
| A6 | 6 | 12 | 3 | 5 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| A7 | 6 | 12 | 3 | 5 | 4 | 9 | 7 | 8 | 1 | 1 | |||
| A8 | 6 | 12 | 3 | 5 | 4 | 9 | 7 | 10 | 2 | 1 | 1 | ||
| A9 | 6 | 13 | 3 | 5 | 4 | 9 | 7 | 3 | 2 | 1 | 1 | ||
| A10 | 6 | 13 | 3 | 5 | 4 | 9 | 7 | 9 | 2 | 1 | 1 | ||
| A11 | 6 | 13 | 3 | 5 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| A12 | 11 | 13 | 3 | 5 | 4 | 9 | 7 | 9 | 2 | 2 | |||
| A13 | 5 | 14 | 3 | 5 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| A14 | 6 | 14 | 3 | 5 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| A15 | 10 | 14 | 3 | 5 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| A16 | 6 | 15 | 3 | 5 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| A17 | 6 | 3 | 3 | 5 | 4 | 9 | 3 | 3 | 1 | 1 | |||
| A18 | 5 | 3 | 7 | 5 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| A19 | 6 | 3 | 7 | 5 | 4 | 9 | 7 | 10 | 2 | 2 | |||
| A20 | 3 | 7 | 7 | 5 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| A21 | 6 | 13 | 7 | 5 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| A22 | 6 | 15 | 7 | 5 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| A23 | 5 | 19 | 7 | 5 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| B1 | 5 | 2 | 3 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B2 | 6 | 3 | 3 | 7 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| B3 | 6 | 3 | 3 | 7 | 4 | 9 | 7 | 10 | 2 | 1 | 1 | ||
| B4 | 6 | 3 | 3 | 7 | 4 | 9 | 7 | 11 | 1 | 1 | |||
| B5 | 10 | 3 | 3 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B6 | 11 | 3 | 3 | 7 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| B7 | 6 | 7 | 3 | 7 | 4 | 9 | 7 | 3 | 4 | 4 | |||
| B8 | 6 | 7 | 3 | 7 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| B9 | 6 | 7 | 3 | 7 | 4 | 9 | 7 | 10 | 4 | 3 | 1 | ||
| B10 | 5 | 8 | 3 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B11 | 5 | 12 | 3 | 7 | 4 | 9 | 7 | 3 | 2 | 2 | |||
| B12 | 6 | 12 | 3 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B13 | 6 | 12 | 3 | 7 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| B14 | 6 | 12 | 3 | 7 | 4 | 9 | 7 | 10 | 3 | 2 | 1 | ||
| B15 | 10 | 12 | 3 | 7 | 4 | 9 | 7 | 2 | 1 | 1 | |||
| B16 | 5 | 13 | 3 | 7 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| B17 | 6 | 13 | 3 | 7 | 4 | 9 | 7 | 3 | 2 | 1 | 1 | ||
| B18 | 6 | 13 | 3 | 7 | 4 | 9 | 7 | 9 | 3 | 1 | 2 | ||
| B19 | 6 | 13 | 3 | 7 | 4 | 9 | 7 | 10 | 2 | 1 | 1 | ||
| B20 | 6 | 13 | 3 | 7 | 4 | 9 | 7 | 11 | 1 | 1 | |||
| B21 | 6 | 17 | 3 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B22 | 6 | 7 | 3 | 7 | 4 | 9 | 3 | 10 | 1 | 1 | |||
| B23 | 10 | 3 | 3 | 7 | 4 | 9 | 4 | 3 | 1 | 1 | |||
| B24 | 6 | 7 | 3 | 7 | 4 | 9 | 4 | 3 | 1 | 1 | |||
| B25 | 11 | 3 | 3 | 7 | 4 | 9 | 4 | 9 | 1 | 1 | |||
| B36 | 6 | 20 | 3 | 7 | 4 | 3 | 4 | 10 | 1 | 1 | |||
| B26 | 5 | 3 | 7 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B27 | 11 | 3 | 7 | 7 | 4 | 9 | 7 | 3 | 2 | 2 | |||
| B28 | 11 | 3 | 7 | 7 | 4 | 9 | 7 | 4 | 1 | 1 | |||
| B29 | 6 | 14 | 7 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B30 | 6 | 14 | 7 | 7 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| B31 | 6 | 15 | 7 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B32 | 5 | 13 | 7 | 7 | 4 | 9 | 4 | 3 | 1 | 1 | |||
| B33 | 5 | 13 | 9 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B34 | 6 | 13 | 9 | 7 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| B35 | 6 | 13 | 9 | 7 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| C1 | 3 | 3 | 3 | 4 | 4 | 9 | 7 | 2 | 1 | 1 | |||
| C2 | 6 | 3 | 3 | 4 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| C3 | 10 | 3 | 3 | 4 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| C4 | 6 | 7 | 3 | 4 | 4 | 9 | 7 | 2 | 1 | 1 | |||
| C5 | 11 | 11 | 3 | 4 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| C6 | 5 | 12 | 3 | 4 | 4 | 9 | 7 | 10 | 2 | 2 | |||
| C7 | 6 | 12 | 3 | 4 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| C8 | 6 | 12 | 3 | 4 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| C9 | 5 | 13 | 3 | 4 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| C10 | 6 | 13 | 3 | 4 | 4 | 9 | 7 | 8 | 2 | 2 | |||
| C11 | 6 | 13 | 3 | 4 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| C20 | 6 | 13 | 3 | 4 | 4 | 6 | 7 | 3 | 2 | 2 | |||
| C21 | 6 | 13 | 3 | 4 | 4 | 7 | 7 | 3 | 2 | 2 | |||
| C12 | 7 | 18 | 3 | 4 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| C13 | 6 | 3 | 7 | 4 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| C14 | 11 | 3 | 7 | 4 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| C15 | 5 | 12 | 7 | 4 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| C16 | 6 | 13 | 7 | 4 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| C17 | 11 | 13 | 7 | 4 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| C18 | 5 | 14 | 7 | 4 | 4 | 9 | 7 | 2 | 1 | 1 | |||
| D1 | 6 | 3 | 3 | 6 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| D2 | 11 | 12 | 3 | 6 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| D3 | 11 | 12 | 7 | 6 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| D4 | 6 | 13 | 7 | 6 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| D5 | 5 | 12 | 1 | 6 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| E1 | 6 | 3 | 3 | 8 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| E2 | 6 | 12 | 3 | 8 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| E3 | 11 | 12 | 3 | 8 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| E4 | 6 | 13 | 3 | 8 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| E5 | 6 | 15 | 3 | 8 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| E6 | 5 | 17 | 7 | 8 | 4 | 9 | 7 | 9 | 1 | 1 | |||
| E7 | 5 | 16 | 7 | 8 | 4 | 6 | 4 | 9 | 1 | 1 | |||
| F1 | 5 | 13 | 7 | 10 | 4 | 9 | 7 | 8 | 2 | 2 | |||
| F2 | 6 | 13 | 7 | 10 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| G1 | 5 | 12 | 7 | 11 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| G2 | 5 | 13 | 7 | 11 | 4 | 9 | 7 | 10 | 1 | 1 | |||
| H1 | 5 | 12 | 7 | 13 | 4 | 9 | 7 | 10 | 7 | 4 | 3 | ||
| I1 | 5 | 13 | 3 | ? | 4 | 9 | 7 | 3 | 1 | 1 | |||
| J1 | 6 | 2 | 9 | 12 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| K1 | 5 | 12 | 3 | 3 | 4 | 9 | 7 | 3 | 1 | 1 | |||
| L1 | 5 | 12 | 7 | 12 | 4 | 9 | 6 | 2 | 1 | 1 | |||
| M1 | 6 | 17 | 3 | 5 | 4 | 11 | 7 | 10 | 1 | 1 | |||
| M2 | 6 | 3 | 3 | 7 | 4 | 11 | 7 | 10 | 1 | 1 | |||
| M3 | 5 | 12 | 7 | 13 | 4 | 11 | 7 | 10 | 1 | 1 | |||
Table C.
Frequency of Unique HMSNL Disease Haplotypes in the Gypsies
|
Frequency of Haplotype in |
||||||||||||||||||
|
Allele at Marker |
Vlax Migrational Category |
|||||||||||||||||
| Haplotype | D8S284 | D8S557 | D8S1835 | D8S558 | SLAP | D8S529 | D8S256 | D8S1708 | D8S1796 | D8S1746 | D8S537 | Overall Sample | Balkan Migrational Category | Kalderash | Rudari | Lom | Romanian | Western European Migrational Category |
| A1 | 13 | 1 | 2 | 6 | 8 | 2 | 2 | 4 | 10 | 3 | 1 | 1 | 1 | |||||
| A2 | 13 | 1 | 2 | 6 | 1 | 8 | 8 | 2 | 5 | 3 | 13 | 2 | 2 | |||||
| A3 | 13 | 1 | 2 | 6 | 8 | 2 | 3 | 1 | 12 | 1 | 1 | |||||||
| A4 | 13 | 1 | 2 | 6 | 8 | 3 | 1 | 1 | 11 | 6 | 6 | |||||||
| A5 | 13 | 1 | 2 | 6 | 8 | 3 | 1 | 1 | 12 | 1 | 1 | |||||||
| A6 | 13 | 1 | 2 | 6 | 8 | 3 | 3 | 1 | 13 | 4 | 4 | |||||||
| A7 | 13 | 1 | 2 | 6 | 8 | 3 | 14 | 6 | 12 | 2 | 2 | |||||||
| A8 | 13 | 1 | 2 | 6 | 8 | 4 | 5 | 3 | 12 | 2 | 1 | 1 | ||||||
| A9 | 13 | 1 | 2 | 6 | 1 | 8 | 8 | 4 | 5 | 1 | 13 | 1 | 1 | |||||
| A10 | 13 | 1 | 2 | 6 | 1 | 8 | 5 | 3 | 1 | 1 | 11 | 3 | 3 | |||||
| A11 | 13 | 1 | 2 | 6 | 1 | 8 | 5 | 3 | 1 | 1 | 4 | 1 | 1 | |||||
| A12 | 13 | 1 | 2 | 6 | 16 | 4 | 14 | 1 | 14 | 1 | 1 | |||||||
| A13 | 13 | 1 | 2 | 6 | 13 | 2 | 1 | 1 | 11 | 1 | 1 | |||||||
| A14 | 13 | 1 | 2 | 6 | 17 | 4 | 14 | 2 | 11 | 1 | 1 | |||||||
| A15 | 14 | 1 | 2 | 6 | 1 | 8 | 8 | 2 | 2 | 4 | 10 | 10 | 10 | |||||
| A16 | 14 | 1 | 2 | 6 | 1 | 8 | 8 | 2 | 3 | 1 | 12 | 1 | 1 | |||||
| A17 | 14 | 1 | 2 | 6 | 1 | 8 | 2 | 1 | 3 | 2 | 11 | 1 | 1 | |||||
| A18 | 14 | 1 | 2 | 6 | 8 | 3 | 14 | 4 | 13 | 1 | 1 | |||||||
| A19 | 14 | 1 | 2 | 6 | 1 | 8 | 8 | 3 | 3 | 2 | 14 | 1 | 1 | |||||
| A20 | 14 | 1 | 2 | 6 | 17 | 4 | 14 | 2 | 11 | 1 | 1 | |||||||
| A21 | 15 | 1 | 2 | 6 | 1 | 8 | 8 | 3 | 1 | 1 | 11 | 1 | 1 | |||||
| A22 | 15 | 1 | 2 | 6 | 1 | 8 | 8 | 3 | 1 | 1 | 14 | 1 | 1 | |||||
| A23 | 15 | 1 | 2 | 6 | 1 | 8 | 8 | 3 | 3 | 2 | 14 | 1 | 1 | |||||
| A24 | 16 | 1 | 2 | 6 | 8 | 2 | 2 | 4 | 10 | 2 | 2 | |||||||
| A25 | 1 | 1 | 2 | 6 | 8 | 3 | 1 | 1 | 11 | 2 | 2 | |||||||
| A26 | 1 | 1 | 2 | 6 | 8 | 3 | 14 | 1 | 11 | 1 | 1 | |||||||
| A27 | 2 | 1 | 2 | 6 | 1 | 8 | 8 | 3 | 1 | 1 | 11 | 1 | 1 | |||||
| A28 | 9 | 1 | 2 | 6 | 8 | 3 | 3 | 1 | 13 | 1 | 1 | |||||||
| A29 | 11 | 1 | 2 | 6 | 13 | 2 | 1 | 1 | 11 | 1 | 1 | |||||||
| A30 | 14 | 6 | 2 | 6 | 1 | 8 | 8 | 2 | 2 | 4 | 10 | 3 | 3 | |||||
| A31 | 13 | 6 | 2 | 6 | 1 | 8 | 8 | 2 | 2 | 1 | 11 | 1 | 1 | |||||
| A32 | 13 | 3 | 2 | 6 | 8 | 3 | 14 | 4 | 13 | 1 | 1 | |||||||
| B1 | 13 | 2 | 1 | 6 | 1 | 8 | 8 | 2 | 2 | 4 | 10 | 9 | 8 | 1 | ||||
| B2 | 13 | 2 | 1 | 6 | 1 | 8 | 8 | 2 | 2 | 4 | 11 | 4 | 1 | 3 | ||||
| B3 | 13 | 2 | 1 | 6 | 1 | 8 | 8 | 2 | 2 | 1 | 11 | 6 | 6 | |||||
| B4 | 13 | 2 | 2 | 6 | 1 | 8 | 8 | 2 | 2 | 1 | 11 | 1 | 1 | |||||
| B5 | 13 | 2 | 1 | 6 | 1 | 8 | 8 | 2 | 2 | 1 | 12 | 1 | 1 | |||||
| B6 | 13 | 2 | 1 | 7 | 8 | 2 | 2 | 3 | 14 | 1 | 1 | |||||||
| B7 | 13 | 2 | 1 | 6 | 8 | 2 | 1 | 11 | 1 | 1 | ||||||||
| B8 | 13 | 2 | 1 | 6 | 8 | 4 | 5 | 3 | 12 | 1 | 1 | |||||||
| B9 | 13 | 2 | 1 | 6 | 1 | 8 | 8 | 3 | 1 | 1 | 11 | 1 | 1 | |||||
| B10 | 13 | 2 | 1 | 6 | 1 | 8 | 8 | 4 | 3 | 2 | 11 | 1 | 1 | |||||
| B11 | 13 | 2 | 1 | 6 | 1 | 8 | 5 | 2 | 3 | 2 | 13 | 3 | 3 | |||||
| B12 | 13 | 2 | 1 | 6 | 1 | 8 | 5 | 2 | 3 | 2 | 10 | 1 | 1 | |||||
| B13 | 13 | 2 | 1 | 6 | 1 | 8 | 5 | 2 | 3 | 2 | 15 | 1 | 1 | |||||
| B14 | 13 | 2 | 1 | 6 | 1 | 8 | 2 | 4 | 3 | 3 | 13 | 2 | 2 | |||||
| B15 | 13 | 2 | 1 | 6 | 1 | 8 | 2 | 4 | 3 | 3 | 11 | 1 | 1 | |||||
| B16 | 13 | 2 | 1 | 6 | 8 | 14 | 2 | 3 | 1 | 1 | 1 | 1 | ||||||
| B17 | 13 | 2 | 1 | 6 | 1 | 8 | 14 | 4 | 3 | 4 | 12 | 1 | 1 | |||||
| B18 | 16 | 2 | 1 | 6 | 8 | 2 | 2 | 4 | 11 | 3 | 1 | 2 | ||||||
| B19 | 14 | 2 | 1 | 6 | 1 | 8 | 8 | 2 | 2 | 4 | 10 | 1 | 1 | |||||
| B20 | 14 | 2 | 1 | 6 | 8 | 2 | 2 | 4 | 13 | 1 | 1 | |||||||
| B21 | 14 | 2 | 1 | 6 | 1 | 8 | 8 | 1 | 4 | 1 | 9 | 1 | 1 | |||||
| B22 | 14 | 2 | 1 | 6 | 1 | 8 | 8 | 1 | 4 | 1 | 13 | 1 | 1 | |||||
| B23 | 5 | 2 | 1 | 6 | 1 | 8 | 5 | 2 | 3 | 2 | 14 | 1 | 1 | |||||
| B24 | 1 | 3 | 1 | 6 | 8 | 8 | 3 | 1 | 11 | 1 | 1 | |||||||
| C1 | 1 | 3 | 13 | 6 | 8 | 3 | 14 | 2 | 11 | 2 | 2 | |||||||
| C2 | 1 | 3 | 13 | 6 | 8 | 2 | 3 | 1 | 14 | 1 | 1 | |||||||
| C3 | 1 | 3 | 13 | 6 | 8 | 3 | 14 | 6 | 12 | 1 | 1 | |||||||
| C4 | 11 | 5 | 13 | 6 | 1 | 8 | 8 | 2 | 2 | 4 | 10 | 1 | 1 | |||||
| C5 | 11 | 5 | 13 | 6 | 8 | 2 | 2 | 1 | 11 | 1 | 1 | |||||||
| C6 | 11 | 5 | 13 | 6 | 1 | 8 | 8 | 2 | 2 | 1 | 12 | 1 | 1 | |||||
| C7 | 15 | 5 | 13 | 6 | 8 | 2 | 2 | 4 | 14 | 1 | 1 | |||||||
| C8 | 14 | 5 | 13 | 6 | 8 | 3 | 14 | 4 | 10 | 1 | 1 | |||||||
| D1 | 13 | 3 | 9 | 6 | 8 | 2 | 2 | 4 | 10 | 1 | 1 | |||||||
| E1 | 1 | 4 | 2 | 1 | 1 | 8 | 8 | 2 | 2 | 4 | 10 | 4 | 4 | |||||
| E2 | 1 | 4 | 2 | 1 | 1 | 8 | 8 | 2 | 2 | 4 | 11 | 1 | 1 | |||||
| F1 | 9 | 6 | 1 | 7 | 5 | 2 | 8 | 4 | 14 | 2 | 14 | 2 | 2 | |||||
| F2 | 9 | 6 | 1 | 7 | 5 | 2 | 8 | 4 | 14 | 4 | 14 | 1 | 1 | |||||
| G1 | 5 | 3 | 2 | 9 | 1 | 8 | 8 | 2 | 2 | 4 | 10 | 2 | 2 | |||||
| H1 | 14 | 3 | 1 | 3 | 5 | 8 | 8 | 3 | 3 | 1 | 14 | 2 | 2 | |||||
| I1 | 14 | 2 | 13 | 2 | 3 | 2 | 8 | 2 | 2 | 10 | 1 | 1 | ||||||
| I2 | 14 | 2 | 13 | 2 | 3 | 2 | 8 | 3 | 14 | 3 | 12 | 1 | 1 | |||||
| J1 | 1 | 5 | 2 | 8 | 1 | 8 | 8 | 2 | 2 | 4 | 10 | 1 | 1 | |||||
| K1 | 13 | 1 | 2 | 4 | 1 | ?? | 8 | 4 | 5 | 3 | 12 | 1 | 1 | |||||
| L1 | 14 | 3 | 1 | 5 | 2 | 2 | 13 | 2 | 1 | 2 | 11 | 1 | 1 | |||||
| M1 | 3 | 1 | 5 | 5 | 1 | 8 | 2 | 2 | 4 | 1 | 1 | |||||||
| N1 | 5 | 3 | 2 | 3 | 5 | 2 | 8 | 2 | 2 | 4 | 10 | 1 | 1 | |||||
| O1 | 5 | 3 | 2 | 3 | 5 | 8 | 8 | 2 | 2 | 4 | 4 | 1 | 1 | |||||
| P1 | 13 | 5 | 13 | 4 | 5 | 6 | 8 | 3 | 3 | 1 | 13 | 1 | 1 | |||||
| Q1 | 14 | 1 | 2 | 6 | 1 | 6 | 8 | 4 | 3 | 1 | 1 | 1 | 1 | |||||
| R1 | 1 | 1 | 1 | 6 | 3 | 6 | 2 | 2 | 1 | 1 | 11 | 1 | 1 | |||||
| S1 | 13 | 6 | 3 | 2 | 3 | 5 | 8 | 5 | 3 | 3 | 13 | 1 | 1 | |||||
| T1 | ?6 | 2 | 4 | 2 | 8 | 2 | 1 | 10 | 1 | 1 | ||||||||
Table D.
Frequency of Unique CCFDN Disease Haplotypes in Geographically Diverse Rudari Gypsies.
|
Frequency of Haplotype in |
|||||||||||||||||||||
|
Allele at Marker |
Rudari from |
||||||||||||||||||||
| Haplotype | D18S462 | D18S461 | D18S1122 | 23090ta1 | 1908ca1 | D18S1095 | LOC125267 (5′UTR) | 21594at1 | LOC125261 (5′UTR) | DIM1 | D18S1390 | D18S70 | 68530gt1 | 68530ca1 | 68530gt2 | PAR-6 G (5′UTR) | Overall Sample | Bulgaria | Serbia | Romania | Europe |
| A1 | 4 | 5 | 3 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 11 | 1 | 6 | 2 | 2 |
| A2 | 4 | 5 | 3 | 6 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 5 | 5 | |||
| A3 | 5 | 5 | 3 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 2 | 2 | |||
| A4 | 3 | 3 | 3 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 2 | 1 | 1 | |||
| A5 | 4 | 2 | 3 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 3 | 3 | |||
| A6 | 2 | 4 | 3 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A7 | 2 | 6 | 3 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A8 | 5 | 3 | 1 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 8 | 6 | 2 | ||
| A9 | 4 | 3 | 1 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 3 | 2 | 1 | ||
| A10 | 2 | 3 | 1 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 3 | 3 | |||
| A11 | 2 | 6 | 1 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A12 | 3 | 1 | 4 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A13 | 4 | 2 | 4 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 2 | 2 | |||
| A14 | 5 | 4 | 4 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A15 | 2 | 7 | 4 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A16 | 4 | 3 | 5 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 4 | 4 | |||
| A17 | 2 | 3 | 5 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 3 | 3 | |||
| A18 | 5 | 3 | 5 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A19 | 2 | 2 | 5 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A20 | 6 | 5 | 5 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | ||||
| A21 | 3 | 2 | 1 | 3 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 2 | 2 | |||
| A22 | 4 | 3 | 2 | 3 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A23 | 2 | 6 | 2 | 3 | 6 | 1 | 1 | 4 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | |||||
| A24 | 2 | 1 | 3 | 3 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A25 | 4 | 1 | 3 | 3 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A26 | 3 | 4 | 4 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | ||||
| A27 | 4 | 3 | 5 | 4 | 6 | 1 | 1 | 4 | 2 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | |||
| A28 | 4 | 2 | 3 | 2 | 3 | 2 | 1 | 4 | 2 | 5 | 1 | 3 | 4 | 2 | 1 | 1 | 1 | ||||
| B1 | 4 | 5 | 3 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 25 | 20 | 4 | 1 | |
| B2 | 4 | 5 | 4 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 2 | 2 | |||
| B3 | 2 | 5 | 3 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 2 | 1 | 1 | ||
| B4 | 3 | 5 | 3 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 2 | 2 | |||
| B5 | 5 | 5 | 3 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 5 | 5 | |||
| B6 | 5 | 5 | 4 | 5 | 6 | 1 | 4 | 5 | 1 | 3 | 5 | 2 | 1 | 1 | |||||||
| B7 | 5 | 3 | 3 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 3 | 3 | |||
| B8 | 4 | 3 | 2 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 2 | 2 | |||
| B9 | 2 | 3 | 4 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 3 | 3 | |||
| B10 | 5 | 6 | 4 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 1 | 1 | |||
| B11 | 6 | 6 | 4 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 1 | 1 | |||
| B12 | 4 | 3 | 1 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 1 | 1 | |||
| B13 | 2 | 6 | 3 | 2 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 1 | 1 | |||
| B14 | 3 | 3 | 4 | 4 | 6 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 1 | 1 | |||
| B15 | 2 | 3 | 3 | 5 | 3 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 1 | 1 | |||
| B16 | 4 | 5 | 3 | 2 | 14 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 1 | 1 | 1 | 1 | |||
| B17 | 5 | 3 | 4 | 2 | 14 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 1 | 1 | 1 | 1 | |||
| B18 | 2 | 3 | 2 | 2 | 12 | 1 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 1 | 1 | |||
| B19 | 6 | 6 | 2 | 3 | 6 | 13 | 1 | 4 | 1 | 1 | 5 | 1 | 3 | 5 | 2 | 1 | 1 | 1 | |||
| C1 | 4 | 5 | 3 | 5 | 6 | 1 | 1 | 4 | 2 | 1 | 4 | 5 | 4 | 7 | 8 | 2 | 4 | 1 | 3 | ||
| C2 | 2 | 3 | 3 | 5 | 6 | 1 | 1 | 4 | 2 | 1 | 4 | 5 | 4 | 7 | 8 | 2 | 1 | 1 | |||
| C3 | 5 | 4 | 4 | 5 | 6 | 1 | 1 | 4 | 2 | 1 | 4 | 5 | 4 | 7 | 8 | 2 | 1 | 1 | |||
| C4 | 5 | 2 | 3 | 3 | 6 | 1 | 1 | 4 | 2 | 1 | 4 | 5 | 4 | 7 | 8 | 2 | 1 | 1 | |||
| D1 | 5 | 4 | 4 | 5 | 6 | 1 | 1 | 4 | 2 | 1 | 4 | 4 | 4 | 6 | 4 | 2 | 1 | 1 | |||
| E1 | 4 | 5 | 3 | 5 | 6 | 1 | 1 | 4 | 2 | 1 | 3 | 5 | 6 | 3 | 3 | 1 | 1 | 1 | |||
| F1 | 4 | 5 | 3 | 5 | 6 | 1 | 1 | 4 | 1 | 1 | 3 | 5 | 7 | 3 | 3 | 1 | 1 | 1 | |||
Table E.
Frequency of Unique LGMD2C Disease Haplotypes in Balkan and Western European Gypsies
|
Allele at Marker |
Frequency of Haplotype in |
|||||||
| Haplotype | D13S115 | D13S232 | D13S292 | D13S787 | D13S283 | Overall Sample | Balkan Migrational Category | Western European Migrational Category |
| A1 | 2 | 3 | 3 | 3 | 10 | 34 | 29 | 5 |
| A2 | 3 | 3 | 3 | 3 | 10 | 7 | 7 | |
| A3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | |
| A4 | 2 | 3 | 3 | 2 | 10 | 1 | 1 | |
| A5 | 7 | 3 | 3 | 2 | 2 | 1 | 1 | |
| A6 | 5 | 3 | 3 | 2 | 2 | 2 | 1 | 1 |
| A7 | 5 | 3 | 3 | 3 | 10 | 1 | 1 | |
| A8 | 2 | 3 | 3 | 3 | 14 | 3 | 1 | 2 |
| A9 | 2 | 3 | 3 | 3 | 2 | 20 | 4 | 16 |
| A10 | 2 | 3 | 3 | 3 | 6 | 1 | 1 | |
| A11 | 2 | 3 | 1 | 3 | 2 | 1 | 1 | |
| A12 | 7 | 3 | 3 | 3 | 2 | 1 | 1 | |
| A13 | 1 | 3 | 3 | 3 | 2 | 1 | 1 | |
| A14 | 2 | 3 | 3 | 2 | 2 | 1 | 1 | |
| A15 | 5 | 3 | 3 | 3 | 2 | 1 | 1 | |
| A16 | 2 | 3 | 1 | 2 | 2 | 2 | 2 | |
Haplotype diversity and coalescence times
In the CMS region (table B [online only]), we identified 91 haplotypes among 134 Gypsy chromosomes and 103 haplotypes in the entire sample of 146 chromosomes (including the Indian/Pakistani), with a mean ± SD haplotype diversity index (HDI) of 0.987±0.003. Haplotypes were assigned to 13 groups, of which groups A, B, and C accounted for ∼80% of chromosomes.
The 140 HMSNL chromosomes (table C [online only]) presented with a total of 84 haplotypes (HDI 0.984±0.004), which fell into 20 different haplotype groups, with groups A and B accounting for 75% of disease chromosomes. Analysis of the CCFDN region revealed 54 haplotypes among 129 chromosomes (HDI 0.947±0.012), with haplotype groups A and B occurring in >90% of chromosomes (table D [online only]).
The LGMD2C region showed limited diversity (HDI 0.741±0.039), with only 16 haplotypes observed among 78 disease chromosomes. Haplotypes fell into a single group, with A1, A2, and A9 accounting for >75% of chromosomes (table E [online only]).
To obtain a direct comparison among the four loci, we reexamined haplotype diversity in similar ∼3-cM intervals. Diversity at the HMSNL, CMS, and CCFDN loci decreased by ∼10% (HDI ∼0.850 for all) and remained considerably higher than that for LGMD2C (HDI 0.741±0.039), indicating that differences in haplotype length introduced minimal bias.
We used the haplotype data at the different loci to estimate coalescence times in the entire Gypsy population, in migrational categories, and in individual Gypsy groups (table 3). Haplotypes surrounding the widely represented CMS 1267delG and HMSNL R148X mutations were used to examine coalescence times for the entire population. The dates obtained with the DMLE+ software were 800 (95% CI 650–1,025) years for 1267delG and 850 (700–1,075) years for R148X. An alternative method (Stephens et al. 1998) produced similar estimates of ∼900 years for both mutations. The LGMD2C mutation C283Y, limited to the Balkan and western European Gypsies, showed an overall haplotype coalescence time of ∼600 (525–775) years, and a time of 725 years was obtained with the Stephens et al. (1998) method. DMLE+ estimates of haplotype coalescence times for the CMS, HMSNL, and LGMD2C mutations in the different migrational categories ranged between 475 (375–700) years for CMS in the Vlax Gypsies and 600 (475–825) years for HMSNL in the Vlax and western European Gypsies. The dating in individual Gypsy groups produced values between 425 (300–650) years for HMSNL in the Lom and LGMD2C in the Turgovzi and 600 (500–775) years for HMSNL in the Rudari.
Table 3.
DMLE+ Estimates of Mutation Ages Obtained Using a Growth Rate of 1.32
|
Mean (95% CI) Age of Mutationa(years) |
||||
| Population | CHRNE(CMS) | NDRG1(HMSNL) | SGCG(LGMD2C) | CTDP1(CCFDN) |
| Entire sample | 800 (650–1,025) | 850 (700–1,075) | 600 (525–775)b | … |
| Migrational category: | ||||
| Balkan | 550 (450–775) | 550 (450–750) | … | … |
| Vlax | 475 (375–700) | 600 (525–825) | … | … |
| Western European | 550 (450–775) | 600 (475–825) | 525 (450–750) | … |
| Individual group: | ||||
| Rudari | … | 600 (500–775) | … | 500 (400–650) |
| Kalderash | … | 500 (375–675) | … | … |
| Lom | … | 425 (300–650) | … | … |
| Turgovzi | … | … | 425 (375–650) | … |
A generation time of 25 years was used in all estimates.
The “entire” Gypsy sample in this case does not include the Vlax Gypsies, in whom LGMD2C does not occur.
Haplotype genealogy and LD mapping
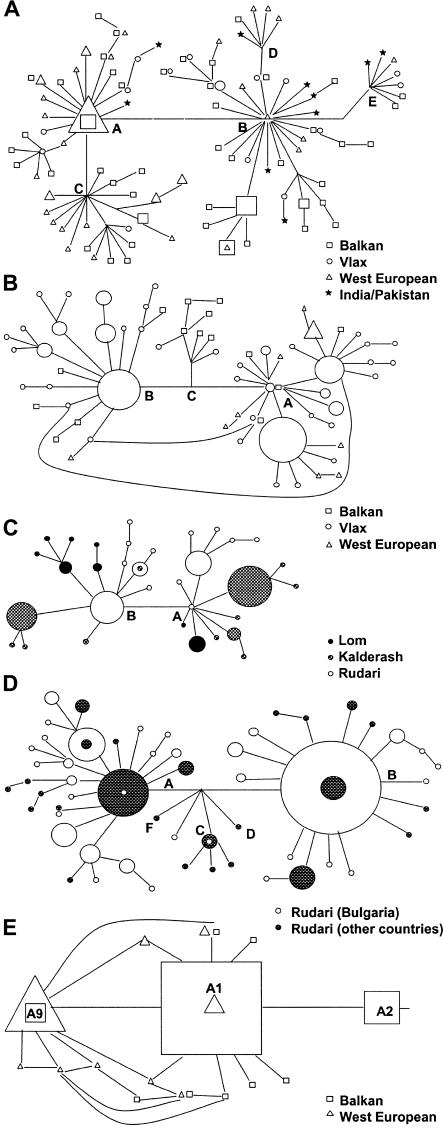
We observed different patterns of the haplotype networks, reflecting the age and history of the mutations. The CMS 1267delG mutation, whose presence in subjects from many parts of the Indian subcontinent pointed to its old age and wide dispersal, presented with a complex genealogy (fig. 4A). The major haplotype groups were represented and nearly evenly distributed in all Gypsy migrational categories and in Indian/Pakistani chromosomes, and unique haplotypes were shared between migrational categories (11% were shared by Balkan and Vlax Gypsies, and 6% were shared between Balkan and western European Gypsies). Haplotype distribution showed no difference between the Balkan and Vlax migrational categories but was significantly different (P<.001 for FST and Fisher’s exact test) between these two and the western European category.
Figure 4.
Networks showing the genealogical relationships of disease haplotypes. The size of the symbols is proportional to the frequency of each haplotype. A, CMS haplotypes in the CHRNE region. B, HMSNL haplotypes in the NDRG1 region. C, HMSNL haplotypes in Gypsy groups from the Vlax migrational category. D, CCFDN haplotypes in the CTDP1 region. E, LGMD2C haplotypes in the SGCG region. For CMS and HMSNL, only haplotype groups comprising more than five unique haplotypes are shown.
In contrast to CMS, the HMSNL haplotype network showed clustering by migrational categories (fig. 4B) and very limited sharing of haplotype groups and unique haplotypes. No haplotype occurred in all migrational categories, and <3% (2 rare haplotypes out of 76) were shared by the Balkan and Vlax categories (FST analysis P<.05; Fisher’s exact test P<.001). We also observed a striking lack of haplotype sharing between Gypsy groups in the Vlax category; no haplotype was common to all three groups, and only one occurred in two groups (fig. 4C). The differences were highly significant, with P<.001 for FST comparisons involving the Lom, P<.05 for the Rudari and Kalderash, and P<.001 for all comparisons using Fisher’s exact test.
The younger CCFDN and LGMD2C mutations, confined to individual Gypsy groups, presented with simple haplotype genealogy (fig. 4D and 4E). In the CCFDN region, the two major haplotype groups occurred in Rudari communities from different European countries, with sharing of ∼10% of unique haplotypes yet significant geographic differences in haplotype distribution (P<.05 for FST analysis and P<.001 for Fisher’s exact test). LGMD2C haplotypes showed a relatively high degree of sharing, with 25% of unique haplotypes occurring in both the Balkan and western European categories and an obvious secondary founder effect in western European Gypsies. Again, haplotype frequency distributions differed markedly (P<.001 for FST analysis and for Fisher’s exact test).
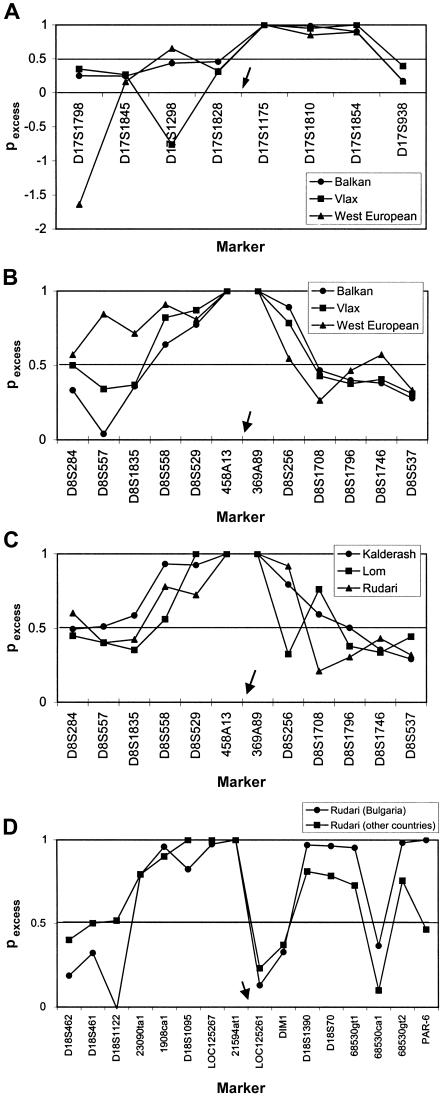
Haplotype divergence and limited sharing were reflected in the LD analysis of the CMS, HMSNL, and CCFDN gene regions (fig. 5A–5D). The pexcess analysis (Hastbacka et al. 1992, 1994) revealed different patterns in the different migrational categories and Gypsy groups, pointing to a history of independent recombinations and marker mutations, and the distance where pexcess was >0.5 in all populations varied from 0.07 to 2.12 cM. In all cases, the comparison between migrational categories and Gypsy groups pinpointed the location of the disease-causing mutation through the identification of a shared interval of complete LD (pexcess=1).
Figure 5.
LD estimated by pexcess (Hastbacka et al. 1992, 1994) for markers surrounding the founder disease mutations. Markers are shown as equidistant from one another. Intermarker distances are shown in figures 2 and 6. The arrows indicate the approximate location of the disease mutation. A, LD in the CMS gene region in the three migrational categories. B, LD in the HMSNL region in the three migrational categories. C, LD in the HMSNL region in Gypsy groups from the Vlax migrational category. D, LD in the CCFDN region for geographically separated Rudari groups.
Normal Haplotypes
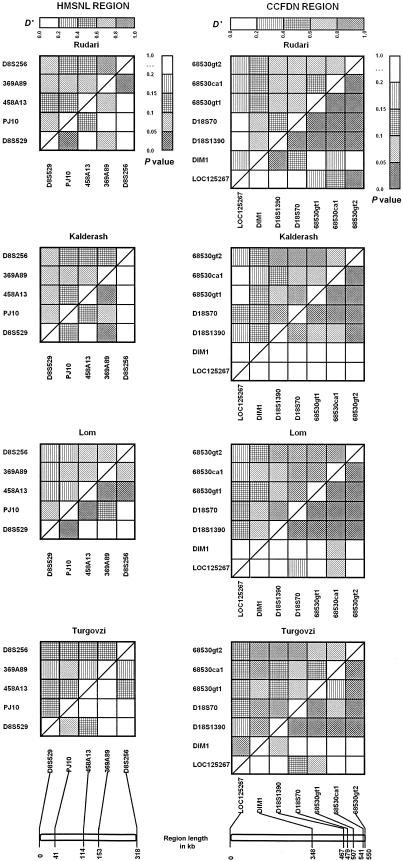
Mean D′ levels were >0.5 for marker pairs separated by as many as ∼150 kb in all Gypsy groups (fig. 6) except in the Turgovzi for the HMSNL region (D′=0.45). Within this range, we did not observe a trend toward decreasing D′ values with increasing distances. For the entire length of both regions, mean D′ values were in excess of 0.5 in three Vlax groups and 0.47 in the Turgovzi. The significance, presented as P values, showed a more variable pattern, especially for the HMSNL region on 8q (fig. 6). The patterns observed were distinctly different in the four Gypsy groups, reflecting an independent history of random recombinations. They showed notably greater similarity in the small D18S70-to-68530gt2 interval in the CCFDN region, spanning a total of ∼70 kb.
Figure 6.
Pairwise LD between markers in the HMSNL and CCFDN disease gene regions in four Gypsy groups. A, Rudari. B, Kalderash. C, Lom. D, Turgovzi. Marker loci are listed across the bottom and down the left side. A scale indicating physical distances (in kb) between markers is shown at the bottom. D′ values are shown in the upper left-hand triangle, and P values are shown in the lower right-hand triangle. The shading patterns used to illustrate the scale of D′ and P values are shown above and at the right-hand side of the top panel. P values <.05 have been corrected for multiple comparisons through use of the Holm-Sidak approach (Lautenberger et al. 2000).
Discussion
In this study, the detailed characterization of several disease loci was used to address hitherto unanswered questions about Gypsy population history. This is also, to our knowledge, the first systematic investigation of mutation carrier rates in a sample representative of the traditional social structure of this population.
Mutation screening revealed high carrier rates, with an average of one in eight subjects heterozygous for one of the mutations tested. Within individual Gypsy groups, carrier rates >5% for specific mutations were observed in 14 cases. These frequencies, which suggest strong founder effects, fall within the range of estimates for five of the most common mutations in the Finnish and Ashkenazi Jewish populations (Pastinen et al. 2001; Risch et al. 2003). The data indicate that recessive Mendelian disorders represent a considerable health burden in the Gypsy population and, given the observed allelic homogeneity, that carrier-testing programs in Gypsy communities are likely to be highly beneficial.
Sharing of the five mutations and related disease haplotypes further supported the genetic relatedness of geographically and socially separated Gypsy groups. Moreover, we have demonstrated the identity of the CMS 1267delG mutation in Gypsy and in Indian/Pakistani subjects, thus providing the strongest evidence to date for the Indian origins of the Gypsies. At the same time, the observed highly significant differences in the distribution of the ancestral mutations and of their associated chromosomal haplotypes indicate that the population fissions that led to the formation of migrational categories and individual Gypsy groups have played the role of profound secondary and tertiary bottleneck events, generating internal differentiation and multiple genetic subisolates. The time frame within which the observed population structure originated and evolved is unclear. Although the founding of the proto-Gypsy population is likely to coincide with the exodus from India, proposed dates range from 900 to 1,500 years before present, and there have been suggestions of multiple migrations (Turner 1926; Sampson 1927; Hancock 1987, 1999, 2000; Fraser 1992). There are also no records of historical events that could explain and date the splits into divergent migration routes and numerous endogamous groups. These could have been nonrandom and based on preexisting internal structure.
To address these unresolved yet important issues of population history, we used haplotype coalescence times at independent disease loci. We reasoned that the two most common and widespread mutations (1267delG and R148X) are likely to be derived from the ancestral population, an assumption supported by the identity of the CMS 1267delG mutation in Gypsy and Indian/Pakistani chromosomes. Their haplotype coalescence times, estimated for the entire set of Gypsy disease chromosomes, would thus correspond to the founding of the proto-Gypsy population. The results for both loci, obtained by two different approaches, support a single recent founding event ∼800 years ago (range 650–1,075 years) when the Reeve and Rannala (2002) approach is used and ∼900 years ago when the method of Stephens et al. (1998) is used. Although such dates should be regarded as an upper limit and are based on a number of assumptions (such as generation time and growth rate), the mean of 800–900 years coincides closely with the time of the exodus proposed by Hancock (2000), who links it historically to the early Islamic invasions of India in the 11th century a.d. Further comparisons of the CMS, HMSNL, and LGMD2C haplotype coalescence times were performed, to date the separation into migrational categories. The estimated period, ∼500–600 years ago, coincides with a previous dating of the founding Y chromosome haplogroup in the Vlax Roma, ∼500 years ago (Kalaydjieva et al. 2001a). Similar and somewhat more recent estimates of 425–600 years were obtained for individual Gypsy groups. The overlap between the time of the founding of the overall Vlax migrational category and of its constituent groups is plausible, since historical records suggest that the enslavement of Gypsies and their forced separation into groups based on ownership and trade took place very soon after their arrival in Romania (Hancock 1987). Our previous analyses have produced more recent dates, closer to the periods of migration of these groups out of Romania and into Bulgaria (Kalaydjieva et al. 2001a; Chaix et al. 2004). As more information on Mendelian disorders in both Gypsies and populations in the Indian subcontinent becomes available, it will be important to perform further studies to corroborate the present findings. Although more-accurate dating may be achieved with larger samples and numbers of loci, the current results clearly point to very recent splits and, hence, to rapidly occurring genetic divergence.
Our earlier observations on the limited diversity of the ancestral Indian Y chromosome and mtDNA lineages have led us to suggest that the European Gypsy population was founded by a small group of related individuals (Gresham et al. 2001). In a subsequent study using coalescence-based methods (Kuhner et al. 1998; Wilson and Balding 1998; Beerli and Felsenstein 2001), we estimated the effective size of the male founding population in individual Vlax Roma groups at ∼100 individuals per group (Chaix et al. 2004). The genetic data are supported by the limited available information on historical demography: according to the Ottoman tax registry, in 1522–1523, there were a total of 5,700 Gypsies in Bulgaria (Marushiakova and Popov 1997). The numerous extant groups of the Balkan category are derived from this small population. The number of mutation carriers among the founders would have been very small, and random drift would have played a major role in the genetic divergence of Gypsy groups. A surprising finding of the present study is the independent evolution of disease haplotypes and the lack of sharing, pointing to a limited gene flow. The present-day rules of endogamy proscribe intermarriage between most Gypsy groups; however, there is no historical information on marriage patterns, and it is unclear when these strict rules were established (Liegeois 1994). Our data suggest that the history of endogamy is as old as Gypsy groups, and the exchange of migrants has been very limited throughout.
Tentative evidence of substructure in the European Gypsy population has been provided by numerous early studies, using classical protein markers such as blood groups (reviewed and reanalyzed by Kalaydjieva et al. [2001b]), that examined genetic affinities between Gypsies residing in different countries but failed to place the findings in the context of the social organization of the Gypsies and the cultural divergence of Gypsy groups. Further support has come from recent studies of Y chromosome and mtDNA diversity (Gresham et al. 2001; Kalaydjieva et al. 2001a). Internal differentiation of a founder population is not unique to the Gypsies; it has been well investigated and exploited in genetic research in Finland and Sardinia (Peltonen et al. 2000; Pastinen et al. 2001; Angius et al. 2002). On a condensed time scale, the population history of the Gypsies replicates that of the Jews, with its exodus, diaspora, and subsequent fragmentation into small, geographically dispersed and isolated communities. The ages of disease mutations in the Gypsies can thus be likened to the scenario described by Risch et al. (2003) for the Jewish population. The widely distributed CMS and HMSNL mutations were already present at the time of the founding of the proto-Gypsy population, 32–40 generations before present. LGMD2C and galactokinase deficiency (Hunter et al. 2002) are more restricted and represent the founding and expansion of the Gypsy migrational categories 20+ generations ago, whereas CCFDN falls into the most recent “age group,” coinciding with the founding of individual subisolates. However, unlike the Ashkenazi Jewish population, the amalgamation of which has already been going on for several generations, social organization and marriage customs have maintained the genetic structure in the Gypsies to the present day.
Our data on single-gene disorders point to the Gypsies as a population offering unique opportunities for the mapping of genes involved in complex disorders. The strong primary founder effect raises expectations of a relatively homogeneous genetic basis of complex disorders, similar to all founder populations. The observed haplotype divergence in migrational categories and Gypsy groups illustrates the limitations of the concept of universal haplotype blocks and the complexity that can be revealed through a detailed, “magnifying glass” examination of population history. It also provides a powerful tool for the fine mapping of disease genes. The best opportunities are presented by older genetic variants with a wide distribution, in which initial analyses aimed at the crude mapping of susceptibility genes can be performed in traditional extended families originating from a single Gypsy group, with no specific advantage of any Gypsy group revealed by our current data on background LD. Fine mapping would be greatly facilitated by wider sampling, based on recognition of the divergent history of multiple Gypsy groups, where the disease locus can be placed within the small genomic region in which all subisolates share the highest LD values. The full potential of this interesting founder population can be met only by informed study designs that acknowledge traditional social organization and its genetic impact.
Acknowledgments
We are indebted to the patients, their families, and the members of many Gypsy communities, for making this study possible. We thank the Laboratory of Molecular Pathology of the Medical University in Sofia, Bulgaria, for infrastructure support; Lyle Palmer, for critical reading of the manuscript; Elena Marushiakova and Vesselin Popov, for consultations and guidance in cultural anthropology; Ursula Klutzny, for expert technical assistance; and Dr. Andrew G. Engel (Rochester, NY), for initial mutation testing in Bulgarian patients with CMS. This work was supported by the Australian Research Council, the National Health and Medical Research Council, and The Wellcome Trust of the U.K. We acknowledge support from the Deutsche Forschungsgemeinschaft (to H.L. and A.A.), from the Medical Research Council (U.K.) and the Muscular Dystrophy Campaign/Myasthenia Gravis Association of Great Britain (to D.B.), and from Hungarian-German Oktatasi Miniszterium–Bundesministerium für Bildung und Forschung (OM-BMBF) and Magyar Tudomanyos Akademia–Deutsche Forschungsgemeinschaft (MTA-DFG) Intergovernmental Scientific Cooperation Grants (to V.K. and H.L.).
Electronic-Database Information
The URLs for data presented herein are as follows:
- National Center for Biotechnology Information Genome View, http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=9606 (for the deCODE genetic map)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HMSNL, CCFDN, CMS, LGMD2C, and galactokinase deficiency)
References
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 10.1093/bioinformatics/16.2.182 [DOI] [PubMed] [Google Scholar]
- Abicht A, Stucka R, Karcagi V, Herczegfalvi A, Horvath R, Mortier W, Schara U, Ramaekers V, Jost W, Brunner J, Janssen G, Seidel U, Schlotter B, Muller-Felber W, Pongratz D, Rudel R, Lochmuller H (1999) A common mutation (epsilon1267delG) in congenital myasthenic patients of Gypsy ethnic origin. Neurology 53:1564–1569 [DOI] [PubMed] [Google Scholar]
- Agarwala R, Biesecker LG, Tomlin JF, Schaffer AA (1999) Towards a complete North American Anabaptist genealogy: a systematic approach to merging partially overlapping genealogy resources. Am J Med Genet 86:156–161 [DOI] [PubMed] [Google Scholar]
- Angelicheva D, Turnev I, Dye D, Chandler D, Thomas PK, Kalaydjieva L (1999) Congenital cataracts facial dysmorphism neuropathy (CCFDN) syndrome: a novel developmental disorder in Gypsies maps to 18qter. Eur J Hum Genet 7:560–566 [DOI] [PubMed] [Google Scholar]
- Angius A, Bebbere D, Petretto E, Falchi M, Forabosco P, Maestrale B, Casu G, Persico I, Melis PM, Pirastu M (2002) Not all isolates are equal: linkage disequilibrium analysis on Xq13.3 reveals different patterns in Sardinian sub-populations. Hum Genet 111:9–15 10.1007/s00439-002-0753-z [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Muenke M (2002) Genetics of population isolates. Clin Genet 61:233–247 10.1034/j.1399-0004.2002.610401.x [DOI] [PubMed] [Google Scholar]
- Beerli P, Felsenstein J (2001) Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA 98:4563–4568 10.1073/pnas.081068098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix R, Austerlitz F, Morar B, Kalaydjieva L, Heyer E (2004) Vlax Roma history: what do coalescent-based methods tell us? Eur J Hum Genet 12:285–292 10.1038/sj.ejhg.5201126 [DOI] [PubMed] [Google Scholar]
- Chandler D, Angelicheva D, Heather L, Gooding R, Gresham D, Yanakiev P, de Jonge R, et al (2000) Hereditary motor and sensory neuropathy–Lom (HMSNL): refined genetic mapping in Romani (Gypsy) families from several European countries. Neuromuscul Disord 10:584–591 10.1016/S0960-8966(00)00148-6 [DOI] [PubMed] [Google Scholar]
- Croxen R, Newland C, Betty M, Vincent A, Newsom-Davis J, Beeson D (1999) Novel functional epsilon-subunit polypeptide generated by a single nucleotide deletion in acetylcholine receptor deficiency congenital myasthenic syndrome. Ann Neurol 46:639–647 [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Wright FA (1998) Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc Natl Acad Sci USA 95:12416–12423 10.1073/pnas.95.21.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A (1992) The Gypsies. Blackwell Publishers, Oxford [Google Scholar]
- Gresham D, Morar B, Underhill PA, Passarino G, Lin AA, Wise C, Angelicheva D, Calafell F, Oefner PJ, Shen P, Tournev I, de Pablo R, Kucinskas V, Perez-Lezaun A, Marushiakova E, Popov V, Kalaydjieva L (2001) Origins and divergence of the Roma (Gypsies). Am J Hum Genet 69:1314–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulcher J, Stefansson K (1998) Population genomics: laying the groundwork for genetic disease modeling and targeting. Clin Chem Lab Med 36:523–527 [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA (1992) Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48:361–372 [PubMed] [Google Scholar]
- Hancock I (1987) The pariah syndrome. Karoma Publishers, Ann Arbor [Google Scholar]
- ——— (1999) The origins and westward migration of the Romani people. Occasional Paper of the International Romani Archives No 5:675, 989 [Google Scholar]
- ——— (2000) The emergence of Romani as a koine outside of India. In: Acton T (ed) Scholarship and Gypsy struggle: commitment in Romani studies. University of Hertfordshire Press, Hatfield, England, pp 1–13 [Google Scholar]
- Hastbacka J, de la Chapelle A, Kaitila I, Sistonen P, Weaver A, Lander E (1992) Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nat Genet 2:204–211 10.1038/ng1192-204 [DOI] [PubMed] [Google Scholar]
- Hastbacka J, de la Chapelle A, Mahtani MM, Clines G, Reeve-Daly MP, Daly M, Hamilton BA, Kusumi K, Trivedi B, Weaver A, Coloma A, Lovett M, Buckler A, Kaitila I, Lander ES (1994) The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell 78:1073–1087 10.1016/0092-8674(94)90281-X [DOI] [PubMed] [Google Scholar]
- Heutink P, Oostra BA (2002) Gene finding in genetically isolated populations. Hum Mol Genet 11:2507–2515 10.1093/hmg/11.20.2507 [DOI] [PubMed] [Google Scholar]
- Hunter M, Heyer E, Austerlitz F, Angelicheva D, Nedkova V, Briones P, Gata A, de Pablo R, Laszlo A, Bosshard N, Gitzelmann R, Tordai A, Kalmar L, Szalai C, Balogh I, Lupu C, Corches A, Popa G, Perez-Lezaun A, Kalaydjieva LV (2002) The P28T mutation in the GALK1 gene accounts for galactokinase deficiency in Roma (Gypsy) patients across Europe. Pediatr Res 51:602–606 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Calafell F, Jobling MA, Angelicheva D, de Knijff P, Rosser ZH, Hurles ME, Underhill P, Tournev I, Marushiakova E, Popov V (2001a) Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages. Eur J Hum Genet 9:97–104 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Calafell F (2001b) Genetic studies of the Roma (Gypsies): a review. BMC Med Genet 2:5 10.1186/1471-2350-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Gresham D, Gooding R, Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D, Chandler D, Worsley P, Rosenthal A, King RH, Thomas PK (2000) N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy–Lom. Am J Hum Genet 67:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Hallmayer J, Chandler D, Savov A, Nikolova A, Angelicheva D, King RH, Ishpekova B, Honeyman K, Calafell F, Shmarov A, Petrova J, Turnev I, Hristova A, Moskov M, Stancheva S, Petkova I, Bittles AH, Georgieva V, Middleton L, Thomas PK (1996) Gene mapping in Gypsies identifies a novel demyelinating neuropathy on chromosome 8q24. Nat Genet 14:214–217 10.1038/ng1096-214 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, King R, Gresham D, Molnar M, Tournev I, Angelicheva D, Butinar D, Colomer J, Corches A, Lupu C, Merlini L, Zanetti M, Bergonzoni C, Thomas PK (2001c) Hereditary motor and sensory neuropathy Lom. Acta Myologica XX:192–201 [Google Scholar]
- Kalaydjieva L, Morar B (2003) Roma (Gypsies): genetic studies. In: Cooper DN (ed) Nature encyclopedia of the human genome. Vol 5. Nature Publishing Group, London, pp 160–165 [Google Scholar]
- Kalaydjieva L, Nikolova A, Turnev I, Petrova J, Hristova A, Ishpekova B, Petkova I, Shmarov A, Stancheva S, Middleton L, Merlini L, Trogu A, Muddle JR, King RH, Thomas PK (1998) Hereditary motor and sensory neuropathy–Lom, a novel demyelinating neuropathy associated with deafness in Gypsies: clinical, electrophysiological and nerve biopsy findings. Brain 121:399–408 10.1093/brain/121.3.399 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Perez-Lezaun A, Angelicheva D, Onengut S, Dye D, Bosshard NU, Jordanova A, Savov A, Yanakiev P, Kremensky I, Radeva B, Hallmayer J, Markov A, Nedkova V, Tournev I, Aneva L, Gitzelmann R (1999) A founder mutation in the GK1 gene is responsible for galactokinase deficiency in Roma (Gypsies). Am J Hum Genet 65:1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 10.1038/9642 [DOI] [PubMed] [Google Scholar]
- Kuhner MK, Yamato J, Felsenstein J (1998) Maximum likelihood estimation of population growth rates based on the coalescent. Genetics 149:429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger JA, Stephens JC, O’Brien SJ, Smith MW (2000) Significant admixture linkage disequilibrium across 30 cM around the FY locus in African Americans. Am J Hum Genet 66:969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegeois J-P (1994) Roma, Gypsies, travellers. Council of Europe Press, Strasbourg [Google Scholar]
- Marushiakova E, Popov V (1997) Gypsies (Roma) in Bulgaria. In: Studien zur Tsiganologie und Folkloristik. Peter Lang, Frankfurt and Main, pp 15–122 [Google Scholar]
- Merlini L, Kaplan JC, Navarro C, Barois A, Bonneau D, Brasa J, Echenne B, Gallano P, Jarre L, Jeanpierre M, Kalaydjieva L, Leturcq F, Levi-Gomes A, Toutain A, Tournev I, Urtizberea A, Vallat JM, Voit T, Warter JM (2000) Homogeneous phenotype of the Gypsy limb-girdle MD with the gamma-sarcoglycan C283Y mutation. Neurology 54:1075–1079 [DOI] [PubMed] [Google Scholar]
- Motulsky AG (1995) Jewish diseases and origins. Nat Genet 9:99–101 10.1038/ng0295-99 [DOI] [PubMed] [Google Scholar]
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York [Google Scholar]
- Norio R (2003) Finnish disease heritage II: population prehistory and genetic roots of Finns. Hum Genet 112:457–469 [DOI] [PubMed] [Google Scholar]
- Ostrer H (2001) A genetic profile of contemporary Jewish populations. Nat Rev Genet 2:891–898 10.1038/35098506 [DOI] [PubMed] [Google Scholar]
- Ota T (1993) Dispan: genetic distance and phylogenetic analysis. Institute of Molecular Evolutionary Genetics, The Pennsylvania State University, University Park, PA [Google Scholar]
- Pastinen T, Perola M, Ignatius J, Sabatti C, Tainola P, Levander M, Syvanen AC, Peltonen L (2001) Dissecting a population genome for targeted screening of disease mutations. Hum Mol Genet 10:2961–2972 10.1093/hmg/10.26.2961 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Jalanko A, Varilo T (1999) Molecular genetics of the Finnish disease heritage. Hum Mol Genet 8:1913–1923 10.1093/hmg/8.10.1913 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 10.1038/35042049 [DOI] [PubMed] [Google Scholar]
- Petulengro G (1915–1916) Report on the Gypsy tribes of north-east Bulgaria. J Gypsy Lore Soc 9:1–109 [Google Scholar]
- Piccolo F, Jeanpierre M, Leturcq F, Dode C, Azibi K, Toutain A, Merlini L, Jarre L, Navarro C, Krishnamoorthy R, Tome FM, Urtizberea JA, Beckmann JS, Campbell KP, Kaplan JC (1996) A founder mutation in the gamma-sarcoglycan gene of gypsies possibly predating their migration out of India. Hum Mol Genet 5:2019–2022 10.1093/hmg/5.12.2019 [DOI] [PubMed] [Google Scholar]
- Rannala B, Slatkin M (1998) Likelihood analysis of disequilibrium mapping, and related problems. Am J Hum Genet 62:459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve JP, Rannala B (2002) DMLE+: Bayesian linkage disequilibrium gene mapping. Bioinformatics 18:894–895 10.1093/bioinformatics/18.6.894 [DOI] [PubMed] [Google Scholar]
- Risch N, de Leon D, Ozelius L, Kramer P, Almasy L, Singer B, Fahn S, Breakefield X, Bressman S (1995) Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat Genet 9:152–159 10.1038/ng0295-152 [DOI] [PubMed] [Google Scholar]
- Risch N, Tang H, Katzenstein H, Ekstein J (2003) Geographic distribution of disease mutations in the Ashkenazi Jewish population supports genetic drift over selection. Am J Hum Genet 72:812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T, Chandler D, Angelicheva D, Thomas PK, Youl B, Tournev I, Gergelcheva V, Kalaydjieva L (2000) A novel locus for autosomal recessive peripheral neuropathy in the EGR2 region on 10q23. Am J Hum Genet 67:664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425 [DOI] [PubMed] [Google Scholar]
- Sampson J (1927) Notes on Professor RL Turner’s “The position of Romani in Indo-Aryan.” J Gypsy Lore Soc 6:57–68 [Google Scholar]
- Schneider S, Roessli D, Excoffier L (2000) Arlequin version 2.000: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva [Google Scholar]
- Scriver CR (2001) Human genetics: lessons from Quebec populations. Annu Rev Genomics Hum Genet 2:69–101 10.1146/annurev.genom.2.1.69 [DOI] [PubMed] [Google Scholar]
- Sheffield VC, Stone EM, Carmi R (1998) Use of isolated inbred human populations for identification of disease genes. Trends Genet 14:391–396 10.1016/S0168-9525(98)01556-X [DOI] [PubMed] [Google Scholar]
- Shifman S, Darvasi A (2001) The value of isolated populations. Nat Genet 28:309–310 10.1038/91060 [DOI] [PubMed] [Google Scholar]
- Stephens JC, Reich DE, Goldstein DB, Shin HD, Smith MW, Carrington M, Winkler C, et al (1998) Dating the origin of the CCR5-Delta32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet 62:1507–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournev I, Kalaydjieva L, Youl B, Ishpekova B, Guergueltcheva V, Kamenov O, Katzarova M, Kamenov Z, Raicheva-Terzieva M, King RH, Romanski K, Petkov R, Schmarov A, Dimitrova G, Popova N, Uzunova M, Milanov S, Petrova J, Petkov Y, Kolarov G, Aneva L, Radeva O, Thomas PK (1999) Congenital cataracts facial dysmorphism neuropathy syndrome, a novel complex genetic disease in Balkan Gypsies: clinical and electrophysiological observations. Ann Neurol 45:742–750 [DOI] [PubMed] [Google Scholar]
- Turner RL (1926) The position of Romani in Indo-Aryan. J Gypsy Lore Soc 5:145–189 [Google Scholar]
- Varon R, Gooding R, Steglich C, Marns L, Tang H, Angelicheva D, Yong KK, et al (2003) Partial deficiency of the C-terminal-domain phosphatase of RNA polymerase II is associated with congenital cataracts facial dysmorphism neuropathy syndrome. Nat Genet 35:185–189 10.1038/ng1243 [DOI] [PubMed] [Google Scholar]
- Wilson IJ, Balding DJ (1998) Genealogical inference from microsatellite data. Genetics 150:499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M (1999) Population choice in mapping genes for complex diseases. Nat Genet 23:397–404 10.1038/70501 [DOI] [PubMed] [Google Scholar]