Abstract
We completed fine mapping of nine positional candidate regions for attention-deficit/hyperactivity disorder (ADHD) in an extended population sample of 308 affected sibling pairs (ASPs), constituting the largest linkage sample of families with ADHD published to date. The candidate chromosomal regions were selected from all three published genomewide scans for ADHD, and fine mapping was done to comprehensively validate these positional candidate regions in our sample. Multipoint maximum LOD score (MLS) analysis yielded significant evidence of linkage on 6q12 (MLS 3.30; empiric P=.024) and 17p11 (MLS 3.63; empiric P=.015), as well as suggestive evidence on 5p13 (MLS 2.55; empiric P=.091). In conjunction with the previously reported significant linkage on the basis of fine mapping 16p13 in the same sample as this report, the analyses presented here indicate that four chromosomal regions—5p13, 6q12, 16p13, and 17p11—are likely to harbor susceptibility genes for ADHD. The refinement of linkage within each of these regions lays the foundation for subsequent investigations using association methods to detect risk genes of moderate effect size.
Attention-deficit/hyperactivity disorder (ADHD [MIM 143465]) is one of the most commonly diagnosed neurobehavioral disorders of childhood, affecting ∼5%–7% of children and ∼3% of adults (Wolraich et al. 1996; McCracken 1998; Swanson et al. 1998). ADHD is defined as the childhood onset of multiple symptoms of inattention and/or hyperactivity-impulsivity leading to significant impairment in at least two settings (American Psychiatric Association 1994). Heritability estimates in the range of 60%–90% (Levy et al. 1997; Faraone and Doyle 2001), sibling relative risk estimates (λs) in the range of 4–8 (Smalley 1997; Faraone et al. 2000), and consistent prevalence rates across diverse regions of the world (Anderson et al. 1987; Gomez et al. 1999; Tahir et al. 2000; Wilens et al. 2002) suggest a strong genetic etiology.
The vast majority of molecular genetic studies of ADHD have been predicated on the detection of association between ADHD and the allelic variants of functional candidates. The selection of functional candidates has relied on assumptions about the molecular mechanisms underlying ADHD and has been generally constrained to the most obvious genes integral to the dopaminergic, serotonergic, and adrenergic pathways. This approach is fundamentally limited by our current understanding of the pathways involved in this disorder, as well as the molecular components of a given pathway. Although positive associations with polymorphisms near or within the dopamine transporter gene (DAT-1), dopamine receptor D4 gene (DRD4), and dopamine receptor D5 gene (DRD5) have been reproduced in independent studies, failures to replicate these results in adequately sized populations are also evident (Palmer et al. 1999; Holmes et al. 2000), and the purported effect sizes are small (Cook et al. 1995; LaHoste et al. 1996; Gill et al. 1997; Smalley et al. 1998; Daly et al. 1999; McCracken et al. 2000; Faraone et al. 2001; Mill et al. 2001; Lowe et al. 2004). Other candidate genes have been the subject of fewer investigations, and both positive and negative findings are reported (Barr et al. 2000; Brophy et al. 2002; Kustanovich et al. 2003).
Although the pooling of resources within the ADHD-genetics community may improve the power to detect subtle associations, the effect of such alleles is proving to be small (Lowe et al. 2004). Genomewide linkage methods provide a complementary and powerful approach to candidate-gene studies, because they do not rely on prior knowledge of the molecular etiology, and novel genes may be identified through positional cloning. Three genomewide linkage scans of ADHD have been published to date with the use of affected sibling pair (ASP) sampling (Fisher et al. 2002; Bakker et al. 2003; Ogdie et al. 2003). The first genomewide scan was conducted on 126 ASPs (Fisher et al. 2002) and identified four nominal regions (5p12, 10q26, 12q23, and 16p13) with multipoint maximum LOD scores (MLSs) >1.5, but none exceeded recommended thresholds for suggestive or significant linkage. A second genomewide scan in an independent set of 101 families was pooled with this initial group of 126 ASPs (270 ASPs total), and six chromosomal regions were identified with MLS values >1 (5p13, 6q14, 11q25, 16p13, 17p11, and 20q13), with one region exceeding suggestive evidence of linkage (17p11; MLS 2.98) (Ogdie et al. 2003). An independent team of investigators (Bakker et al. 2003) completed a genomewide scan of 164 Dutch ASPs with ADHD and presented evidence for four strong-candidate chromosomal locations (7p13, 9q33, 13q33, and 15q15), with overlap of only one nominally significant region (5p13) across the scans.
Fine mapping of the 16p13 region highlighted in our genomewide scans was completed and described elsewhere in an extended sample, and results of that analysis demonstrated genomewide significance under both theoretical and empirical criteria (MLS 3.73; empiric P=.01) (Ogdie et al. 2003; Smalley et al. 2002). In this report, we describe fine mapping of nine additional regions in an extended sample of 308 ASPs. The nine regions included five selected on the basis of our genomewide linkage analyses (5p13, 6q12/6q14, 11q25, 17p11, and 20q13) and the four strongest regions identified in the Dutch sample (7p13, 9q33, 13q33, and 15q15) (Bakker et al. 2003).
The sample of 308 ASPs with ADHD is derived from 226 multiplex families (194 two-sib families, 26 three-sib families, and 6 four-sib families), containing a total of 490 affected children and 308 ASPs (all possible pairs). The current sample contains 269 of the 270 ASPs reported in the previous genomewide scans (Fisher et al. 2002; Ogdie et al. 2003) and an independent set of 39 ASPs. Parents of all ASPs were genotyped, with both parents available for 289 (94%) ASPs and one parent available for 19 (6%) ASPs. The affected individuals in the sample are 73% male, 78% white, and fairly representative of the subtype proportions evident in epidemiological studies of ADHD (49% combined, 44% inattentive, and 7% hyperactive-impulsive) (Wolraich et al. 1996). ADHD was diagnosed in accordance with DSM-IV criteria, by use of a best-estimate procedure (American Psychiatric Association 1994). A definite ADHD diagnosis was made for 95% of the ASP members, whereas a probable ADHD diagnosis (i.e., the individual showed one less symptom than the diagnosis requirement but met criteria of impairment and age at onset) was made in 5% of the sample. All ASPs have at least one member with a definite diagnosis. Details of the sample characteristics are published elsewhere (Smalley et al. 2000). Expected psychiatric comorbidity is present in the sample and includes oppositional-defiant disorder (46%), conduct disorder (11%), mood disorders (major depression, dysthymia, and bipolar disorder) (17%), and anxiety disorders (8%). Although five of the ASP members had a comorbid diagnosis of bipolar disorder, none came from the same family. We tested the impact these cases might have had on the MLS results by removing them from the data set and rerunning the MLS analysis. There were no significant changes in the MLS values for any of the chromosomes of interest in the sample from which the five cases were excluded.
The nine chromosomal regions selected for fine mapping (table 1) were defined by the 1-LOD support intervals of the linkage peaks presented in the genomewide scans. For the five candidate regions identified in our sample (5p13, 6q14, 11q25, 17p11, and 20q13), we selected microsatellite markers to create an ∼2-cM grid across the 1-LOD support intervals (12–19 markers per region). For the four candidate regions identified in the Dutch cohort, we selected microsatellite markers to create an ∼3-cM grid across the 1-LOD support intervals (8–10 markers per region). The genetic positions of all markers were initially determined using the deCODE high-resolution map (Kong et al. 2002) and Marshfield (Center for Medical Genetics) genetic maps and were validated by mapping procedures within our own data set by use of ASPEX v. 2.3 sib_map (Hinds and Risch 1996). In addition, the physical-mapping positions and order of all markers were verified with both the University of California–Santa Cruz (UCSC) genome database build hg16 and the National Center for Biotechnology Information (NCBI) build 34. Genotyping was performed in accordance with standard procedures(Ogdie et al. 2003). Mendelian inheritance errors were identified and removed using GAS 2.0 (A. Young, Oxford University), and improbable genotypes, as determined by the presence of unlikely recombination events, were identified and removed using Simwalk2 (Sobel and Lange 1996).
Table 1.
Multipoint MLS Values in Chromosomal Regions of Fine Mapping
|
Location |
P |
|||||
| Chromosome | Markera | Cytogeneticb | Geneticc(cM) | MLS | Nominald | Empirice |
| 5 | D5S418 | 5p13 | 59 | 2.55 | .00058 | .091 |
| 6 | D6S430 | 6q12 | 83 | 3.30 | .000098 | .024 |
| 7 | D7S1818 | 7p13 | 69 | .00 | .60 | 1.000 |
| 9 | D9S1825 | 9q33 | 136 | .00 | .60 | 1.000 |
| 11 | D11S4126 | 11q25 | 173 | 1.00 | .026 | .931 |
| 13 | D13S796 | 13q33 | 94 | .84 | .039 | .972 |
| 15 | D15S146 | 15q15 | 35 | .28 | .18 | 1.000 |
| 16f | D16S3114 | 16p13 | 23 | 3.73 | .000035 | .012 |
| 17 | D17S947 | 17p11 | 38 | 3.63 | .000045 | .015 |
| 20 | D20S1106 | 20q13 | 101 | 1.09 | .020 | .871 |
Marker nearest to the center of the 1-LOD support interval.
Approximate cytogenetic position, as determined by physical mapping of markers under a 1-LOD support interval.
The approximate Marshfield genetic position of the center of the 1-LOD support interval. For the four Utrecht regions presenting MLSs <1 (7p, 9q, 13q, and 15q), the listed genetic position was determined by the center of the fine-mapping marker panel.
LOD scores were converted into nominal pointwise P values: P(LOD)=.5×(χ21>2ln10×LOD)+.098×(χ22>2ln10×LOD) (Nyholt 2000).
Approximate empiric P values were determined from 1,000 replicates of the genomewide data set under the null hypothesis at an artificially high density of one marker every 2 cM across the genome. Calculated as (x+1)/(n+1), where x is the number of independent regions of linkage presenting an MLS greater than or equal to the threshold and n is the number of replicates. For peaks yielding MLS values that occurred more frequently than once per replicate, the P value was calculated as (r+1)/(n+1), where r is the number of replicates yielding an MLS greater than or equal to the threshold (7p, 9q, 11q, 13q, 15q, and 20q).
Data for 16p13 was published elsewhere by Ogdie et al. (2003) and is presented here for reference.
Multipoint MLS analysis (Risch 1990) was conducted by ASPEX sib_ibd v. 2.3 (Hinds and Risch 1996), under the multiplicative model with parameters restricted to the possible triangle (Holmans 1993). MLSs were calculated at 1-cM increments, with all possible pairs treated as independent. Single-point MLS analysis was performed by Genehunter v. 2.1 with the use of Mapmaker/Sibs commands (Kruglyak and Lander 1995; see Kruglyak Laboratory and Mapmaker/Sibs Web sites). ASPEX sib_ibd uses only fully informative transmissions, whereas single-point MLS analysis in Genehunter uses all inheritance information for the reconstruction of missing parental genotypes and for the determination of identity-by-descent (IBD) and thus maximizes the number of ASPs contributing to a single-point MLS. MLS values were converted to pointwise P values by the derivation for MLS calculated under the possible triangle: P(LOD)=.5×(χ21>2ln10×LOD)+.098×(χ22>2ln10×LOD) (Holmans 1993; Nyholt 2000). In accordance with convention, we have defined the nominal threshold as a pointwise P value <.05. To estimate empiric P values, simulations were performed using the unique parameters of the current data set, including marker parameters, pedigree structure, and missing genotypes. The purpose of the current simulations is to estimate a threshold of significance that accounts for genomewide sampling (i.e., an MLS equivalent to P=.05 or an event likely to randomly occur once in 20 genomewide scans) and to estimate how often one would encounter an independent region of linkage with an MLS exceeding a particular level in a genomewide scan. Theoretically derived thresholds of significance do not account for missing data points, uninformative markers, or the variable density of markers employed in linkage studies. A total of 1,000 replicates of the genomewide data were generated under the null hypothesis of no linkage, by use of Simulate 2.4 (Terwilliger et al. 1993), and were analyzed by the same procedures as the actual data set. Simulate 2.4 distributes alleles among the founders of each pedigree on the basis of the allele frequencies present in the real data set, creates recombination events on the basis of the probabilities defined by the marker maps, and randomly transmits chromosomes to progeny. We generated replicates of the data set at an artificially high genomewide density of one marker every 2 cM, accounting for the assumption that fine mapping within this sample will continue in the future. The empiric P values presented in this study are intended to estimate the likelihood of observing a given MLS value in a 2-cM genomewide scan performed in a sample of 308 ASPs without linkage to a disease.
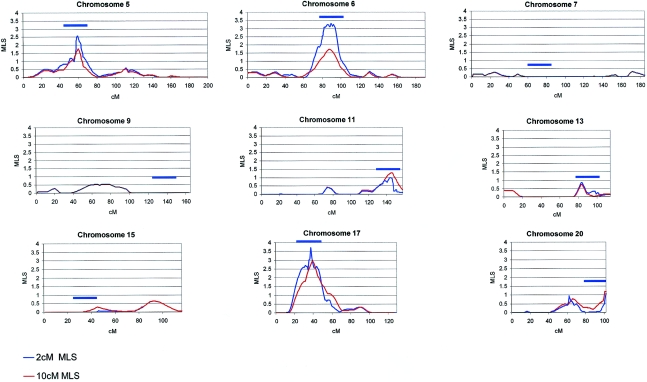
Multipoint MLS analysis yielded significant evidence of linkage for the regions on 6q12-6q14 (MLS 3.30; empiric P=.024) and 17p11 (MLS 3.63; empiric P=.015) (table 1 and fig. 1). Region 5p13, the only region presenting evidence of linkage in both our sample and the Dutch sample, yielded suggestive evidence (MLS 2.55; empiric P=.091). Evidence of linkage to 11q25 (MLS 1.00; empiric P=.931) and 20q13 (MLS 1.09; empiric P=.871) remained below the suggestive threshold and did not increase above that of the 10-cM data set. Region 13q33 yielded evidence of linkage above the nominal threshold (MLS 0.84; pointwise P=.039; empiric P=.972), whereas analysis of the other three regions identified by Bakker et al. (2003) failed to yield even nominal evidence of linkage in our sample and did not vary appreciably from the evidence provided by our 10-cM data set, indicating that the 10-cM genomewide scan provided an accurate estimate of IBD sharing in these unlinked regions (table 1 and fig. 1). To assess the evidence of linkage present in the independent set of 39 ASPs—included in the current study and absent from the previously reported genomewide scans—we performed a separate multipoint MLS analysis of this independent set. Although the sample of 39 independent ASPs is not large enough to demonstrate statistical significance comparable to the samples described in the genomewide scans (Fisher et al. 2002; Ogdie et al. 2003), the trend of excess sharing continues, and evidence of linkage is present: 5p13 (sharing 63.55%, MLS 0.22), 6q12 (sharing 70.09%, MLS 1.27), and 17p11 (sharing 59.79%, MLS 0.18). Thus, the increased evidence of linkage is the result of both greater marker density and the expansion of the sample size. Results from single-point MLS analyses were highly consistent with multipoint findings (table 2). Of note, in fine-mapping regions on chromosomes 5, 6, and 17, several markers yielded MLSs >1 under each peak (three markers in 5p13, seven markers in 6q12, and nine markers in 17p11).
Figure 1.
Multipoint MLS values for nine candidate regions in 308 ASPs with ADHD. The X-axis values are distances from the p-telomere, in Kosambi cM. The MLS values for the 10-cM genomewide scan of 270 ASPs (Ogdie et al. 2003) are shown in red. The fine-mapping values, at ∼ 2-cM marker density in candidate regions, are shown in blue. The blue bar indicates the approximate fine-mapping interval. The identical MLS values for the 2-cM and 10-cM analyses for chromosomes 7 and 9 and a large portion of chromosome 15 give the appearance of a single line. MLS analysis was performed by ASPEX sib_ibd, under the possible triangle.
Table 2.
Markers Yielding Single-Point MLS >1 in Fine-Mapping Regions
|
Location |
|||||
| Chromosomeand Marker | Cytogeneticc | Geneticd(cM) | Heterozygositya(%) | MLS | Nominal Pb |
| 5: | |||||
| D5S418 | 5p13 | 58.6 | 80 | 3.24 | .00011 |
| D5S1958 | 5p12 | 59.9 | 58 | 1.24 | .014 |
| D5S1968 | 5q11 | 60.9 | 77 | 1.26 | .013 |
| 6: | |||||
| D6S465 | 6p12 | 74.3 | 75 | 2.46 | .00072 |
| D6S1960 | 6p12 | 76.6 | 74 | 1.05 | .022 |
| D6S257 | 6p12 | 79.9 | 87 | 1.83 | .0033 |
| D6S430 | 6q12 | 81.5 | 83 | 1.72 | .0043 |
| D6S1557 | 6q13 | 82.6 | 76 | 1.97 | .0023 |
| D6S460 | 6q14 | 89.8 | 81 | 1.06 | .022 |
| D6S1609 | 6q14 | 92.3 | 80 | 1.50 | .0074 |
| 11: | |||||
| D11S4126 | 11q25 | 138.6 | 54 | 1.19 | .016 |
| 16e: | |||||
| D16S3114 | 16p13 | 23.3 | 79 | 1.74 | .0041 |
| D16S3060 | 16p13 | 28.3 | 81 | 1.42 | .0090 |
| 17: | |||||
| D17S947 | 17p12 | 32.0 | 76 | 1.72 | .0043 |
| D17S799 | 17p12 | 32.1f | 90 | 1.54 | .0067 |
| D17S1856 | 17p12 | 35.6 | 81 | 2.51 | .00064 |
| D17S839 | 17p11 | 37.8 | 64 | 1.84 | .0032 |
| D17S953 | 17p11 | 43.0 | 68 | 1.23 | .014 |
| D17S1857 | 17p11 | 43.1f | 71 | 2.06 | .0019 |
| D17S2196 | 17p11 | 44.6 | 84 | 1.70 | .0045 |
| D17S1824 | 17q11 | 49.7 | 82 | 1.35 | .011 |
| D17S798 | 17q11 | 53.4 | 74 | 1.41 | .0092 |
| 20: | |||||
| D20S1106 | 20q13 | 101.2f | 71 | 2.11 | .0017 |
Heterozygosity calculated from the entire study sample.
LOD scores were converted into nominal pointwise P values: P(LOD)=.5×(χ21>2ln10×LOD)+.098×(χ22>2ln10×LOD) (Nyholt 2000).
Approximate cytogenetic position, as determined by physical mapping of markers under 1-LOD support interval.
The approximate Marshfield genetic position of the marker.
Data for 16p13 was published elsewhere by Ogdie et al. (2003), and we have presented two markers here for reference.
The genetic position was estimated from physical-mapping position and/or deCODE.
The MLS analysis presented here, in conjunction with previously published work (Smalley et al. 2002), strongly supports four chromosomal regions (5p13, 6q12, 16p13, and 17p11) as likely candidate locations of susceptibility loci for ADHD. The 5p13 region is highlighted, despite not reaching genomewide significance, because of the overlap of this region with that presented by the independent scan in the sample of Dutch ASPs (MLS 1.43) (Bakker et al. 2003). The estimated IBD sharing parameters for all four regions indicate loci of moderate effect size, with λs in the range of 1.4–1.6, under a multiplicative model with a recombination fraction of zero (θ=0) (Risch 1990). The current analysis eliminates the possibility that our failure to detect linkage in the four major regions identified in the Dutch study is the result of poor coverage. The present study does not address the lack of linkage detected in the Dutch sample for 6q12, 16p13, and 17p11. To assess the likelihood of observing three linkage peaks above the threshold of significance, we assumed a Poisson distribution (Lander and Kruglyak 1995; Wiltshire et al. 2002) and derived a posterior probability of this event under the null hypothesis. The cumulative Poisson distribution indicates that the probability of observing the three significant linkage peaks within this sample is <2.3 ×10−6, strongly suggesting that at least one susceptibility gene is located in one of these three regions (6p12, 16p13, and 17p12).
The region on 5p13 is centered at D5S418 (58 cM, Marshfield; 40 Mb, UCSC hg16), with a 1-LOD support interval spanning ∼7 cM (15 Mb) from D5S2105 to D5S1968. Fine mapping of 5p13 resulted in an increase from an MLS of 1.77 to an MLS of 2.55 and significantly narrowed the 1-LOD support interval from ∼20 cM to ∼7 cM. The region on 6q12-6q14 is centered at D6S430 (81 cM, Marshfield; 67 Mb, UCSC hg16), with a 1-LOD support interval spanning ∼18 cM (33 Mb) from D6S465 to D6S1609 and a maximum MLS value on 6q14 (∼89 cM, D6S460). The fine-mapping data increased the MLS from 1.75 to 3.30 and reduced the 1-LOD support interval by 12 cM. Note that the gene encoding serotonin receptor 1B (HTR1B) is directly under the maximum MLS (89 cM), and the gene encoding serotonin receptor 1E (HTR1E) resides just outside of the 1-LOD q-boundary. Elsewhere, Quist et al. (2003) have reported a trend toward excess transmission of a polymorphism in HTR1B in a sample of 115 families with ADHD (P=.09). The region on 16p13 is centered at D16S3060 (28 cM, Marshfield; 12 Mb, UCSC hg16), with a 1-LOD support interval spanning ∼12 cM (7 Mb) from D16S519 to D16S499, and overlaps a region highlighted in genomewide scans for autism (Smalley et al. 2002). The 1-LOD support interval on 17p11, centered at D17S839 (37 cM, Marshfield; 14 Mb, UCSC hg16) and spanning ∼20 cM (25 Mb) across the centromere from D17S947 to D17S798, also overlaps with two genomewide scans in autism (International Molecular Genetic Study of Autism Consortium 2001; Yonan et al. 2003). Fine mapping of this region increased the evidence of linkage from an MLS of 2.98 to an MLS of 3.63 and refined the 1-LOD support interval by 5 cM. The gene encoding serotonin transporter (5-HTT), a commonly cited functional candidate for both ADHD and autism, is located on 17q11 within the q-boundary of the 1-LOD support interval. Manor et al. (2001) reported an association between ADHD and a promoter polymorphism in 5-HTT (5-HTTLPR) in 98 trios (P=.008). In addition, Seeger et al. (2001) have reported an association between hyperkinetic disorder and the 5-HTTLPR long variant (P=.009). These four regions do not contain the most commonly studied functional candidate genes, highlighting the importance of genomewide linkage strategies for complex traits.
In conclusion, we have defined three genomic regions yielding empirically significant linkage to ADHD (6q12, 16p13, and 17p11) and a fourth region yielding suggestive evidence (5p13). Association studies (e.g., with the use of high-density–SNP data) constitute a viable and realistic strategy for the identification of causal polymorphisms in these regions. From the predicted λs values (1.4–1.6) observed in our fine-mapping studies, the genotype relative risks are estimated to be >3 (Risch and Merikangas 1996), indicating that a sample of 170 trios would provide adequate power for the detection of common effect alleles. Replication of linkage in these regions in an independent sample would provide an important validation. Thus, ongoing collection of ADHD samples is critical for the replication of both linkage and association, given the genetic heterogeneity demonstrated by the discordance of linkage studies and the small population–attributable risks found for previously associated polymorphisms. Finally, the refinement of phenotypes and trait measures more closely reflecting the true biological underpinnings may greatly facilitate efforts to identify susceptibility genes for ADHD.
Acknowledgments
This work was supported by National Institute of Mental Health grants MH58277 (to S.L.S.), MH01969 (to J.J.M.), and MH01805 (to J.T.M.); by University of California Life Sciences Informatics grant L9808 (to S.F.N.); and by U.S. Public Health Service National Research Service Award GM07104 (to M.N.O.). S.E.F. is a Royal Society Research Fellow. Thanks to all the families who participated in this research. Thanks to Professor Anthony Monaco for his support throughout. Thanks to Jaana Hartiala at the UCLA Genotyping Core, for technical assistance, and to Dennis Cantwell, M.D., who inspired our work on ADHD.
Electronic-Database Information
The URLs for data presented herein are as follows:
- ASPEX Linkage Analysis Package, http://hpcio.cit.nih.gov/lserver/ASPEX.html
- GAS, http://hpcio.cit.nih.gov/lserver/GAS.html (for genotype linkage analysis)
- Kruglyak Laboratory, http://www.fhcrc.org/labs/kruglyak/Downloads/ (for Genehunter)
- Mapmaker/Sibs software, http://www-genome.wi.mit.edu/ftp/distribution/software/sibs/
- Marshfield Clinic Research Foundation (Center for Medical Genetics) http://research.marshfieldclinic.org/genetics/
- NCBI, http://www.ncbi.nlm.nih.gov/genome/guide/human/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ADHD) [PubMed]
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/
References
- American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders. 4th ed (DSM-IV). American Psychiatric Association, Washington, DC [Google Scholar]
- Anderson JC, Williams S, McGee R, Silva PA (1987) DSM-III disorders in preadolescent children: prevalence in a large sample from the general population. Arch Gen Psychiatry 44:69–76 [DOI] [PubMed] [Google Scholar]
- Bakker SC, van der Meulen EM, Buitelaar JK, Sandkuijl LA, Pauls DL, Monsuur AJ, van ’t Slot R, Minderaa RB, Gunning WB, Pearson PL, Sinke RJ (2003) A whole-genome scan in 164 Dutch sib pairs with attention-deficit/hyperactivity disorder: suggestive evidence for linkage on chromosomes 7p and 15q. Am J Hum Genet 72:1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr CL, Feng Y, Wigg K, Bloom S, Roberts W, Malone M, Schachar R, Tannock R, Kennedy JL (2000) Identification of DNA variants in the SNAP-25 gene and linkage study of these polymorphisms and attention-deficit hyperactivity disorder. Mol Psychiatry 5:405–409 10.1038/sj.mp.4000733 [DOI] [PubMed] [Google Scholar]
- Brophy K, Hawi Z, Kirley A, Fitzgerald M, Gill M (2002) Synaptosomal-associated protein 25 (SNAP-25) and attention deficit hyperactivity disorder (ADHD): evidence of linkage and association in the Irish population. Mol Psychiatry 7:913–917 10.1038/sj.mp.4001092 [DOI] [PubMed] [Google Scholar]
- Cook EH Jr, Stein MA, Krasowski MD, Cox NJ, Olkon DM, Kieffer JE, Leventhal BL (1995) Association of attention-deficit disorder and the dopamine transporter gene. Am J Hum Genet 56:993–998 [PMC free article] [PubMed] [Google Scholar]
- Daly G, Hawi Z, Fitzgerald M, Gill M (1999) Mapping susceptibility loci in attention deficit hyperactivity disorder: preferential transmission of parental alleles at DAT1, DBH and DRD5 to affected children. Mol Psychiatry 4:192–196 10.1038/sj.mp.4000510 [DOI] [PubMed] [Google Scholar]
- Faraone SV, Biederman J, Monuteaux MC (2000) Toward guidelines for pedigree selection in genetic studies of attention deficit hyperactivity disorder. Genet Epidemiol 18:1–16 [DOI] [PubMed] [Google Scholar]
- Faraone SV, Doyle AE (2001) The nature and heritability of attention-deficit/hyperactivity disorder. Child Adolesc Psychiatr Clin N Am 10:299–316 [PubMed] [Google Scholar]
- Faraone SV, Doyle AE, Mick E, Biederman J (2001) Meta-analysis of the association between the 7-repeat allele of the dopamine D(4) receptor gene and attention deficit hyperactivity disorder. Am J Psychiatry 158:1052–1057 10.1176/appi.ajp.158.7.1052 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Francks C, McCracken JT, McGough JJ, Marlow AJ, MacPhie IL, Newbury DF, Crawford LR, Palmer CG, Woodward JA, Del’Homme M, Cantwell DP, Nelson SF, Monaco AP, Smalley SL (2002) A genomewide scan for loci involved in attention-deficit/hyperactivity disorder. Am J Hum Genet 70:1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill M, Daly G, Heron S, Hawi Z, Fitzgerald M (1997) Confirmation of association between attention deficit hyperactivity disorder and a dopamine transporter polymorphism. Mol Psychiatry 2:311–313 10.1038/sj.mp.4000290 [DOI] [PubMed] [Google Scholar]
- Gomez R, Harvey J, Quick C, Scharer I, Harris G (1999) DSM-IV AD/HD: confirmatory factor models, prevalence, and gender and age differences based on parent and teacher ratings of Australian primary school children. J Child Psychol Psychiatry 40:265–274 10.1017/S0021963098003321 [DOI] [PubMed] [Google Scholar]
- Hinds D, Risch N (1996) The ASPEX package: affected sibpair mapping. Available at: http://hpcio.cit.nih.gov/lserver/ASPEX.html. Accessed on 30 July 2004.
- Holmes J, Payton A, Barrett JH, Hever T, Fitzpatrick H, Trumper AL, Harrington R, McGuffin P, Owen M, Ollier W, Worthington J, Thapar A (2000) A family-based and case-control association study of the dopamine D4 receptor gene and dopamine transporter gene in attention deficit hyperactivity disorder. Mol Psychiatry 5:523–530 10.1038/sj.mp.4000751 [DOI] [PubMed] [Google Scholar]
- Holmans P (1993) Asymptotic properties of affected–sib-pair linkage analysis. Am J Hum Genet 52:362–374 [PMC free article] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (2001) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69:570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Kustanovich V, Merriman B, McGough J, McCracken JT, Smalley SL, Nelson SF (2003) Biased paternal transmission of SNAP-25 risk alleles in attention-deficit hyperactivity disorder. Mol Psychiatry 8:309–315 10.1038/sj.mp.4001247 [DOI] [PubMed] [Google Scholar]
- LaHoste GJ, Swanson JM, Wigal SB, Glabe C, Wigal T, King N, Kennedy JL (1996) Dopamine D4 receptor gene polymorphism is associated with attention deficit hyperactivity disorder. Mol Psychiatry 1:121–124 [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 10.1038/ng1195-241 [DOI] [PubMed] [Google Scholar]
- Levy F, Hay DA, McStephen M, Wood C, Waldman I (1997) Attention-deficit hyperactivity disorder: a category or a continuum? genetic analysis of a large-scale twin study. J Am Acad Child Adolesc Psychiatry 36:737–744 10.1097/00004583-199706000-00009 [DOI] [PubMed] [Google Scholar]
- Lowe N, Kirley A, Hawi Z, Sham P, Wickham H, Kratochvil CJ, Smith SD, et al (2004) Joint analysis of the DRD5 marker concludes association with attention-deficit/hyperactivity disorder confined to the predominantly inattentive and combined subtypes. Am J Hum Genet 74:348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor I, Eisenberg J, Tyano S, Sever Y, Cohen H, Ebstein RP, Kotler M (2001) Family-based assocation study of the serotonin transporter promoter region polymorphism (5-HTTLPR) in attention hyperactivity disorder. Am J Med Genet 105:91–95 [DOI] [PubMed] [Google Scholar]
- McCracken JT (1998) Attention-deficit disorder II: neuropsychiatric aspects. In: Brumbach R, Coffey E (eds) Textbook of pediatric neuropsychiatry. American Association Press, New York, pp 483–502 [Google Scholar]
- McCracken JT, Smalley SL, McGough JJ, Crawford L, Del’Homme M, Cantor RM, Liu A, Nelson SF (2000) Evidence for linkage of a tandem duplication polymorphism upstream of the dopamine D4 receptor gene (DRD4) with attention deficit hyperactivity disorder (ADHD). Mol Psychiatry 5:531–536 10.1038/sj.mp.4000770 [DOI] [PubMed] [Google Scholar]
- Mill J, Curran S, Kent L, Richards S, Gould A, Virdee V, Huckett L, Sharp J, Batten C, Fernando S, Simanoff E, Thompson M, Zhao J, Sham P, Taylor E, Asherson P (2001) Attention deficit hyperactivity disorder (ADHD) and the dopamine D4 receptor gene: evidence of association but no linkage in a UK sample. Mol Psychiatry 6:440–444 10.1038/sj.mp.4000881 [DOI] [PubMed] [Google Scholar]
- Nyholt DR (2000) All LODs are not created equal. Am J Hum Genet 67:282–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogdie MN, Macphie IL, Minassian SL, Yang M, Fisher SE, Francks C, Cantor RM, McCracken JT, McGough JJ, Nelson SF, Monaco AP, Smalley SL (2003) A genomewide scan for attention-deficit/hyperactivity disorder in an extended sample: suggestive linkage on 17p11. Am J Hum Genet 72:1268–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CG, Bailey JN, Ramsey C, Cantwell D, Sinsheimer JS, Del’Homme M, McGough J, Woodward JA, Asarnow R, Asarnow J, Nelson S, Smalley SL (1999) No evidence of linkage or linkage disequilibrium between DAT1 and attention deficit hyperactivity disorder in a large sample. Psychiatr Genet 9:157–160 [DOI] [PubMed] [Google Scholar]
- Quist JF, Barr CL, Schachar R, Roberts W, Malone M, Tannock R, Basile VS, Beitchman J, Kennedy JL (2003) The serotonin 5-HT1B receptor gene and attention deficit hyperactivity disorder. Mol Psychiatry 8:98–102 10.1038/sj.mp.4001244 [DOI] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–224 [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Seeger G, Schloss P, Schmidt MH (2001) Functional polymorphism within the promoter of the serotonin transporter gene is associated with severe hyperkinetic disorders. Mol Psychiatry 6:235–238 10.1038/sj.mp.4000820 [DOI] [PubMed] [Google Scholar]
- Smalley SL (1997) Genetic influences in childhood-onset psychiatric disorders: autism and attention-deficit/hyperactivity disorder. Am J Hum Genet 60:1276–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, Bailey JN, Palmer CG, Cantwell DP, McGough JJ, Del’Homme MA, Asarnow JR, Woodward JA, Ramsey C, Nelson SF (1998) Evidence that the dopamine D4 receptor is a susceptibility gene in attention deficit hyperactivity disorder. Mol Psychiatry 3:427–430 10.1038/sj.mp.4000457 [DOI] [PubMed] [Google Scholar]
- Smalley SL, Kustanovich V, Minassian SL, Stone JL, Ogdie MN, McGough JJ, McCracken JT, MacPhie IL, Francks C, Fisher SE, Cantor RM, Monaco AP, Nelson SF (2002) Genetic linkage of attention-deficit/hyperactivity disorder on chromosome 16p13, in a region implicated in autism. Am J Hum Genet 71:959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, McGough JJ, Del’Homme M, NewDelman J, Gordon E, Kim T, Liu A, McCracken JT (2000) Familial clustering of symptoms and disruptive behaviors in multiplex families with attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 39:1135–1143 10.1097/00004583-200009000-00013 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Swanson JM, Sergeant JA, Taylor E, Sonuga-Barke EJ, Jensen PS, Cantwell DP (1998) Attention-deficit hyperactivity disorder and hyperkinetic disorder. Lancet 351:429–433 10.1016/S0140-6736(97)11450-7 [DOI] [PubMed] [Google Scholar]
- Tahir E, Yazgan Y, Cirakoglu B, Ozbay F, Waldman I, Asherson PJ (2000) Association and linkage of DRD4 and DRD5 with attention deficit hyperactivity disorder (ADHD) in a sample of Turkish children. Mol Psychiatry 5:396–404 10.1038/sj.mp.4000744 [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Speer M, Ott J (1993) Chromosome-based method for rapid computer simulation in human genetic linkage analysis. Genet Epidemiol 10:217–224 [DOI] [PubMed] [Google Scholar]
- Wilens TE, Biederman J, Spencer TJ (2002) Attention deficit/hyperactivity disorder across the lifespan. Annu Rev Med 53:113–131 10.1146/annurev.med.53.082901.103945 [DOI] [PubMed] [Google Scholar]
- Wiltshire S, Cardon LR, McCarthy MI (2002) Evaluating the results of genomewide linkage scans of complex traits by locus counting. Am J Hum Genet 71:1175–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolraich ML, Hannah JN, Pinnock TY, Baumgaertel A, Brown J (1996) Comparison of diagnostic criteria for attention-deficit hyperactivity disorder in a county-wide sample. J Am Acad Child Adolesc Psychiatry 35:319–324 10.1097/00004583-199603000-00013 [DOI] [PubMed] [Google Scholar]
- Yonan AL, Alarcon M, Cheng R, Magnusson PK, Spence SJ, Palmer AA, Grunn A, Juo SH, Terwilliger JD, Liu J, Cantor RM, Geschwind DH, Gilliam TC (2003) A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet 73:886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]