Abstract
The Cornelia de Lange syndrome (CdLS) is a multisystem developmental disorder characterized by facial dysmorphia, upper-extremity malformations, hirsutism, cardiac defects, growth and cognitive retardation, and gastrointestinal abnormalities. Both missense and protein-truncating mutations in NIPBL, the human homolog of the Drosophila melanogaster Nipped-B gene, have recently been reported to cause CdLS. The function of NIPBL in mammals is unknown. The Drosophila Nipped-B protein facilitates long-range enhancer-promoter interactions and plays a role in Notch signaling and other developmental pathways, as well as being involved in mitotic sister-chromatid cohesion. We report the spectrum and distribution of NIPBL mutations in a large well-characterized cohort of individuals with CdLS. Mutations were found in 56 (47%) of 120 unrelated individuals with sporadic or familial CdLS. Statistically significant phenotypic differences between mutation-positive and mutation-negative individuals were identified. Analysis also suggested a trend toward a milder phenotype in individuals with missense mutations than in those with other types of mutations.
Introduction
Cornelia de Lange syndrome (CdLS [MIM 122470]), which was recognized as a distinct entity >70 yearsago, is a clinically heterogeneous developmental disorder characterized by facial dysmorphia, upper-extremity malformations, hirsutism, cardiac defects, growth and cognitive retardation, and gastrointestinal abnormalities (Brachmann 1916; de Lange 1933). The distinctive facial features include synophrys, long eyelashes, depressed nasal bridge with an uptilted nasal tip and anteverted nares, thin upper lip with downturned corners of the mouth, and posteriorly rotated low-set ears. Abnormalities in the upper extremities range from subtle changes in the phalanges and metacarpal bones and small hands to oligodactyly and severe reduction defects. Gastrointestinal abnormalities include gastroesophageal reflux, intestinal malrotation, and pyloric stenosis. Additional relatively frequent features include hearing loss, ophthalmologic findings (ptosis and myopia), palatal abnormalities, genitourinary abnormalities (cryptorchidism and hypospadias), cardiac septal defects, and congenital diaphragmatic hernias. Growth retardation is an almost universal finding in CdLS and typically has a prenatal onset. Standard growth curves have been established for height, weight, and head circumference (Kline et al. 1993a). The mental retardation in CdLS is often severe, with a mean IQ of 53 (range 30–86) (Kline et al. 1993b). Many patients also demonstrate autistic-like behavior and self-injurious behavior (Jackson et al. 1993).
The clinical features seen in individuals with classic CdLS are striking and easily recognizable; however, there is marked variability, and a milder phenotype has been consistently described (Ireland et al. 1993; Saul et al. 1993; Selicorni et al. 1993; Van Allen et al. 1993). Indeed, even the first reported descriptions of CdLS were markedly discrepant in phenotype; Brachmann (1916) described major upper-limb–reduction abnormalities, whereas de Lange (1933) reported no limb-reduction defects. This phenotypic variability and the lack of a diagnostic marker have complicated the diagnosis and genetic counseling for CdLS.
The prevalence of CdLS is estimated to be as high as 1/10,000 (Opitz 1985), and most cases appear to be sporadic. Pedigree analyses of several families demonstrate autosomal dominant inheritance with both maternal and paternal transmission (Robinson et al. 1985; Bankier et al. 1986; Halal and Silver 1992; Feingold and Lin 1993; Chodirker and Chudley 1994; Kozma 1996; Russell et al. 2001; McConnell et al. 2003). Given autosomal dominant inheritance, cases of apparently unaffected parents who have multiple children with CdLS were hypothesized to be the result of germline mosaicism (Beratis et al. 1971; Lieber et al. 1973; Fryns et al. 1987; Naguib et al. 1987; Krajewska-Walasek et al. 1995). This hypothesis of germline mosaicism was further supported by the identification of several families in which an unaffected parent had multiple affected children through different partners (Krantz et al. 2001).
Two recent reports document that mutations in NIPBL cause CdLS (Krantz et al. 2004; Tonkin et al. 2004). The types of mutations identified in NIPBL included missense, splice site, nonsense, and frameshift. Severe protein-truncating mutations likely lead to haploinsufficiency of the NIPBL protein. Haploinsufficiency of NIPBL was documented as a disease mechanism in the report of a child with classic features of CdLS who was stillborn but was found prenatally to have a large, cytogenetically visible deletion of chromosome 5p13.1-14.2 (Hulinsky et al. 2003). This deletion would be predicted to encompass the NIPBL gene. To our knowledge, there have not been any other reported cases of constitutional deletions of this region. The hypothesis of germline mosaicism was also validated by the identification of the same NIPBL mutation in affected siblings born to unaffected mutation-negative parents (Krantz et al. 2004).
The prevalence of NIPBL mutations in a large population of individuals with CdLS and the correlation of specific mutations with phenotypic characteristics have not previously been formally addressed. This article reports the systematic molecular and cytogenetic evaluation of 120 individuals with CdLS for disruptions in the NIPBL gene. We have identified mutations in 47% of tested probands and have examined this cohort for genotype-phenotype correlations.
Subjects and Methods
Clinical Evaluation
All patients and family members were enrolled in the study under an institutional review board–approved protocol of informed consent at The Children’s Hospital of Philadelphia. All subjects were evaluated by one or more clinical dysmorphologists (I.D.K, A.D.K, and L.G.J) with experience with CdLS. Clinical histories and photographs were obtained routinely for all probands, as well as for any other affected family members. Clinical records were reviewed for the presence of other CdLS-associated anomalies, such as deafness; cleft palate; and cardiac, ophthalmologic, gastrointestinal, genitourinary, and renal anomalies. For the purposes of the genotype-phenotype studies, only probands (no other family members) were included. This may result in a bias toward the more severe phenotype; however, as familial recurrences are extremely rare, it was not possible to perform a separate analysis on the small number of affected family members. Although all probands had characteristic facial features that were part of the criteria for their inclusion into the study, we chose to further stratify the CdLS cohort on the basis of the severity of three phenotypic parameters: limb differences, growth, and cognitive function (summarized in table 1).
Table 1.
Phenotypic Classifications
|
Characteristics of |
|||
| Phenotypic Parameter | Class I (Mild) | Class II (Moderate) | Class III (Severe) |
| Limb reduction | No reduction defect | Partial defect, oligodactyly (>2 digits on each hand) | Severe defect (⩽2 digits on either hand) |
| Development and cognitive abilities | Motor milestones <2 years delayed; speech and communication skills present | Motor milestones >2 years delayed; limited speech and communication | Profound delay in motor milestones; lack of meaningful communication |
| Growtha | >75th percentile | 25th–75th | <25th percentile |
Average of percentiles for weight, height, and head circumference, plotted on CdLS standard growth curves.
Limb malformations were classified by the presence or absence of reduction defects in the upper extremities, as follows: class I (mild), no reduction defect; class II (moderate), partial reduction defect/oligodactyly (>2 digits on each hand); and class III (severe), severe reduction defect/oligodactyly (⩽2 digits on either hand). A score for severity of the physical growth parameters was calculated by averaging the percentiles for weight, height, and head circumference that were plotted on sex- and age-standardized growth curves for individuals with CdLS (Kline et al. 1993a). Growth parameters were classified as follows: class I (mild), average growth parameters and >75th percentile on CdLS growth curves; class II (moderate), average growth parameters and 25th–75th percentile on CdLS growth curves; and class III (severe), average growth parameters and <25th percentile on CdLS growth curves. Cognitive function was the most difficult parameter to standardize, because most individuals with CdLS who enrolled in the study had not received formal developmental evaluations and because of the inherent difficulty in comparing the developmental abilities of individuals of various ages. Wedetermined classifications of developmental/cognitive abilities on the basis of deviation from age-appropriate standards as follows: class I (mild), motor milestones <2 years delayed from normal standards, with development of speech and communication skills in older individuals; class II (moderate), delay in reaching motor milestones >2 years behind normal developmental standards, with limited speech and communication; class III (severe), profound delay in achieving motor milestones, with a lack of meaningful communication. Clinical stratification of all probands was performed without knowledge of mutational status.
Mutational Analysis
Genomic DNA was isolated from peripheral blood lymphocytes (Gentra Systems). Parental DNA was available for 41 (85%) of 48 patients with sporadic CdLS who had NIPBL mutations. DNA from both parents was available in 25 (52%) of 48 patients, and DNA from only one parent was available in 16 (33%) of 48 patients. The entire NIPBL coding region (exons 2–47) was screened for mutations. Primer sequences, annealing temperatures, and sizes of PCR products are listed in table A1 (online only). Primer pairs were designed to amplify exons, exon/intron boundaries, and short flanking intronic sequences. Larger exons were subdivided to allow for optimal product lengths. All PCR reactions were performed in a 25-μl reaction volume containing 75 ng of genomic DNA, 1 U of AmpliTaq Gold (Applied Biosystems), 20 pmol of each primer, 75 μM of each dNTP, 10× PCR buffer II (Applied Biosystems), and 1.0 mM or 1.5 mM of MgCl2 (Applied Biosystems). Cycling parameters were as follows: 36 cycles at 94°C for 30 s, at 51°C–60°C for 45 s, and at 72°C for 30 s, with a last extension step at 72°C for 5 min. Amplifications for exons 6, 11, 21, 26, 30, 44, and 45 were performed with 10 cycles at 95°C for 30 s, at 51°C–62°C for 30 s, and at 72°C for 35 s, followed by 25 cycles at 95°C for 30 s, at 51°C–62°C for 30 s, and at 72°C for 45 s, increasing by 5 s for each cycle (table A1 [online only]). Mutational analysis of the amplimers was performed using conformation-sensitive gel electrophoresis (CSGE) with standard protocols (Ganguly et al. 1993). PCR products corresponding to all altered migration patterns (shifts) on CSGE were purified using QIAquick PCR purification kit (Qiagen) and were sequenced bidirectionally on an ABI 377 sequencer.
Genotype-Phenotype Correlations
Genotype-phenotype correlations were assessed using contingency-table analysis. This was performed for the three categories (mild, moderate, and severe) of each phenotypic parameter (limb defect, growth, and development), with a focus on the presence versus the absence of a mutation in NIPBL and on individuals with missense mutations versus those with other types of mutations. For analysis of the mutation-positive versus mutation-negative individuals, the χ2 test with 2 df was used. For the analysis of missense versus other types of mutations, Fisher’s exact test was used. The significance threshold was set at P⩽.05.
FISH Analysis
FISH studies were performed on metaphase chromosomes prepared from peripheral blood lymphocytes by use of standard techniques (Krantz et al. 1997). FISH was performed with the NIPBL-containing BAC RP11-14I21 (NCBI accession number AC018853.3) on 28 mutation-negative individuals (4 familial and 24 sporadic CdLS cases) to evaluate for the possibility of a large but submicroscopic deletion encompassing the NIPBL gene. BAC DNA was isolated (Perfect Prep Plasmid XL [Eppendorf]) and labeled by nick translation in the presence of Spectrum Red dUTP (Vysis). The labeled BAC probe was dissolved in LSI/WCP hybridization buffer (Vysis), and 10 μg of Human Cot-1 DNA (Invitrogen) was added per 1 μg of labeled BAC RP11-14I21 probe. TelVysion probe 5p and/or 5q (Vysis) (in accordance with the manufacturer's instructions) and 100 ng of labeled BAC probe per micoscope slide were codenatured under a coverslip for 2 min on a 75°C slide warmer and were hybridized at 37°C for ∼16 h in a humid chamber. Slides were subjected to two posthybridization washes—wash 1 (0.4 × saline-sodium citrate [SSC]; 0.3% NP-40) at 73°C for 2 min and wash 2 (2 × SSC; 0.1% NP-40) at room temperature for 1 min—and were counterstained with DAPI II (Vysis). A Nikon microscope, equipped with the appropriate filters, was used to visualize each slide. CytoVision software version 3.1, build 10 (Applied Imaging), and a CCD camera were used to capture FISH images.
Results
Study Population
The study population consisted of 120 subjects with CdLS, including 106 sporadic and 14 familial cases. Linkage to the NIPBL locus at 5p13.1 was reported elsewhere in 11 of the 14 families, with the identification of mutations in NIPBL in 2 of these families (Krantz et al. 2004). In one family, a missense mutation in the first codon (M1K) was identified in three affected half-siblings who each had a different father. The mutation was not present in DNA extracted from lymphocytes in their mother or in the two fathers from whom samples were available. In the second family, a splice site mutation (6763+5G→T) in the intron between exons 39 and 40 was identified in two affected siblings but not in DNA isolated from lymphocytes in either parent. The study population also included four previously reported, unrelated patients with CdLS with unique de novo mutations in NIPBL (Krantz et al. 2004).
Spectrum of NIPBL Mutations Detected
The 120 subjects with CdLS were screened for NIPBL coding-region mutations. Exons 2–47 and flanking intron sequences were amplified by PCR and were analyzed by use of CSGE. All products with variant migration profiles (band shifts) on CSGE were sequenced bidirectionally. NIPBL mutations were identified in 56 patients (47%) with CdLS (7 familial and 49 sporadic) (table 2 and fig. 1). A total of 51 different mutations were identified and comprised 21 frameshift, 12 missense, 10 nonsense, and 8 splice-site mutations. All identified mutations were private, except for four (8%): a 2-bp deletion of exon 10 (2479delAG), in two unrelated patients; a nonsense mutation (R1536X) of exon 22, in three unrelated patients; a splice-site mutation in the intron upstream of exon 35 (6109−3T→C), in two unrelated patients; and a missense mutation (R2298H) of exon 40, in two unrelated patients.
Table 2.
Summary of Mutations in NIPBL Identified in Probands with CdLS
| Exon and Mutation | Type | Effect on Protein | Fathera | Mothera | Number |
| 2: | |||||
| 2 T→A; M1K | Missense | No initiating methionine | Negative | Negative | Familial |
| 6+1A→G | Splice site | … | NA | Negative | 1 |
| 3: | |||||
| 150delG | Frameshift | Truncates protein 27 aa downstream | Negative | Negative | 1 |
| 65−5A→G | Splice site | … | Negative | Negative | 1 |
| 199del10; 199ins13b | Complex | Truncates protein 9 aa into insertion | NA | NA | 1 |
| 7: | |||||
| 611−2A→G | Splice site | … | NA | Negative | 1 |
| 742delCT | Frameshift | Truncates protein 8 aa downstream | NA | NA | 1 |
| 9: | |||||
| 961delA | Frameshift | Truncates protein 7 aa downstream | NA | NA | 1 |
| R479X | Nonsense | Truncates protein | Negative | Negative | 1 |
| 10: | |||||
| 1546insG | Frameshift | Truncates protein 5 aa downstream | Negative | Negative | 1 |
| 1669insC | Frameshift | Truncates protein 11 aa downstream | NA | Negative | 1 |
| 1902insA | Frameshift | Truncates protein 2 aa downstream | Negative | Negative | 1 |
| 2479delAG | Frameshift | Truncates protein 2 aa downstream | Negative | Negative | 2 |
| 2520delT | Frameshift | Truncates protein 6 aa downstream | Negative | Negative | 1 |
| 2969delG | Frameshift | Truncates protein 1 aa downstream | NA | NA | 1 |
| 3023delTGTCT | Frameshift | Truncates protein 2 aa downstream | NA | Negative | 1 |
| 3057delTAGA | Frameshift | Truncates protein 23 aa downstream | Negative | Negative | 1 |
| 3060delAGAG | Frameshift | Truncates protein 22 aa downstream | Negative | Negative | 1 |
| R797X | Nonsense | Truncates protein | NA | Negative | 1 |
| R832X | Nonsense | Truncates protein | Negative | NA | 1 |
| E977X | Nonsense | Truncates protein | Negative | Negative | 1 |
| S1024X | Nonsense | Truncates protein | NA | Negative | 1 |
| 15: | |||||
| 3736C→G; A1246G | Missense | … | Negative | Negative | 1 |
| 17: | |||||
| 3969insG | Frameshift | Truncates protein 6 aa downstream | Negative | Negative | 1 |
| 3935T→C; L1312P | Missense | … | NA | Negative | 1 |
| 18: | |||||
| S1398X | Nonsense | Truncates protein | NA | Negative | 1 |
| 20: | |||||
| S1459X | Nonsense | Truncates protein | Negative | Negative | 1 |
| 21: | |||||
| 4556delAAAAA | Frameshift | Truncates protein 1 aa downstream | Negative | Negative | 1 |
| 22: | |||||
| R1536X | Nonsense | Truncates protein | Negative (1), NA (2) | Negative (2), NA (1) | 3 |
| 4567delC | Frameshift | Truncates protein 1 aa downstream | NA | NA | 1 |
| 26: | |||||
| R1723X | Nonsense | Truncates protein | Negative | Negative | Familial |
| 27: | |||||
| R1758X | Nonsense | Truncates protein | Negative | Negative | 1 |
| 28: | |||||
| 5366G→T; R1789L | Missense | … | NA | NA | 1 |
| 5408A→T; D1803V | Missense | … | NA | Negative | 1 |
| 29: | |||||
| 5567G→C; R1856T | Missense | … | Negative | Negative | Familial |
| 5574+1G→T | Splice site | … | Negative | Negative | 1 |
| 35: | |||||
| 6109−3T→C | Splice site | … | NA (2) | Negative (1) | 2 |
| 39: | |||||
| 6763+5G→T | Splice site | … | Negative | Negative | Familial |
| 40: | |||||
| 6892C→T; R2298C | Missense | … | NA | Negative | 1 |
| 42: | |||||
| 6893G→A; R2298H | Missense | … | Negative | Negative | 2 |
| 6934G→C; G2312R | Missense | … | Negative | Negative | 1 |
| 7151delAAGAC | Frameshift | Truncates protein 3 aa downstream | NA | Negative | Familial |
| 43: | |||||
| 7210delC | Frameshift | Truncates protein 21 aa downstream | NA | Negative | 1 |
| 7142G→C; G2381A | Missense | … | NA | Negative | 1 |
| 7168G→A; A2390T | Missense | … | Negative | Negative | 1 |
| 7318T→C; Y2440H | Missense | … | Negative | Negative | 1 |
| 44: | |||||
| 7321+4A→G | Splice site | … | Negative | Positive | Familial |
| 7431delG | Frameshift | Truncates protein 30 aa downstream | NA | Negative | 1 |
| 45: | |||||
| 7780delC | Frameshift | Truncates protein 16 aa downstream | Negative | Negative | Familial |
| 7825insG | Frameshift | Truncates protein 22 aa downstream | Negative | Negative | 1 |
| 46: | |||||
| 7861−1G→C | Splice site | … | Negative | Negative | 1 |
NA = sample not available.
Insertion of atcaacaggtgac.
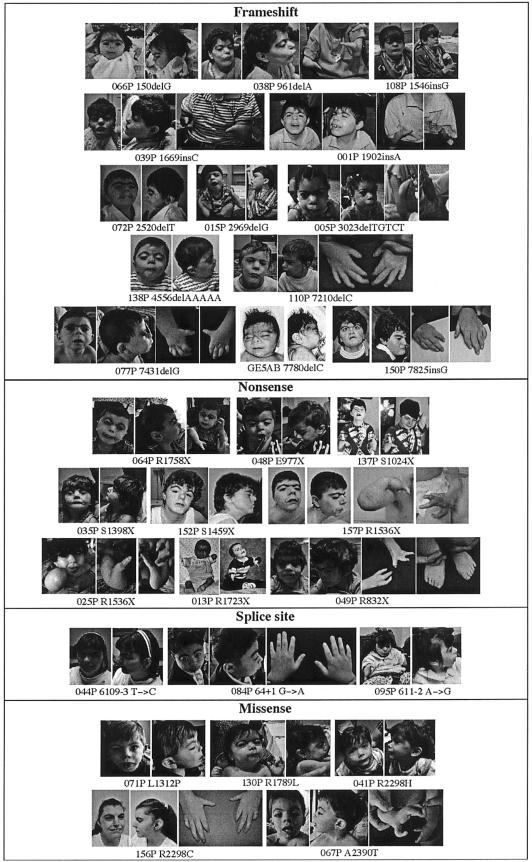
Figure 1.
Facial features and limb findings in mutation-positive individuals with CdLS. Note the variability of features even among individuals with similar mutation types.
In 49 (46%) of the 106 individuals with sporadic CdLS, 44 different mutations were identified. Of these, 14 (32%) were small deletions, and 6 (14%) were insertions—5 were single–base-pair insertions, and 1 was a complex deletion/insertion mutation with a net insertion of 3 bp. All deletions and insertions led to frameshifts that resulted in a prematurely truncated protein product. The deletion 2479delAG in exon 10 was seen in two unrelated patients with sporadic CdLS. Nine (20%) of the different mutations had single–base-pair changes that led to immediate stop codons. Four (44%) of the nonsense mutations were found in exon 10. The nonsense mutation R1536X in exon 22 was identified in three unrelated patients with sporadic CdLS. In one patient with sporadic CdLS with a de novo t(14q;21q)(q32;q11) (Wilson et al. 1983), the nonsense mutation S1459X in exon 20 of NIPBL was identified and was not present in either patient's parents, suggesting that the balanced de novo translocation may represent an unrelated event.
Seven patients (16%) with sporadic CdLS had different mutations predicted to lead to alterations in splicing. The six splice-site mutations were not identified in available parental samples (six mothers and three fathers) or in the 150 control patient samples.
Of the 44 mutations identified in patients with sporadic CdLS, 10 (23%) resulted in the substitution of a single amino acid. These substitutions were predicted to result in missense mutations by three criteria: absence in parental samples, absence in control samples, and evolutionary conservation of the altered amino acid. One missense mutation—R2298H in exon 40—was identified in two unrelated patients. The missense mutations identified included A1246G (exon 15), L1312P (exon 17), R1789L (exon 28), D1803V (exon 28), R2298C (exon 40), R2298H (exon 40), G2312R (exon 40), G2381A (exon 42), A2390T (exon 42), and Y2440H (exon 43). These amino acids were, in general, highly conserved throughout evolution (fig. 2). These mutations were not identified in available parental samples or in the 150 control patient samples.
Figure 2.
Evolutionary conservation of amino acid residues altered by missense mutations in NIPBL. A comparison of amino acids and the flanking sequence altered by 11 of the unique missense mutations in human (NIPBL), rat, mouse, and Drosophila is depicted. The mutated amino acid residue is shaded gray. Amino acid residue 2298 was mutated in three individuals—two had an R2298H change, and one had an R2298C change. The missense mutation M1K in the initiation codon is not depicted, since it is conserved in all species.
In one apparent familial case of CdLS (reported elsewhere as family XII [Krantz et al. 2001])—which was excluded from subsequent linkage analyses, since it showed an atypical inheritance pattern with two affected male first cousins born to unaffected sisters—the two affected males were found to carry different de novo mutations in NIPBL. In one child, an A1246G missense change in exon 15 was identified, whereas, in his affected cousin, a 7861−1G→C splice-site change was identified in the intron upstream of exon 46. Neither mutation was identified in either of the two sets of parents or in the other cousin. The maternal 5p13 regions flanking NIPBL (including intragenic SNP markers) in the affected individuals were not shared (data not shown). Paternity was confirmed in both cases.
Mutations were identified in 7 (50%) of 14 familial cases of CdLS. Mutations in NIPBL were previously reported in two of these families. In the first family, a missense mutation in the first codon (M1K) was identified in three affected siblings, all of whom had different fathers, and was not present in the mother or in the two fathers available for testing (in all familial cases, mutational analysis of parental samples was performed on DNA extracted from lymphocytes, and mosaicism in other tissues cannot be excluded). In the second family, a splice-site mutation (6763+5G→T) segregated with the CdLS phenotype in two affected siblings and was not identified in either parent (Krantz et al. 2004). Four of the five remaining mutation-positive families had been linked to the NIPBL locus in a previous study (Krantz et al. 2004). In the first family, a nonsense mutation, R1723X in exon 26, was identified in two affected brothers; neither the parents nor the unaffected brother have the mutation. In the second family, the two affected siblings share a unique 5-bp deletion, 7151delAAGAC in exon 42, resulting in protein truncation 3 aa downstream. The mother did not carry the change, and there was no sample available from the father. An affected brother and sister in the third family share a single–base-pair deletion, 7780delC of exon 45; this deletion results in premature protein truncation 16 aa downstream. Neither parent carried this mutation. In the fourth family, with four affected siblings and a mildly affected mother, a splice-site mutation, 7321+4A→G in exon 43, was identified in the two affected siblings from whom samples were available, as well as in the affected mother. In the final family—which was not included in the initial linkage studies, since the affected female sibling of the proband was deceased and no sample was available—a missense mutation, R1856T in exon 29, was identified in the affected male child but was not present in either parent or in an unaffected sibling. Paternity was confirmed in all familial cases (as part of the genomewide and high-resolution linkage analysis using multiple polymorphic markers) for which a paternal sample was available.
None of the 51 different mutations were observed in 150 ethnically matched control subjects. Forty-two sequence variants that are likely to represent neutral polymorphisms were observed in subjects with CdLS, unaffected family members, and/or control individuals (table 3). Three of the polymorphisms (N674S, N1994S, and I1206V) in the coding region of NIPBL led to an altered amino acid residue, whereas three (D817D, L1591L, and S1958S) were silent. Thirty-six polymorphisms were identified in intronic sequences flanking the exons.
Table 3.
Polymorphisms in NIPBL Identified in Affected Individuals, Family Members, and/or Controls
| Polymorphism | Located in Exon |
| 230+61C→Aa,b | 3 |
| 611+102A→Gc | 6 |
| 2021A→G; N674Sa,b | 10 |
| 2451C→T; D817Dc | 10 |
| 3575−14A→Ga | 14 |
| 3616A→G; I1206V | 14 |
| 3855+52A→Ga,b | 16 |
| 4088+53T→Cc | 17 |
| 3586−59G→A | 17 |
| 4239+53T→Cc | 18 |
| 4239+152C→Gc | 18 |
| 4240−48C→Tc | 19 |
| 4321−96C→T | 20 |
| 4321−35T→Cc | 20 |
| 4560+77A→Ga | 21 |
| 4560+108delTa | 21 |
| 4561−9T→Ac | 22 |
| 4561−106C→Tc | 22 |
| 4634+24G→Ac | 22 |
| 4773G→T; L1591Lc | 23 |
| 4777−108delAa | 24 |
| 4921−58G→Aa,b | 25 |
| 5575−193T→Ca | 30 |
| 5575−92G→C | 30 |
| 5575−18G→Cc | 30 |
| 5710−59A→Gc | 31 |
| 5710−78G→Ac | 31 |
| 5862+74delTTa,b | 32 |
| 5863−12delATa | 33 |
| 5863−30delATa,b | 33 |
| 5863−52delTc | 33 |
| 5874C→T; S1958Sa,b | 33 |
| 5971A→G; N1994Sc | 34 |
| 6109−54insA | 35 |
| 6109−3T→Cc | 35 |
| 6498−94T→Cc | 38 |
| 6499−80A→Gc | 39 |
| 6764−35C→Ga | 40 |
| 6954+62A→Gc | 40 |
| 6955−9delTa | 41 |
| 7861+39G→A | 45 |
| 8698−8701delACAA | 47d |
Found in patients, family members, and/or normal controls.
Previously reported by Krantz et al. (2004).
Found in patients and family members.
In the 3′ UTR.
FISH Analysis
To evaluate the possibility of a submicroscopic deletion encompassing the NIPBL gene, 28 probands with CdLS (4 familial and 24 sporadic) in whom a NIPBL mutation was not identified were analyzed by FISH with an NIPBL-containing BAC probe (RP11-14I21). RP11-14I21 encompasses ∼16 kb of sequence 5′ of the NIPBL gene, nearly through exon 10 of NIPBL. No deletionof RP11-14I21 was detected in any of the probands analyzed.
Genotype-Phenotype Correlation
Because of the clinical heterogeneity observed in CdLS (fig. 1), we evaluated possible associations between the NIPBL genotype and the severity of the phenotype (severity of limb, growth, and developmental involvement). The distribution of several major clinical features in our study cohort is shown in table 4. The results of genotype-phenotype correlation analysis, performed for each phenotypic parameter, with a focus on the presence versus the absence of a mutation in NIPBL and on missense mutations versus other types of mutations, are also summarized in table 4. Statistically significant (P<.05) differences were observed in the distribution of severity of growth retardation and developmental delay among the mutation-positive and mutation-negative groups, with the mutation-positive group displaying a more severe phenotype for these parameters. A similar trend was also observed in the severity of limb defects, although, in this case, the difference was not statistically significant. In comparing individuals with missense mutations versus those with all other mutation types, it appeared that the latter showed more severe phenotypes in all categories (except possibly in growth retardation), although the number of missense mutations was small.
Table 4.
Distribution of Clinical Severity and Results of Genotype-Phenotype Correlation Analysis[Note]
|
No. of Mutation-PositivePatients (%) [n=56] |
P |
||||
| Phenotype andClassificationa | Missense | Frameshift, Splice Site, and Nonsense | No. of Mutation-NegativePatients(%) [n=64] | Mutation-Positive vs.Mutation-Negative(χ2 Test) | Missense vs.All Other Mutations(Fisher’s Exact Test) |
| Limb reduction: | |||||
| I | 12 (92) | 25 (60) | 47 (75) | ||
| II | 1 (8) | 3 (7) | 9 (14) | ||
| III | 0 | 14 (33) | 7 (11) | ||
| NA | 0 | 1 | 1 | .085 | .029 |
| Developmental delay: | |||||
| I | 3 (23) | 1 (3) | 12 (20) | ||
| II | 6 (46) | 10 (25) | 27 (44) | ||
| III | 4 (31) | 29 (73) | 22 (36) | ||
| NA | 0 | 3 | 3 | .014 | .008 |
| Growth retardation: | |||||
| I | 3 (33) | 2 (7) | 17 (47) | ||
| II | 4 (44) | 11 (38) | 13 (36) | ||
| III | 2 (22) | 16 (55) | 6 (17) | ||
| NA | 4 | 14 | 28 | .002 | .057 |
Note.— n=56 for mutation-positive, and n=64 for mutation-negative individuals.
NA = Not assessed.
Discussion
Through the combined use of genomewide linkage-exclusion analysis and the mapping of a chromosomal rearrangement on chromosome 5p13, NIPBL was identified as a CdLS disease gene (Krantz et al. 2004; Tonkin et al. 2004). In the present study, we have identified mutations in 47% of a well-characterized cohort of 120 unrelated probands with sporadic and familial CdLS. Mutation-detection rates among the sporadic and familial cases were comparable: 49 (46%) of 106 sporadic cases and 7 (50%) of 14 familial cases had identifiable mutations. We expected to detect NIPBL mutations in all of the familial cases shown elsewhere to be positively linked to the 5p13 region (Krantz et al. 2004); however, in our present analysis, we have identified mutations in only 6 of the 11 families available for mutational analysis. This indicates either that the methods used for screening are not identifying all mutations in individuals with CdLS or that additional genes in 5p13 may potentially be responsible for the phenotype. Alternatively, the linkage to 5p could be coincidental in the mutation-negative families, and genes from elsewhere in the genome may be causative when mutated.
If NIPBL is the only CdLS disease gene, then our mutation detection rate of only 47% may be partly the result of the large size of the NIPBL gene and the use of CSGE for mutational analysis. Factors that may account for missed mutations in the gene include (1) variations in sequence beyond the immediate intron/exon boundaries (such as regulatory regions or intronic sequence), (2) large intragenic deletions, (3) subtle sequence variations (such as point mutations), and (4) difficulty in amplifying and sequencing several NIPBL exons (e.g., exon 33) as a result of numerous polymorphisms. The multiple splice variants of this gene have made it difficult to screen cDNA accurately for mutations, at this time, although this testing is currently being optimized and will allow for improved detection of intronic variations that lead to splice mutations as well as complete exonic deletions. Large-scale deletions of NIPBL were assayed in those individuals in whom an NIPBL mutation was not identified, and, in the 28 mutation-negative individuals studied by FISH, no deletion of the region was seen.
Alternatively, it is possible that the linkage established in some of these small families was coincidental and that a second CdLS gene may yet be identified elsewhere in the genome to account for the phenotype in those individuals in whom NIPBL mutations were not identified. In our initial genomewide linkage exclusion analysis, three other regions were not excluded—chromosome 2q37, chromosome 10p13, and chromosome 14q24. These additional loci may contain a second CdLS gene (Krantz et al. 2004). Likewise, several individuals with CdLS have been found to carry an apparently balanced de novo translocation, suggesting possible additional loci for a CdLS disease gene. The child with the t(5;13)(p13.1;q12.1) was critical in the identification of NIPBL on chromosome 5p13 as the cause, when mutated, of CdLS (Hulinsky et al. 2003; Krantz et al. 2004; Tonkin et al. 2004). Two other de novo balanced translocations have been reported. A de novo t(3;17)(q26.3;q23.1) (Ireland et al. 1991) has been extensively evaluated in a child with sporadic CdLS, and, to date, no CdLS disease genes have been identified (Tonkin et al. 2001, 2004). In the present study, a child with a de novo t(14q;21q)(q32;q11) described elsewhere (Wilson et al. 1983) has been found to carry a de novo S1459X mutation in exon 20 of NIPBL. This may indicate that this translocation is an unrelated event. Although not all breakpoints in these rare translocation cases have been completely evaluated, they do not appear to lend additional support to a potential second locus, at this time.
To date, no mutations have been identified in exons 4–6, 8, 11–14, 16, 19, 23–25, 30–34, 36, 37, 41, and 47. Several exons have been found to have multiple mutations, including exons 2, 3, 7, 9, 10, 17, 22, 28, 29, 40, 42, 43, and 45. There is a preponderance of mutations identified in exon 10; however, this 1,625-bp exon is ∼8 times the size of the average exon (∼200 bp) in the NIPBL gene. Exon 42, 200 bp in length, was found to contain four different mutations in this cohort. The majority of the 56 mutations identified are frameshift mutations (22 [∼39%; 16 deletions, 5 insertions, and 1 complex]), followed by missense mutations (13 [∼23%]), nonsense mutations (12 [∼21%]), and splice-site mutations (9 [∼16%]). The frameshift, nonsense, and splice-site mutations are likely to result in a prematurely truncated protein causing haploinsufficiency of NIPBL (a disease mechanism that has been documented in the study of a child with CdLS and a large cytogenetically visible deletion of chromosome 5p13.1-14.2 that encompasses the NIPBL gene [Hulinsky et al. 2003]).
The missense mutations are important, in that they may indicate residues of the NIPBL protein that are functionally important. Of the 12 unique missense mutations identified, 8 are in amino acid residues that are evolutionarily conserved back to Drosophila (including the M1K change in the initiation codon), and 3 are evolutionarily conserved back to the mouse. One missense mutation, Y2440H, is present in an amino acid located in a stretch of the human NIPBL protein not seen in rat, mouse, or Drosophila.
As mentioned above, four mutations were identified in unrelated individuals: 2479delAG in exon 10, R1536X in exon 22, 6109−3T→C in the intronic sequence upstream of exon 35, and R2298H in exon 40. An additional missense mutation in amino acid residue 2298 (R2298C) was seen in another individual. Even among individuals with the same mutation, the phenotype demonstrated some variability. The three individuals with the R1536X mutation are all severely affected in terms of growth and development; however, two have severe limb-reduction defects, whereas the third did not have a reduction defect. The two children with the 2479delAG mutation are also severely affected in terms of growth and development; however, one has significant limb-reduction defects, whereas the other does not have a reduction defect. The two children with the 6109−3T→C mutation and the two children with the R2298H missense mutation are all moderately affected in terms of growth and development, and none have limb-reduction defects. This variability in severity of phenotypes associated with identical mutations indicates that mutations in NIPBL are not the sole determinants of phenotype and that other factors (genetic and/or environmental) can modify the clinical picture.
In six of the seven familial cases in which an NIPBL mutation has been identified, germline mosaicism is the most likely mechanism. In five of these families, DNA from lymphocytes of both parents was available for testing, and neither parent carried the mutations identified in the affected siblings (paternity was confirmed in all cases). In one family, with a 7151delAGAC, the father was not available for testing; however, he reportedly has no clinical features of CdLS, and the mutation was not seen in the mother. Autosomal dominant transmission was demonstrated in the seventh family; a 7321+4A→G mutation in exon 43 was identified in the mildly affected mother and in the two of her four affected daughters from whom samples were available.
In the 25 sporadic cases in which both parents were available for screening, all mutations were found to have arisen de novo, and, in the 17 sporadic cases in which only one parent was available for screening, none of these parents was found to carry the change seen in the child. This would indicate that the vast majority of mutations in individuals with CdLS arise as new events, and, in the rare cases of familial recurrence in which neither parent is affected, germline mosaicism is the likely explanation. In the family in which two male first cousins have CdLS and their mothers, who are sisters, are unaffected, the two affected males were each found to carry a different de novo mutation (neither mutation was seen in either set of parents, and direct sequencing of the two cousins confirmed that they did not share the same mutation).
A large number of polymorphisms also have been identified in NIPBL (table 3). There were three polymorphisms (N674S, I1206V, and N1994S) that resulted in an amino acid substitution. All three of these were identified in probands who had mutations in other exons, and, in the case of N1994S, the mutation was present in one of the unaffected parents as well (for the other two variants, both parents were not available for screening). One polymorphism, N674S, was seen in25 unrelated probands, 11 of whom had identifiable NIPBL mutations, and in several controls. This amino acid residue is conserved back to the mouse but is not conserved in Drosophila. If the polymorphism was on the nonmutant allele and had a mild functional effect on the protein, it is possible that it could be a modifier of the phenotype. In the cohort of 11 probands with a mutation and this polymorphism, there did not appear to be a marked effect on phenotype. It is of interest that the one individual with a missense mutation (A2390T) who also had this polymorphism was the only one of the probands with missense mutations to have limb-reduction defects. Further work is needed to evaluate this and other polymorphisms as potential modifiers of the phenotype, through determination of allelic localization of these changes in relation to the mutation, as well as through functional studies to assess their effects.
Comparisons were made of genotype-phenotype correlations in mutation-positive and mutation-negative individuals, as well as in individuals with different types of mutations. Severity of limb defects and retardation in growth and cognitive development were evaluated (outlined in tables 1 and 4). Mutations in NIPBL were found in mildly and severely affected individuals with CdLS. Similarly, in the group of probands with CdLS and without identifiable mutations, there are also severely and mildly affected individuals. The analysis of genotype-phenotype associations demonstrated a trend toward a more severe phenotype in mutation-positive versus mutation-negative individuals. This indicates that a subset of individuals with “mild” CdLS may have a different genetic etiology causing their phenotype or may have mutations in NIPBL that have not yet been detected by use of CSGE.
We hypothesized that the missense mutations identified in NIPBL may result in either a milder phenotype as a result of a less severe structural effect on the protein, or, conversely, a more severe phenotype if the mutations occurred in critical domains of the protein, causing a dominant-negative effect. For these reasons, a similar analysis was performed to compare genotype-phenotype correlations seen in individuals with missense and all other types of mutations. This analysis suggests that individuals with missense mutations may have a milder phenotype; however, the number of individuals with missense mutations is too small to reach definite conclusions at this time.
The role of NIPBL in mammals has yet to be elucidated, and what is known about its function has come from Drosophila studies. The Drosophila homolog of NIPBL, Nipped-B, was identified through a screen for mutations that reduce activation by the wing margin enhancer in the presence of a gypsy insertion (Rollins et al. 1999); gypsy insertions in the cut gene in Drosophila are known to block a remote wing margin enhancer located 85 kb upstream of the promoter. This long-range effect on transcription as well as its homology to chromosomal adherins (proteins that have a role in chromosome compaction and sister-chromatid cohesion) suggest that the Nipped-B protein performs an architectural role in enhancer-promoter communication (Rollins et al. 1999). These interactions have been demonstrated to be involved in the regulation of multiple developmental pathways in Drosophila, including the Notch signaling pathway (Rollins et al. 1999). Recently, a role for the Drosophila Nipped-B protein in sister-chromatid cohesion has also been demonstrated, and a model of how Nipped-B interacts with the cohesin protein complex to affect gene expression was proposed (Rollins et al. 2004). The ability of distal enhancers to activate promoters and to initiate transcription relies on the coordinated interaction of multiple proteins and protein complexes. The large number of additional proteins that interact in these complexes suggests multiple possibilities for modifiers of NIPBL and potential additional CdLS disease genes.
In this study, we have shown that mutations in NIPBL are detected, at present, in 47% of individuals with either familial or sporadic CdLS. The mutations are spread throughout the gene, and frameshift, nonsense, splice-site, and missense mutations have been identified. The majority of mutations are protein truncating, likely leading to haploinsufficiency of the protein product. The 12 unique missense mutations identified in this screen will be important in characterizing functionally important domains of this novel protein. There appears to be a genotype-phenotype correlation in mutation-positive and mutation-negative individuals and possibly in individuals with missense mutations as compared to those with all other mutation types. To understand the striking variability of the CdLS phenotype, further work is needed to delineate the functionally important regions of the NIPBL protein, to characterize the various splice isoforms in different tissues and during development, to define the role of interacting proteins, and to determine the effect of polymorphisms on the coding and noncoding sequences of mutant and nonmutant alleles.
Acknowledgments
The authors are greatly indebted to the patients and families, for generously donating samples and clinical information. We thank the Cornelia de Lange Syndrome Foundation for their tireless support, as well as the many clinicians and investigators who referred families and sent samples. This work was generously supported by grant 1 RO1 HD39323 from the National Institute of Child Health and Human Development, National Institutes of Health (to I.D.K. and M.D.). L.A.G. is supported by the Florence R. C. Murray Award through The Children’s Hospital of Philadelphia. We especially acknowledge Nancy Spinner, Ph.D., Marie Jackson, Marcia Budarf, Ph.D., Jennifer Morrissette, Ph.D., and Julie Mairano, M.S., for their help and sage advice.
Appendix A: Supplemental Data
Table A1.
Primers and Conditions used to Amplify NIPBL Coding Sequence
|
Primer |
||||
| Exon | Forward | Reverse | Length(bp) | Conditions |
| 2 | ACTGGGTTGTTGTGAGAACTG | GCATTTCAGTTGCTATTTCTG | 470 | 1.5 mM MgCl2 55°C |
| 3 | TTAGGAAGAGGAGGAATGCC | CTGAAATAAAACCAGGAATACGG | 387 | 1.5 mM MgCl2 55°C |
| 4 | TGGGGGACAAGAGTGAGACTTC | GCATAAACATCGCATTCCTGATAG | 532 | 1.5 mM MgCl2 55°C +DMSO |
| 5 | AAGGACACTTTACTGTTAGAAGAA | GCAAATGCAAAGTGGATTACT | 301 | 1.5 mM MgCl2 55°C |
| 6 | CAGTCAGATTTCAAGGAATAGCG | CTCCTTTCACCTCCTAAAATGAC | 429 | 1.5 mM MgCl2 58°Ca |
| 7 | AACTAGTCAGTACATGAGTATCTG | GAAATGGAAATACTAGGTTATATG | 369 | 1.5 mM MgCl2 60°C |
| 8 | CAAGAAGAAAACAGGAAAGTGC | CTGCTTTAGGAAGTCTGAGTTCT | 325 | 1.5 mM MgCl2 55°C |
| 9A | GTGAAACCACCACAACTG | TGAGCAGCATTTAGTGGGC | 429 | 1.5 mM MgCl2 55°C |
| 9B | CAGGACAGACTTCAAAAACACC | CCAAATCTCATATAGTTGTTTCAG | 512 | 1.5 mM MgCl2 55°C |
| 10A | TTGCATTTGCATTTTACTCCA | GTGTCTCAGGATGGTTTTCTGG | 428 | 1.5 mM MgCl2 58°C |
| 10B | TACGGGAAATGGGTCAAGGC | AGGCTCAACTATGGTGCTCTCG | 424 | 1.5 mM MgCl2 55°C +DMSO |
| 10C | TGAGAGCAGAACAACTGAATGC | TGGCTTTCCAGGAATCCCTCC | 352 | 1.5 mM MgCl2 55°C +DMSO |
| 10D | AGGTGAGAGCCGCCCTGAAACTC | CACGAGGACTGTCAGGTCTTGA | 467 | 1.5 mM MgCl2 55°C +DMSO |
| 10E | TGAATCAGGGGACTCAAGGG | AGGGAACTTCTTGATTTGTCCTC | 468 | 1.5 mM MgCl2 55°C +DMSO |
| 10F | AGGAGCTAAGCCTGTAGTTGTG | CTTGAGTAGTGGGTGGGGAAGA | 349 | 1.0 mM MgCl2 55°C +DMSO |
| 11 | TGTCACTTTAGGGTTAAGAGT | GACTGTGCTTTTGCTAAACCC | 439 | 1.5 mM MgCl2 52°Ca |
| 12 | CACTGAATTTCCTAGACCCTATG | ATCACTGCACATAGAAACTAAG | 464 | 1.5 mM MgCl2 55°C +DMSO |
| 13/14 | GTTTCTATGTGCAGTGATTATCG | GATTTCAAGGTAGGACACATCAC | 483 | 1.5 mM MgCl2 58°C |
| 15 | ATTCAGGGTTTACTTGAGGTT | AGTCCATGCCTCTTTCAATGCAG | 486 | 1.5 mM MgCl2 58°C |
| 16 | AGTCATTTAGGGTCGTTGAGT | GCATGGGAAGAGATTAATGAC | 449 | 1.5 mM MgCl2 58°C |
| 17 | CATCATAACACTTTTCCACCAG | TGGTGCCATTTTAAGTCCTAT | 415 | 1.5 mM MgCl2 55°C |
| 18 | CTTCCAGGTTCTGTAGCTAGA | GAGTTTGGAATTTACACTACATT | 483 | 1.5 mM MgCl2 55°C |
| 19 | TGCTAACGTGCTTTGAGGATG | TAGTCCTTAGATTGAAATGAATG | 393 | 1.5 mM MgCl2 55°C |
| 20 | GAGCAGCTTACCTTAGATACTGA | ATGCTGTTCTGATGTAACTGCC | 363 | 1.5 mM MgCl2 60°C |
| 21 | GGCAAACACAGTATCGTGAAAC | GATCGCGCCACTGCACTC | 389 | 1.0 mM MgCl2 55°Ca |
| 22 | TAGTGTGCTAATTTTGGCTTCT | ATTCAAGGTTCAGATTATGGC | 350 | 1.5 mM MgCl2 60°C |
| 23 | CAATTTCAATCATGTTGGTAGAC | GTGTACAGTTATGCACATGC | 359 | 1.0 mM MgCl2 52°C +DMSO |
| 24 | ACAGTTGAGCCTGCATATTTA | ACCATTCAGAAGTCCCTGTTA | 594 | 1.5 mM MgCl2 55°C |
| 25 | AAGGCAAACTTCAGCTATCAA | CCTCTTCATCATGCTACCTCC | 366 | 1.5 mM MgCl2 58°C |
| 26 | TGTATTCCTGTAATGTGAGCACTC | TCATCCTGCAACAAAAAGTCA | 413 | 1.5 mM MgCl2 58°Ca |
| 27 | ACCACACCTTCTCAGTTTAGCA | CTCACAAGCATCCAGAATCAG | 297 | 1.5 mM MgCl2 55°C |
| 28/29A | ACGAAAGGCTCCAAAGTATG | ACTGCTGCTTCTCGGACAC | 473 | 1.5 mM MgCl2 52°C +DMSO |
| 28/29B | GTCTGAGGTTGTTGCTGTAGA | ATGATATTGCAAGGGCTATTC | 423 | 1.5 mM MgCl2 55°C |
| 30 | TTCTAGTCTTGTGTCCAGGGC | ATCAACATTTAGGTGCAATAA | 462 | 1.5 mM MgCl2 55°Ca |
| 31 | TCCTGGCAGTTTGTGTTTTG | CTGGAGGAATAGGAAAATCTCAG | 470 | 1.5 mM MgCl2 55°C +DMSO |
| 32 | GTTCTGTAACGTTGGTAAATGGT | GGTTCTTTTAAATCATACAGTCCA | 321 | 1.5 mM MgCl2 58°C |
| 33 | ACCTTAGGTCTTACACAGCAA | TGTGCTCAACTAGGTTATCAAC | 362 | 1.5 mM MgCl2 60°C |
| 34 | TTGAGGCCTATACTGGACCTA | GGTTGACGCATGTGAACTCTA | 333 | 1.5 mM MgCl2 60°C |
| 35 | TAACTGGACCTTTACGTGCAA | GCTCACACAATGTTGCACTAC | 423 | 1.5 mM MgCl2 55°C |
| 36 | TGGCATGACTGTAAGCACTCA | AGAGGACCACGGTGGATAATC | 381 | 1.5 mM MgCl2 60°C +DMSO |
| 37 | TGGTGGCACACGACTGTAATCC | TCATCCTGGGTCACTACTGTCAT | 467 | 1.5 mM MgCl2 60°C |
| 38 | CTGATACTTTGAATGCCACTG | CACCAAATCCTACTGCTAATA | 378 | 1.5 mM MgCl2 55°C |
| 39 | CTCTAGGTAAGGCCACCAGCAT | TAGACCTCAGCATAAGGACTGC | 466 | 1.5 mM MgCl2 55°C |
| 40 | CAGATTAAGAACCATTGAGCC | GCAGTAATCATAACCCAAGAG | 492 | 1.5 mM MgCl2 58°C |
| 41 | AGTGTGAGAATGCTTTATGTT | ATTATGAATGTGGGCAGAGCA | 474 | 1.5 mM MgCl2 55°C |
| 42 | ATGAAGCTAGCCTCAGAATGT | CAAAATTTCCCCTTCACTTCTGA | 472 | 1.5 mM MgCl2 58°C |
| 43 | GTGAGGTGAAAGTGCCCTGTA | TCCCAAGTCAAGTATTGCCCAG | 401 | 1.5 mM MgCl2 52°C |
| 44 | CAAGCTGTTGAATGGAGCATAC | CATGAGCCACCACACCCAGC | 434 | 1.0 mM MgCl2 58°Ca |
| 45 | TCCAAATACGTTGTTTCCATAG | TCAATGTGAAGGAGATAGTTAT | 329 | 1.5 mM MgCl2 51°Ca |
| 46 | CCACACCAAACTACTGCCATAG | CATTTTACGTAATACGCTGCG | 334 | 1.0 mM MgCl2 60°C |
| 47A | GTCACGGTGCGTCTCATTGC | TAGTGTCTACCCAAGGCACCA | 395 | 1.5 mM MgCl2 58°C |
| 47B | GGCTTCAGTGTTCAGTGGATG | TTTGCCCAACATTTCCTTC | 364 | 1.5 mM MgCl2 58°C |
| 47C | TGAAGAGTAAGTGGAACCTGG | GCTAAAGAAAGCCATCCGC | 274 | 1.5 mM MgCl2 55°C |
GC-rich PCR cycle.
Electronic-Database Information
The URLs for data presented herein are as follows:
- National Center for Biotechnology Information (NCBI),http://www.ncbi.nlm.nih.gov/ (for BAC RP11-14I21 [accession number AC018853.3])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CdLS) [PubMed]
References
- Bankier A, Haan E, Birrell R (1986) Familial occurrence of Brachmann-de Lange syndrome. Am J Med Genet 25:163–165 [DOI] [PubMed] [Google Scholar]
- Beratis NG, Hsu LY, Hirschhorn K (1971) Familial de Lange syndrome: report of three cases in a sibship. Clin Genet 2:170–176 [PubMed] [Google Scholar]
- Brachmann W (1916) Ein Fall von symmetrischer Monodaktylie durch Ulnadefekt, mit symmetrischer Flughautbildung in den Ellenbeugen, sowie anderen Abnormitäten (Zwerghaftigkeit, Halsrippen, Behaarung) [A case of symmetrical monodactyly representing ulnar deficiency, with symmetrical antecubital webbing and other abnormalities (dwarfish, cervical ribs, hirsutism)]. Jahrbuch für Kinderheilkunde und physische Erziehung 84:225–235 [Google Scholar]
- Chodirker BN, Chudley AE (1994) Male-to-male transmission of mild Brachmann-de Lange syndrome. Am J Med Genet 52:331–333 [DOI] [PubMed] [Google Scholar]
- de Lange C (1933) Sur un type nouveau de dégénération (typus Amstelodamnesis) [On a new type of degeneration (type Amstelodamnesis)]. Arch Méd Enfants 36:713–719 [Google Scholar]
- Feingold M, Lin AE (1993) Familial Brachmann-de Lange syndrome: further evidence for autosomal dominant inheritance and review of the literature. Am J Med Genet 47:1064–1067 [DOI] [PubMed] [Google Scholar]
- Fryns JP, Dereymaeker AM, Hoefnagels M, D’Hondt F, Mertens G, van den Berghe H (1987) The Brachmann-de Lange syndrome in two siblings of normal parents. Clin Genet 31:413–415 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA 90:10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halal F, Silver K (1992) Syndrome of microcephaly, Brachmann-de Lange–like facial changes, severe metatarsus adductus, and developmental delay: mild Brachmann-de Lange syndrome? Am J Med Genet 42:381–386 [DOI] [PubMed] [Google Scholar]
- Hulinsky R, Winesette H, Dent K, Silver R, King J, Lowichik A, Chen Z, Viskochil D (2003) Prenatal diagnosis dilemma: fetus with del(5)(p13.1p14.2) diagnosed postnatally with Cornelia de Lange syndrome. Am J Hum Genet Suppl 73:602 [Google Scholar]
- Ireland M, Donnai D, Burn J (1993) Brachmann-de Lange syndrome: delineation of the clinical phenotype. Am J Med Genet 47:959–964 [DOI] [PubMed] [Google Scholar]
- Ireland M, English C, Cross I, Houlsby WT, Burn J (1991) A de novo translocation t(3;17)(q26.3;q23.1) in a child with Cornelia de Lange syndrome. J Med Genet 28:639–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L, Kline AD, Barr MA, Koch S (1993) de Lange syndrome: a clinical review of 310 individuals. Am J Med Genet 47:940–946 [DOI] [PubMed] [Google Scholar]
- Kline AD, Barr M, Jackson LG (1993a) Growth manifestations in the Brachmann-de Lange syndrome. Am J Med Genet 47:1042–1049 [DOI] [PubMed] [Google Scholar]
- Kline AD, Stanley C, Belevich J, Brodsky K, Barr M, Jackson LG (1993b) Developmental data on individuals with the Brachmann-de Lange syndrome. Am J Med Genet 47:1053–1058 [DOI] [PubMed] [Google Scholar]
- Kozma C (1996) Autosomal dominant inheritance of Brachmann-de Lange syndrome. Am J Med Genet 66:445–448 [DOI] [PubMed] [Google Scholar]
- Krajewska-Walasek M, Chrzanowska K, Tylki-Szymanska A, Bialecka M (1995) A further report of Brachmann-de Lange syndrome in two sibs with normal parents. Clin Genet 47:324–327 [DOI] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJM, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36:631–635 10.1038/ng1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, Rand EB, Genin A, Hunt P, Jones M, Louis AA, Graham JM Jr, Bhatt S, Piccoli DA, Spinner NB (1997) Deletions of 20p12 in Alagille syndrome: frequency and molecular characterization. Am J Med Genet 70:80–86 [DOI] [PubMed] [Google Scholar]
- Krantz ID, Tonkin E, Smith M, Devoto M, Bottani A, Simpson C, Hofreiter M, Abraham V, Jukofsky L, Conti BP, Strachan T, Jackson L (2001) Exclusion of linkage to the CDL1 gene region on chromosome 3q26.3 in some familial cases of Cornelia de Lange syndrome. Am J Med Genet 101:120–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber E, Glaser JH, Jhaveri R (1973) Brachmann-de Lange syndrome: report of two cases in a sibship. Am J Dis Child 125:717–718 [DOI] [PubMed] [Google Scholar]
- McConnell V, Brown T, Morrison PJ (2003) An Irish three-generation family of Cornelia de Lange syndrome displaying autosomal dominant inheritance. Clin Dysmorphol 12:241–244 10.1097/00019605-200310000-00006 [DOI] [PubMed] [Google Scholar]
- Naguib KK, Teebi AS, Al-Awadi SA, Marafie MJ (1987) Brachmann-de Lange syndrome in sibs. J Med Genet 24:627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz JM (1985) The Brachmann-de Lange syndrome. Am J Med Genet 22:89–102 [DOI] [PubMed] [Google Scholar]
- Robinson LK, Wolfsberg E, Jones KL (1985) Brachmann-de Lange syndrome: evidence for autosomal dominant inheritance. Am J Med Genet 22:109–115 [DOI] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D (2004) Drosophila Nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol 24:3100–3111 10.1128/MCB.24.8.3100-3111.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D (1999) Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152:577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell KL, Ming JE, Patel K, Jukofsky L, Magnusson M, Krantz ID (2001) Dominant paternal transmission of Cornelia de Lange syndrome: a new case and review of 25 previously reported familial recurrences. Am J Med Genet 104:267–276 10.1002/ajmg.10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul RA, Rogers RC, Phelan MC, Stevenson RE (1993) Brachmann-de Lange syndrome: diagnostic difficulties posed by the mild phenotype. Am J Med Genet 47:999–1002 [DOI] [PubMed] [Google Scholar]
- Selicorni A, Lalatta F, Livini E, Briscioli V, Piguzzi T, Bagozzi DC, Mastroiacovo P, Zampino G, Gaeta G, Pugliese A, Cerutti-Mainaroli P, Guala A, Zelante L, Stabile M, Belli S, Franceschini P, Gianotti A, Scarano G (1993) Variability of the Brachmann-de Lange syndrome. Am J Med Genet 47:977–982 [DOI] [PubMed] [Google Scholar]
- Tonkin E, Smith M, Eichhorn P, Hagan DM, Herrell S, Lusher M, Ireland M, Burn J, Strachan T (2001) A novel gene is disrupted by a Cornelia de Lange–associated translocation breakpoint at 3q26.3. Am J Hum Genet 69:A618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36:636–641 10.1038/ng1363 [DOI] [PubMed] [Google Scholar]
- Van Allen MI, Filippi G, Siegel-Bartelt J, Yong SL, McGillivray B, Zuker RM, Smith CR, Magee JF, Ritchie S, Toi A, Reynolds JF (1993) Clinical variability within Brachmann-de Lange syndrome: a proposed classification system. Am J Med Genet 47:947–958 [DOI] [PubMed] [Google Scholar]
- Wilson WG, Kennaugh JM, Kugler JP, Wyandt HE (1983) Reciprocal translocation 14q;21q in a patient with the Brachmann-de Lange syndrome. J Med Genet 20:469–471 [DOI] [PMC free article] [PubMed] [Google Scholar]