Abstract
The rapid decrease in the cost of DNA sequencing will enable its use for novel applications. Here, we investigate the use of DNA sequencing for simultaneous discovery and genotyping of polymorphisms in family linkage studies. In the proposed approach, short contiguous segments of genomic DNA, regularly spaced across the genome, are resequenced in each pedigree member, and all sequence polymorphisms discovered within a pedigree are used as genetic markers. We use computer simulations consistent with observed human sequence diversity to show that segments of 500–1,000 base pairs, spaced at intervals of 1–2 Mb across the genome, provide linkage information that equals or exceeds that of traditional marker-based approaches. We validate these results experimentally by implementing the sequence-based linkage approach for chromosome 19 in CEPH pedigrees.
Introduction
Since the introduction of a framework for genetic linkage analysis in humans (Botstein et al. 1980), several different types of polymorphisms and genotyping techniques have been used in linkage studies. Originally, restriction fragment-length polymorphisms, assayed by Southern hybridization, served as markers. Currently, short tandem repeats (STRs) are the markers of choice, but difficulties in attempts to fully automate STR genotyping have driven the development of alternatives. In particular, the possibility of using SNPs as markers has received considerable attention (Lander 1996; Kruglyak 1997; Wang et al. 1998; Wilson and Sorant 2000; Sachidanandam et al. 2001; Goddard and Wijsman 2002). SNPs are abundant in the human genome (Sachidanandam et al. 2001), and genotyping large numbers of SNPs can offset their lower polymorphism compared to that of STRs (Kruglyak 1997). The use of SNPs in linkage analysis has been limited to date (John et al. 2004; Middleton et al. 2004) but is poised to grow with the recent publication of the first SNP-based genetic map (Matise et al. 2003) and with the development of high-throughput technologies for SNP genotyping (Jurinke et al. 2002; Oliphant et al. 2002; Hardenbol et al. 2003; Kennedy et al. 2003; Barker et al. 2004; Matsuzaki et al. 2004).
All of these approaches to linkage analysis share the following steps. First, a set of polymorphic markers is discovered in a reference population and is placed on a genetic map. Second, this common set of markers is genotyped in a number of pedigrees collected for a genetic linkage study. Separate techniques may be used for the marker discovery and genotyping steps. An alternative approach is to simultaneously discover and genotype polymorphisms in each pedigree under study. Here, we investigate the use of DNA sequencing in such an approach. DNA sequencing is a mature and accessible technology with very high throughput capacity, and further dramatic improvements in efficiency and cost are predicted to take place in the near future (Braslavsky et al. 2003; Kling 2003; Schlotterer 2004; Shendure et al. 2004). We propose resequencing a number of short contiguous segments of genomic DNA (sequencing blocks [SBs]) for each pedigree member. All sequence polymorphisms in the SBs discovered within a pedigree, whether common or rare in the population, are used as markers. In practice, for current sequencing technologies, a single SB will consist of ⩾1 sequencing reads, with each read typically in the range of 500–1,000 bp.
A key factor in the sequence requirements of this approach (the size of SBs and their spacing) is the amount of sequence variation among the pedigree founders. We use computer simulations consistent with observed human sequence diversity to show that SBs of 500–1,000 bp, spaced at intervals of 1–2 Mb across the genome, provide linkage information that equals or exceeds that of traditional approaches. Thus, sequence-based linkage analysis is practical with a modest amount of resequencing. We validate these results experimentally by implementing the sequence-based linkage approach for chromosome 19 in CEPH pedigrees.
Methods
Simulations
We created pedigree data in two steps: the generation of variant sites in founder chromosomes followed by the transmission of genetic material to offspring. In the case of resequencing data, we first generated founder chromosomes consistent with population-genetics theory and observed human sequence diversity with the use of the MS program (Hudson 2002). The second step was performed using the program Gensim (M. J. Daly, unpublished data, with modifications by one of the authors [I.F.]). In the case of STR and SNP data, both steps were performed using Gensim.
Simulated Resequencing Data
We used a model of a single population with constant effective size, nucleotide diversity θ=8×10-4, and population recombination rate r=4×10-4. The pedigree data consisted of 1,000 sets of eight chromosomes for every combination of SB size and interval length. In each set, the SBs were generated independently of each other. These segments were then positioned at intervals of 1, 2, or 5 cM, to produce resequenced founder chromosomes. This procedure assumes that SBs are in complete linkage equilibrium, which is appropriate for genetic distances ⩾1 cM. Empirical data suggest that linkage disequilibrium (LD) in human populations is higher than that in our population model, as a result of deviation from the assumptions of a constant effective size and a uniform recombination rate (Pritchard and Przeworski 2001; Ardlie et al. 2002), but this is unlikely to influence our conclusions, because both the model and empirical data predict high LD within SBs and little LD between adjacent SBs. For simplicity, all simulations used a constant ratio for conversion between the physical and genetic maps of 1 cM/Mb. In practice, SBs should be chosen at uniform spacing relative to genetic, rather than physical, maps; this can be readily accomplished because genetic markers with known intervals have been mapped to human genome sequence (Kong et al. 2002; Matise et al. 2003).
Simulated STR and SNP Data
Founder genotypes were drawn from maps of equidistant markers. We generated 1-, 2-, and 5-cM SNP maps and a 10-cM STR map. All markers had equally frequent alleles: four alleles for STRs and two alleles for SNPs. For each map, 500 simulations were performed, each time with newly derived founder genotypes.
CEPH Pedigree Resequencing
The chromosome 19 reference sequence (NCBI build 34, April 2003) was downloaded from the University of California–Santa Cruz (UCSC) Genome Bioinformatics Web site, with repetitive regions premasked using RepeatMasker. Chromosome 19 consisted of ∼63 Mb of total sequence. SBs were selected along the euchromatic portions of the chromosome (∼55 Mb) at 1-Mb intervals, for a total of 55 SBs. At each SB, two overlapping PCR amplicons were designed using PCR-Overlap (Rieder et al. 1999). This overlapping-PCR-amplicon strategy provides sequence data on internal PCR primers for each amplicon and allows screening of potential allele-specific priming, which could lead to misgenotyping. All PCR primers were ∼23 bp in length and wereconcatenated with a universal-M13 forward or reverse sequence to standardize the sequencing protocol.Templates were amplified using the Elongase PCR kit (Invitrogen) on MJR Tetrad thermal cyclers. Samples were sequenced using Big-Dye Terminator chemistry (Applied Biosystems) on an ABI 3730XL DNA analyzer. Each ∼900-bp PCR amplicon was sequenced from both ends, providing redundant sequence information at overlapping regions of the two chromatograms. Data analysts assembled the sequence data for each SB (4 chromatograms/individual generated from 2 PCR amplicons) onto a reference genomic sequence with the use of Phred (Ewing and Green 1998; Ewing et al. 1998) and Phrap (P. Green; see Phred, Phrap, and Consed Web site). The resulting alignments were edited for accuracy with the use of Consed (Gordon et al. 1998). Of the 55 loci initially selected, 8 were eliminated prior to polymorphism discovery as a result of failed or nonspecific PCR amplification. Polymorphisms (90% of which were SNPs and 10% of which were small insertions/deletions) were then identified—using PolyPhred 4.0 (Nickerson et al. 1997)—from pairwise comparisons of individual sequence chromatograms within regions with an average quality score >40. Analysts reviewed all polymorphisms identified by PolyPhred, for false positives associated with features of the surrounding sequence or biochemical artifacts. The total percentage of usable chromatograms was >90%, which is a typical success rate for a large-scale sequencing center. Detailed protocols for PCR and sequencing are available at the University of Washington–Fred Hutchinson Cancer Research Center Web site. DNA samples from CEPH pedigrees were purchased from the Coriell Cell Repository. Samples from the parents and six children each from pedigrees 66, 1424, 1349, 1362, 1408, 1423, 1344, 1345, 1346, 1347, 1340, and 1332 were sequenced.
Quality Control of Experimental Data
Of 19,776 potential genotypes (206 variants typed in 96 individuals), 1,829 were not resolved and were coded as unknown. The vast majority of missing genotypes were due to poor-quality chromatograms that were never examined in detail. All remaining chromatograms were examined for non-Mendelian transmission of alleles within each CEPH family. Genotypes showing non-Mendelian inheritance were reviewed and either were resolved on the basis of the interpretation of the raw chromatogram data or were coded as unknown (<1% of the total). The remaining (17,947) genotypes were checked for obligatory recombinations within SBs with the use of a simple script. When such recombination was detected, the genotypes of the entire SB, across all families, were reviewed on the basis of the raw chromatograms. This led to a change in 58 (0.3%) of 19,776 genotypes. Finally, the program Merlin (version 0.9.12b) was used in error-detection mode with default settings (Abecasis et al. 2002) to detect unlikely recombination events across SBs. For simplicity, we discarded all genotypes that were flagged as potential errors by Merlin. The process converged after a second round, with a total of 157 (0.8%) discarded genotypes. The total number of genotypes coded as missing was 1,986 of 19,776. Thus, single-point and multipoint consistency checks led to the removal of <2% of genotypes from high-quality chromatograms, a rate comparable to error rates for other typing technologies.
Estimation of Allele Frequencies
Allele frequencies were assigned in accordance with their relative count among all pedigree founders. In seven cases, an allele was not present in the founders (because of missing genotypes), but, nevertheless, was implied by the offspring in one or more pedigrees. For those alleles, we increased the allele count by one for each pedigree in which the implied allele was detected.
Simulations of Experimental Study
We used the procedure employed in the simulation of the first-cousin pedigrees, in which each data set consisted of 1,000 simulated pedigrees generated independently of each other from the same population model. Pedigree structure and SB size and position matched the experimental setup. We considered two primary cases. In the first case, we used all 55 blocks initially selected for variation discovery and typing. In addition, we assumed no missing genotypes and the same nucleotide diversity that was used in the simulation of first-cousin pedigrees (θ=8×10-4). In the second case, we included only the 47 SBs that entered into the polymorphism-discovery stage, removed 10% of the remaining genotypes at random, and used a nucleotide diversity value appropriate for CEPH families (θ=7.08×10-4) (see University of Washington–Fred Hutchinson Cancer Research Center Web site).
Conversion of Physical to Genetic Distance
In the first-cousin–pedigree simulations we used a constant conversion factor, 1 cM/Mb. For the experimental data and the simulations of the experimental setup, we used a realistic conversion by building a linear interpolation function based on a set of 22 markers on chromosome 19, for which both the sequence position and the linkage map position were known. The data were obtained from the SNP Consortium Web site (Matise et al. 2003). We excluded from the calculations one marker that introduced a local negative conversion factor. A similar conversion function was obtained on the basis of another set of markers from the LDB2000 Web site.
Information Content (IC) Computation
In both simulation and experimental studies, IC was computed every 0.5 cM, to facilitate homogeneous sampling along the chromosome. The computations were performed with Genehunter (Kruglyak et al. 1996; Markianos et al. 2001) running in multipoint mode. Genehunter version 2.1_r3 was used with a slightly modified output format, to augment automated data extraction. Most of the other analyses were performed within the statistical and graphical environment R (see the R Project Web site).
Results
We report all results in terms of IC, which measures the fraction of inheritance information that was extracted from a collection of pedigrees (Kruglyak et al. 1996). IC ranges from 0 (no information was extracted) to 1 (complete inheritance reconstruction). IC is a good predictor of the power to detect linkage (Kruglyak 1997) and depends only on the properties of the linkage marker set and not on any particular trait model.
SB Size and Spacing for High IC
To evaluate the number and spacing of SBs required for linkage analysis, we simulated an extensive set of resequencing scenarios, with SBs of constant size in the range of 100–2,500 bp spaced at constant intervals of 1, 2, and 5 cM. The mapped region was assumed to span 50 cM. Throughout the simulations, we used a pedigree consisting of two first cousins, their parents, and the two grandparents common to both cousins. Segregating sites in the founder chromosomes were generated to be consistent with population-genetics theory and observed levels of human nucleotide diversity (i.e., 1 nucleotide difference in 1,250 bp between two haploid genomes), and then were propagated to the remaining pedigree members. It was assumed that all pedigree members are available for genotyping.
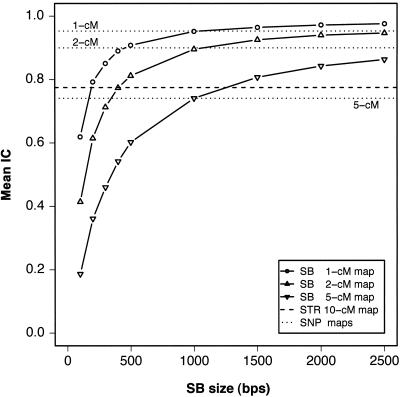
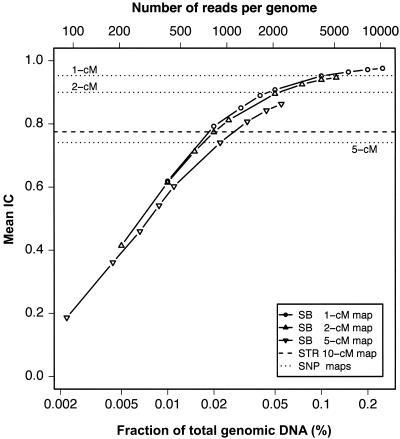
The main consideration for sequence-based linkage analysis is to reach high IC while minimizing the amount of resequencing. As shown in figure 1, an increase in the length of SBs or in their density (and, therefore, an increase in their total number) increases IC, but, at a certain point, more resequencing leads to diminishing returns. The key result of the simulation study is that high IC is reached with an SB size and spacing that is readily achieved with current sequencing technology. With an SB spacing of 1–2 cM, an SB length of 500 bp guarantees IC>0.8, and an SB length of 1,000 bp provides IC>0.9. Thus, a linkage study that uses 1,500–3,000 SBs, each consisting of 500–1,000 bp, will achieve near-complete information extraction. The results can be recast by plotting IC as a function of the total length of resequenced DNA per individual (fig. 2), showing that resequencing 0.02%–0.05% of the genome (∼1 Mb) is sufficient to reach high values of IC. In figure 2, the near overlap of the curves for different SB spacing illustrates that, as long as the genome sampling is sufficiently dense and homogeneous, IC is determined mostly by the total length of resequenced DNA, rather than by the precise details of SB length and spacing (although more densely spaced, shorter SBs achieve the same IC at somewhat shorter total length). This observation allows flexibility in study design. For example, it may be most efficient to minimize the total number of reads, with maximum read length determined by a given technology. In this case, the SB length should be chosen as an integer multiple of the maximum read length, with the spacing dictated by the desired IC. The use of more densely spaced SBs that are shorter than the maximum read length would be inefficient, even if it led to a somewhat shorter total length of resequenced DNA. Although, in figure 2, we show results for SBs that match typical read lengths of Sanger sequencing, we expect the results to remain the same, as long as the same fraction of the genome is sequenced at regular or random intervals. The only requirements are that the resequenced segments represent unique sequences and that there are no large gaps between segments.
Figure 1.
Average IC as a function of SB size and interval length. Curves correspond to data sets with a constant interval and varying SB size; the lines are visual guides. Each point represents the mean of 1,000 simulated instances. The horizontal dotted lines depict the IC expected from conventional genotyping techniques based on STR or SNP markers.
Figure 2.
Average IC as a function of total length of resequenced DNA. (Same data as in fig. 1 but with rescaled X ordinates.) The bottom X-axis shows the total length as a fraction of total genomic DNA, under the assumption of 1 cM/Mb. The top X-axis shows the number of sequencing reads per genome (under the assumption of 800-bp reads and a genome size of 3,300 Mb). Both horizontal axes are logarithmic. Symbols and line types are the same as in figure 1.
We chose the pedigree with two first cousins for comparison with work published elsewhere (Kruglyak 1997). However, we note that very similar results were obtained in simulations and experimental analysis of the two-generation nuclear families from CEPH pedigrees (see “Experimental Validation of Sequence-Based Linkage Analysis” section). Thus, we do not expect major differences in our conclusions when different pedigree structures are used. Removal of genotypes of the grandparents of the cousins produced very similar IC (relative to its maximum value, which is <1 when some individuals are unavailable) as a function of SB size and density.
Comparison of Sequence-Based Approach with STR and SNP Maps
It is important to compare sequence-based linkage analysis with conventional approaches. To provide a baseline for comparison, we computed expected IC for several idealized STR or SNP linkage maps. For STRs, we simulated a standard whole-genome 10-cM scanning set, which provides an average IC of 0.775 for the pedigree structure being considered. A sequence-based approach with equivalent IC requires SBs of ∼200, 400, and 1,250 bp for SB spacing of 1, 2, and 5 cM, respectively (fig. 1). The total amount of resequencing per single STR (SB length times the number of SBs in a 10-cM region) is ∼2 kb. For SNPs, we simulated maps with ideal SNPs (i.e., with 50-50 allele distribution) spaced every 1, 2, and 5 cM, the same as the SB spacing examined for the sequence-based approach. The average IC values for the three maps are shown in figures 1 and 2. For each spacing, the sequencing curve crosses the SNP line at an SB size of 1 kb, showing that the information from a single ideal SNP is equivalent to that from a 1-kb SB (corresponding to one or two sequence reads). IC for SNPs with a more realistic 60-40 allele distribution is expected to be nearly identical (Kruglyak 1997), and thousands of such SNPs have been identified by the HapMap consortium (see HapMap Web site).
Experimental Validation of Sequence-Based Linkage Analysis
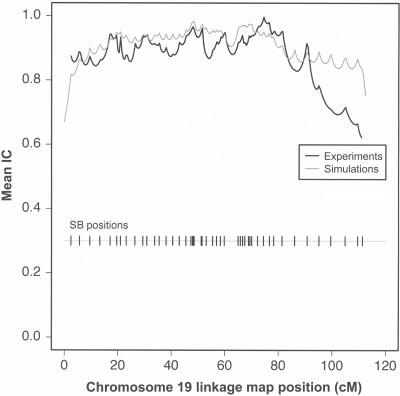
How well do the simulation-based results translate to the laboratory? To answer this question, we performed sequence-based linkage analysis of chromosome 19 in 12 CEPH pedigrees. From each pedigree, we selected the two parents and six children—a total of 96 individuals—for resequencing. On the basis of the prediction that SBs ∼1 kb long spaced at 1–2 cM can produce high IC, the entire chromosome 19 (63 Mb in physical distance and 110 cM in genetic distance [Matise et al. 2003]) of each individual was sampled at a spacing of 1 Mb with SBs of average size 1.6 kb. As noted in the “Methods” section, at each SB we designed two overlapping ∼900-bp PCR amplicons, and each amplicon was sequenced from both ends. The high redundancy of the experimental protocol minimized errors due to allele-specific amplification and base calling at the end of long sequencing reads. Initially, we selected SBs at 55 loci along the chromosome, but 8 failed PCR amplification. We report results for the remaining 47 loci, which encompass ∼75 kb of sequence. In total, we discovered 206 sites that were polymorphic in at least one pedigree; these were used as genetic markers for linkage analysis. The data set included 19,776 potential genotypes, of which 1,986 (∼10%) were coded as missing, either because of low-quality sequence (∼8%) or because they were flagged as potential genotyping errors (<2%). We used the remaining genotypes to reconstruct inheritance and compute IC (fig. 3). As predicted, the average IC was high (0.87) and close to that expected on the basis of simulations for the same pedigree structure and resequencing strategy (0.92). We also ran the simulation with parameters that closely matched the experimental setup—we incorporated the lower sequence diversity observed in CEPH samples, discarded the eight SBs that did not amplify by PCR, and coded 10% of the remaining genotypes as unknown. This resulted in a reduction of IC from 0.92 to 0.90 (see “Methods” section for details). The small difference between the experimental result and theoretical prediction (0.87 vs. 0.90) is primarily due to unexpectedly low sequence diversity at the distal end of chromosome 19 (fig. 3).
Figure 3.
IC obtained from CEPH families as a function of linkage map position along chromosome 19. Curves reflect the average at each position over all pedigrees in the data set (12 in the experiments; 1,000 in the simulations). The bar with vertical lines depicts the locations of the SBs on the genetic map.
Discussion
The rapid decline in the cost of sequencing, along with the availability of reference genome sequences for human and other organisms, makes linkage analysis through simultaneous marker discovery and genotyping by resequencing an approach worth exploring. We have used realistic simulations of human genetic variation and chromosomal segregation to show that SBs of 500–1,000 bp (corresponding to one or two reads, with current sequencing technology), spaced every 1–5 cM, will provide linkage information similar to that obtained from SNP maps with the same density, or from a typical 10-cM STR map. These simulation-based results were validated with proof-of-principle experimental analysis of human chromosome 19 in 12 CEPH pedigrees. In this analysis, observed IC was in close agreement with simulations. Approximately 10% of the SBs were rejected at the PCR-amplification stage, and ∼10% of genotypes were discarded during quality control, but this led to only an ∼5% drop in IC relative to simulations of ideal data. Thus, sequence-based linkage analysis is currently feasible, without the need for further technological or computational developments. The relatively small number and size of sequence segments needed to maximize inheritance information is a reflection of considerable human sequence variation. Sequence-based markers are biallelic SNPs or insertions/deletions. Our results reinforce the fact that such markers, despite low individual heterozygosity, are highly useful for linkage analysis, provided that they are spaced at sufficiently high density.
Given current costs, resequencing is not yet competitive with STR or SNP genotyping for genome scans in human pedigrees. However, even without a high degree of automation, the cost of the sequence-based approach is within an order of magnitude of the cost of currently available high-throughput genotyping technologies. In addition, there are applications in which sequencing is already competitive. For example, the fine-mapping stage of positional-cloning projects could benefit from such an approach. Also, studies in outbred animal populations for which a draft genome sequence is available but for which genetic marker sets are not well developed can readily adopt such a typing strategy. A key advantage of the approach is that a uniform and highly flexible experimental design can be used to meet any desired map resolution and the IC requirements of a study. Finally, it is worth noting that, in sequence-based linkage analysis, every discovered sequence variant is used as a genetic marker. This includes the variants that are individually rare in the population and have very low heterozygosity but whose total number is large enough to provide many polymorphisms in a pedigree. Thus, resequencing provides access to this abundant pool of variation—something that is impossible to achieve with conventional STR and SNP maps. This opens the possibility, for example, of the use of rare variants as unique tags that trace the fate of chromosomal regions in large pedigrees with much-simplified computational algorithms.
Acknowledgments
We thank Elaine Ostrander, for comments on the manuscript, and Mike Eberle, for helpful discussions. We would also like to thank two anonymous readers for constructive comments that helped clarify the presentation. This work was supported by grants from the National Institute of Mental Health (MH59520 to L.K.) and the National Heart, Lung, and Blood Institute (HL66682 to D.A.N. and M.J.R. and HL66642 to L.K.). I.F. is supported by an Immunex Fellowship through the dual-mentor program at Fred Hutchinson Cancer Research Center. K.M. is supported by a National Human Genome Research Institute career development award (HG002491). L.K. is a James S. McDonnell Centennial Fellow.
Electronic-Database Information
The URLs for data presented herein are as follows:
- HapMap project, http://www.hapmap.org
- LDB2000 genetic map, http://cedar.genetics.soton.ac.uk/public_html/LDB2000/release.html
- Phred, Phrap, and Consed Software, http://www.phrap.org/phredphrapconsed.html
- R Project, http://www.r-project.org/
- SNP Consortium, http://snp.cshl.org (for SNP genetic map)
- UCSC Genome Bioinformatics, http://genome.ucsc.edu
- University of Washington–Fred Hutchinson Cancer Research Center, http://pga.gs.washington.edu (for PCR and sequencing protocols and diversity value for CEPH samples)
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Ardlie KG, Kruglyak L, Seielstad M (2002) Patterns of linkage disequilibrium in the human genome. Nat Rev Genet 3:299–309 10.1038/nrg777 [DOI] [PubMed] [Google Scholar]
- Barker DL, Hansen MS, Faruqi AF, Giannola D, Irsula OR, Lasken RS, Latterich M, Makarov V, Oliphant A, Pinter JH, Shen R, Sleptsova I, Ziehler W, Lai E (2004) Two methods of whole-genome amplification enable accurate genotyping across a 2320-SNP linkage panel. Genome Res 14:901–907 10.1101/gr.1949704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, White DL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331 [PMC free article] [PubMed] [Google Scholar]
- Braslavsky I, Hebert B, Kartalov E, Quake SR (2003) Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci USA 100:3960–3964 10.1073/pnas.0230489100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Green P (1998) Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res 8:186–194 [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res 8:175–185 [DOI] [PubMed] [Google Scholar]
- Goddard KA, Wijsman E (2002) Maps for cost-effective genome screens using diallelic markers. Genet Epidemiol 22:205–220 10.1002/gepi.0177 [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202 [DOI] [PubMed] [Google Scholar]
- Hardenbol P, Baner J, Jain M, Nilsson M, Namsaraev EA, Karlin-Neumann GA, Fakhrai-Rad H, Ronaghi M, Willis TD, Landegren U, Davis RW (2003) Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol 21:673–678 10.1038/nbt821 [DOI] [PubMed] [Google Scholar]
- Hudson RR (2002) Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18:337–338 10.1093/bioinformatics/18.2.337 [DOI] [PubMed] [Google Scholar]
- John S, Shephard N, Liu G, Zeggini E, Cao M, Chen W, Vasavda N, Mills T, Barton A, Hinks A, Eyre S, Jones KW, Ollier W, Silman A, Gibson N, Worthington J, Kennedy GC (2004) Whole-genome scan, in a complex disease, using 11,245 single-nucleotide polymorphisms: comparison with microsatellites. Am J Hum Genet 75:54–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurinke C, Van den Boom D, Cantor CR, Koster H (2002) Automated genotyping using the DNA MassArray technology. Methods Mol Biol 187:179–192 [DOI] [PubMed] [Google Scholar]
- Kennedy GC, Matsuzaki H, Dong S, Liu WM, Huang J, Liu G, Su X, Cao M, Chen W, Zhang J, Liu W, Yang G, Di X, Ryder T, He Z, Surti U, Phillips MS, Boyce-Jacino MT, Fodor SP, Jones KW (2003) Large-scale genotyping of complex DNA. Nat Biotechnol 21:1233–1237 10.1038/nbt869 [DOI] [PubMed] [Google Scholar]
- Kling J (2003) Ultrafast DNA sequencing. Nat Biotechnol 21:1425–1427 10.1038/nbt1203-1425 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L (1997) The use of a genetic map of biallelic markers in linkage studies. Nat Genet 17:21–24 10.1038/ng0997-21 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander ES (1996) The new genomics: global views of biology. Science 274:536–539 10.1126/science.274.5287.536 [DOI] [PubMed] [Google Scholar]
- Markianos K, Daly MJ, Kruglyak L (2001) Efficient multipoint linkage analysis through reduction of inheritance space. Am J Hum Genet 68:963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Sachidanandam R, Clark AG, Kruglyak L, Wijsman E, Kakol J, Buyske S, et al (2003) A 3.9-centimorgan-resolution human single-nucleotide polymorphism linkage map and screening set. Am J Hum Genet 73:271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki H, Loi H, Dong S, Tsai YY, Fang J, Law J, Di X, Liu WM, Yang G, Liu G, Huang J, Kennedy GC, Ryder TB, Marcus GA, Walsh PS, Shriver MD, Puck JM, Jones KW, Mei R (2004) Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res 14:414–425 10.1101/gr.2014904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Pato MT, Gentile KL, Morley CP, Zhao X, Eisener AF, Brown A, Petryshen TL, Kirby AN, Medeiros H, Carvalho C, Macedo A, Dourado A, Coelho I, Valente J, Soares MJ, Ferreira CP, Lei M, Azevedo MH, Kennedy JL, Daly MJ, Sklar P, Pato CN (2004) Genomewide linkage analysis of bipolar disorder by use of a high-density single-nucleotide–polymorphism (SNP) genotyping assay: a comparison with microsatellite marker assays and finding of significant linkage to chromosome 6q22. Am J Hum Genet 74:886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 10.1093/nar/25.14.2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A, Barker D, Stuelpnagel JR, Chee MS (2002) BeadArray™ technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques Suppl 32:56–61 [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M (2001) Linkage disequilibrium in humans: models and data. Am J Hum Genet 69:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Clark AG, Nickerson DA (1999) Sequence variation in the human angiotensin converting enzyme. Nat Genet 22:59–62 10.1038/8760 [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, et al (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. The International SNP Map Working Group. Nature 409:928–933 10.1038/35057149 [DOI] [PubMed] [Google Scholar]
- Schlotterer C (2004) Opinion: the evolution of molecular markers—just a matter of fashion? Nat Rev Genet 5:63–69 10.1038/nrg1249 [DOI] [PubMed] [Google Scholar]
- Shendure J, Mitra RD, Varma C, Church GM (2004) Advanced sequencing technologies: methods and goals. Nat Rev Genet 5:335–344 10.1038/nrg1325 [DOI] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, et al (1998) Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280:1077–1082 10.1126/science.280.5366.1077 [DOI] [PubMed] [Google Scholar]
- Wilson AF, Sorant AJ (2000) Equivalence of single- and multilocus markers: power to detect linkage with composite markers derived from biallelic loci. Am J Hum Genet 66:1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]