Abstract
The MAPT H1 haplotype has been associated with four-repeat (4R) tauopathies, including progressive supranuclear palsy, corticobasal degeneration, and argyrophilic grain disease. More controversial is that the same haplotype has been associated with Parkinson disease (PD). Using H1-specific single-nucleotide polymorphisms, we demonstrate that MAPT H1 is a misnomer and consists of a family of recombining H1 alleles. Population genetics, linkage disequilibrium, and association analyses have shown that specific MAPT H1 subhaplotypes are preferentially associated with Parkinson disease. Using a sliding scale of MAPT H1-specific haplotypes—in age/sex-matched PD cases and controls from central Norway—we have refined the disease association to within an ∼90-kb interval of the 5′ end of the MAPT locus.
Introduction
Tau proteins are a group of microtubule-associated proteins that play an important role in promoting the assembly and maintaining the structure of microtubules. They are expressed in neurons, being particularly abundant in axons (Higuchi et al. 2002). Alternate mRNA splicing of exons 2, 3, and 10 of the tau gene (MAPT) on chromosome 17q21 results in the expression of six polypeptides in the human CNS (Higuchi et al. 2002). The predominant isoforms differ by the presence of either three or four microtubule-binding domains (3-repeat [3R] and 4-repeat [4R] isoforms), which result from the exclusion or inclusion of exon 10 (Panda et al. 2003).
Missense and splicing mutations in MAPT were first identified in frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17 [MIM 600274]) (Hutton 2001). This disorder is called, in pathology terms, a “4R tauopathy” because of the presence of fibrillar aggregates comprising the 4R tau isoform. Susceptibility mutations have yet to be identified for other 4R tauopathies, including progressive supranuclear palsy (PSP [MIM 601104]), corticobasal degeneration, and argyrophilic grains disease; however, these disordersare associated with common polymorphic variability in MAPT (Conrad et al. 1997; Houlden et al. 2001; Togo et al. 2002). There are two predominant MAPT haplotypes—termed “H1” and “H2” and extending >500 kb—in which variants appear to be in complete linkage disequilibrium (LD); H1 and H2 haplotypes do not recombine (Pastor et al. 2002). H1/H1 homozygous genotypes are overrepresented in 4R tauopathies.
More recent and far more controversial is that overrepresentation of MAPT H1/H1 genotypes has been associated with idiopathic Parkinson disease (PD [MIM 168600]) (Golbe et al. 2001; Maraganore et al. 2001; Martin et al. 2001; Farrer et al. 2002). PD is clinically characterized by a combination of motor symptoms, including muscular rigidity, bradykinesia, and resting tremor (Lang and Lozano 1998a, 1998b). In pathology terms, PD is classified as an α-synucleinopathy because of the presence of Lewy body inclusions within surviving neurons (Braak et al. 2003).
Since our initial report (Farrer et al. 2002) of a MAPT H1/H1 association with PD (odds ratio [OR] for H1/H1 vs. H1/H2 and H2/H2 is 5.52; 95% CI 2.64–11.10; P<2.1e-6; n=96 cases + 68 controls), we have been collecting additional cases and controls to replicate and refine the evidence for a MAPT H1 association with PD. Studying the homogeneous population of central Norway and H1-specific SNPs, we now demonstrate: (1) evidence of recombination in controls with MAPT H1/H1 genotypes; (2) unusual patterns of LD in PD cases; (3) H1-SNP multilocus association with PD; and (4) that the region associated with PD resides within an ∼90-kb interval of the 5′ end of MAPT. Specific MAPT H1 subhaplotypes are overrepresented in patients with PD and accounts for the magnitude of the H1/H1 association observed in past studies.
Material and Methods
DNA samples and clinical information was available from 296 unrelated Norwegian patients with PD (mean age at onset 59 ± 8 years; mean present age 74 ± 8 years); these were sequential new referrals to the Department of Neurology, University of Trondheim, Norway, between May 1998 and June 2002. All subjects were examined using standardized clinical protocols, including the Unified Parkinson’s Disease Rating Scale and the Mini–Mental State Exam, by a neurologist specialized in movement disorders (J.A.) (Fahn et al. 1987, 1993). Cases with possible PD showed at least two of four cardinal signs (bradykinesia, rigidity, rest tremor, and asymmetric onset). All cases with probable PD showed at least three of four cardinal signs (“definite PD” is a term reserved for autopsy-confirmed cases) and were levodopa responsive. Subjects with minimal or no improvement from levodopa (in combination with carbidopa) at a dosage of ⩾750 mg/d were considered atypical. Patients with other causes of parkinsonism or with unexplained signs of more extensive neurologic involvement were excluded (dementia or mild dysautonomia were allowed if they occurred after the first year of motor symptoms). Our criteria are consistent with those validated elsewhere (Gelb et al. 1999). Unrelated Norwegian controls (mean present age 80 ± 6 years) were volunteers recruited from the Department of Ophthalmology and the Department of Internal Medicine, University of Trondheim, and from the local blood bank. Subjects were recruited from within a 200-mile radius of Trondheim, central Norway, and >80% of them were from within 50 miles of the city. All subjects have Norwegian ancestry dating back >4 generations. Appropriate institutional-review approval and informed consent were obtained. MAPT genotyping in a subset of 96 PD cases and 68 controls has been described elsewhere (Farrer et al. 2002).
Prior analysis from our Norwegian cohort suggests that, on average, “useful” LD (r>0.1) can be detected with as little as 1 SNP/40 kb (unpublished data); our objective was to identify 1 H1-SNP/10 kb of genomic sequence. SNPs were identified in public and Celera databases, and the genotyping of putative variants in H1/H1 and H2/H2 control individuals (n=11+11) demonstrated whether the SNPs were polymorphic and H1 specific. H1-SNPs located within the nonrecombining region are polymorphic in a population of H1 chromosomes; by default, H1-SNPs do not exist on H2 chromosomes. Direct sequencing was used to identify additional H1-SNPs within the 5′ end of the MAPT locus. Approximately 5 kb of resequencing was performedin evolutionarily conserved regions of human-mousesequence identity, highlighted by mVISTA analysis (Mayor et al. 2000). Sequence traces were assessed in 12 Norwegian individuals (6 PD cases and 6 controls) homozygous for the MAPT H1 intron 9 insertion/deletion (in/del) (Baker et al. 1999). Gene Runner v.3.05 (Hastings Software) was used for all primer design; genotyping and sequencing were done with conventional techniques (Farrer et al. 2002; West et al. 2003). All SNPs used in our study have been submitted to dbSNP (tables 1 and 2).
Table 1.
MAPT H1-SNPs
| Assay Type, ID, SNP, and Primers (5′→3′)a | Alleles | Fragments(bp) | Product(bp) | Enzyme |
| Restriction enzyme digest: | ||||
| 8: | ||||
| rs242937: | ||||
| F: ACCCACAGACCACGACCTTCCAAC | G | 279, 192 | ||
| R: CCTGCCTCTTTCTGCCCATTGG | A | 471 | 471 | MnlI |
| 9: | ||||
| rs242935: | ||||
| F: GGGCCACTGGATCACAAGGTTG | T | 258, 181, 59, 30 | ||
| R: CCGGCCCATAATCTGCATTTCTAAC | C | 288, 181, 59 | 528 | Tsp509I |
| 10: | ||||
| rs242928: | ||||
| F: TTAAGGAAGCACCCATGACAGCC | A | 266, 144, 51 | ||
| R: AAACAGTTCTGTGGAATTTCACCCTG | G | 410, 51 | 461 | MboII |
| 12: | ||||
| rs16339368: | ||||
| F: GGTAGAGGCCAGGAATGCTGTTAAAC | C | 277, 202 | ||
| R: GGTCATGCTCCGATTACAGACTCTTG | A | 479 | 479 | AciI |
| 1: | ||||
| rs242562: | ||||
| F: CAGCCTTCCCTGTCCTTGATTC | G | 385 | ||
| R: GCCTTCCCAACAGAGCAACC | A | 287, 98 | 385 | XhoI |
| 2: | ||||
| rs2435207: | ||||
| F: AGCAAGCTGTGTGACCAG | G | 197, 41 | ||
| R: CCCATTCTCTGACAGATTTG | A | 112, 85, 41 | 238 | BclI |
| 14: | ||||
| rs2258689: | ||||
| F: AGACATCCACACGTTCCTC | C | 132, 107, 9 | ||
| R: CAAACCACAGCAGAGCAG | T | 239, 9 | 248 | AflIII |
| 3: | ||||
| rs16339369: | ||||
| F: CGAGTCCTGGCTTCACTCC | G | 257, 54, 26, 20, 6 | ||
| R: CTTCCAGGCACAGCCATACC | A | 201, 56, 54, 26, 20, 6 | 370 | BstNI |
| 4: | ||||
| rs16339370: | ||||
| F: GGCTGGCCCTGCTCCTTCTCTA | T | 247, 105 | ||
| R: TGGCAAGGACGTTGGGGGACAGGG | C | 352 | 352 | TaiI |
| 5: | ||||
| rs16339371: | ||||
| F: GACTGATAGGTGGGAGGTGGCTGC | CT | 228, 226 | ||
| R: CAGCAGCTCGGACGTGAG | AA | 454 | 454 | PvuII |
| SNaPshot: | ||||
| 6: | ||||
| rs110402: | ||||
| F: GTGCACTCTGTACACTCACTGGACC | C | … | ||
| R: GTATGATTCAGGAATAAGGCAGAAGC | T | … | 483 | … |
| S: CACAGAGGACTGGTGTTGC | … | … | … | … |
| 7: | ||||
| rs171440: | ||||
| F: CTGCACAGAACAAAGTACACGTGAC | G | … | ||
| R: TCCTATGCAAAGAAGACACAAGGG | A | … | 433 | … |
| Sb: aactgaGCGAGGGACCAAGAGAAG | … | … | … | … |
| 11: | ||||
| rs2019820: | ||||
| F: GGTCATCTCTAGTGGGCATTAACACG | C | … | ||
| R: TGACAAAGGCAAGAGTACACAAAGGG | T | … | 381 | … |
| Sb: aactCCAGGCTGTTCTCGAACT | … | … | … | … |
| 13: | ||||
| rs3785883: | ||||
| F: CCATCACCTTGTCAGAAACTC | G | … | ||
| R: AGCCATGTGGTAGCCTCAG | A | … | 277 | … |
| Sb: aactgactaaCACTGTCACCACTGGGC | … | … | … | … |
F = forward; R = reverse; S = sequencing.
Sequencing primer plus nonannealing sequence.
Table 2.
SNPs to Determine Extent of the MAPT “Nonrecombining” H2 Haplotype
| SNP and Primers (5′→3′)a | Alleles | Fragments(bp) | Product (bp) | Enzyme | Position |
| Restriction enzyme digest: | |||||
| rs878886: | |||||
| F: GTTAGGTCTCATGCCCACTCCC | G | 225, 126 | |||
| R: GAGTCAGAGGCTGTCACGAGTTG | C | 145, 126, 80 | 351 | BanII | … |
| Sequencing (CRHR1): | |||||
| rs8072451: | |||||
| F: GTTGAGTGTATAGCAGGCCTCCTAAC | G | … | |||
| R: AGGTGGAGGTCACAGTGAGCTG | A | … | 517 | … | Exon 2 −114 bp |
| rs3418: | |||||
| F: AATCCACCTTCTCTCTCTCACAAACC | C | … | |||
| R: TACATCATCTTGCTCGCCTCAGG | T | … | 865 | … | Exon 8 +196 bp |
F = forward; R = reverse.
Tests of departure from Hardy-Weinberg equilibrium were done for each SNP, by use of Arlequin for population genetic analysis (Schneider et al. 1997). Single-marker tests for association were performed with a χ2 test, by use of CLUMP for allele and genotype frequencies (Sham and Curtis 1995). ORs for each genotypic variant were calculated (11 and 12 vs. 22; 11 vs. 12 and 22) using SPSS, release 10.0.0 (SPSS Inc.). Expectation-maximization methods were used to calculate bilocus and multilocus haplotypes and their frequencies. P values were estimated by a Monte Carlo approach with 10,000 simulations. Pairwise measures of LD (D, D′, and r2) were calculated using the Excel macro Genotype Transposer (Cox and Canzian 2001) and were viewed using GOLD (Abecasis and Cookson 2000). The LDMAP program was used to construct LD maps based on the physical positions of H1-SNPs (Maniatis et al. 2002).
Results
Our replication cohort includes 200 cases (median age 70 years; range 40–94 years; male frequency 0.59) and 373 controls (median age 68 years; range 50–93 years; male frequency 0.61) in which the association remains significant (OR=2.3; 95% CI 1.6–3.4; P<1.4e-5). Combined, the data sets also demonstrate that MAPT H1/H1 genotypes are significantly overrepresented in Norwegian cases of PD versus in controls (OR=1.86; 95% CI 1.3–2.6; P<2e-4; n=296 cases + 441 controls) (table 3). Diagnostic stratification of probable, possible, and atypical PD cases does not appreciably alter the OR observed, suggesting that the overrepresentation of H1 homozygotes is unlikely to be explained by misdiagnosis of PSP as PD (Schrag et al. 1999).
Table 3.
MAPT Haplotypes in a Large Community-Based PD Case-Control Series[Note]
|
No. of Genotypes (%) |
H1/H1 vs. H1/H2 and H2/H2 |
H2/H2 vs. H1/H1 and H1/H2 |
||||||||
| Sample | n | H1/H1 | H1/H2 | H2/H2 | OR | 95% CI | P | OR | 95% CI | P |
| Controls | 441 | 282 (63.9) | 143 (32.4) | 16 (3.6) | 1.0 | Reference | … | 1.0 | Reference | … |
| PD cases: | ||||||||||
| Alla | 296 | 227 (76.7) | 62 (20.9) | 7 (2.4) | 1.86 | 1.33–2.59 | .0002 | .64 | .26–1.58 | .33 |
| Probable and possible | 280 | 213 (76.1) | 60 (21.4) | 7 (2.5) | 1.79 | 1.28–2.51 | .001 | .68 | .28–1.68 | .40 |
| Probable | 223 | 169 (75.8) | 50 (22.4) | 4 (1.8) | 1.77 | 1.23–2.54 | .002 | .49 | .16–1.47 | .19 |
Note.— Adjustment for age/sex by logistic regression was not significant.
Includes all probable, possible, and atypical PD cases.
Microsatellite variability within MAPT suggests that the H1 haplotype may be partitioned into H1-specific subhaplotypes (Golbe et al. 2001). Given this assumption, we hypothesized that higher-resolution genetic mapping of MAPT disease association may be feasible by dissecting the genetic architecture of H1 subhaplotypes. mVISTA analysis was used to compare human and mouse MAPT loci (Genbank accession numbers AC091628 and AC091629, respectively) and to highlight conserved regions with >75% sequence identity, which are perhaps functionally important (Mayor et al. 2000). Amplicon sequencing was subsequently prioritized in 12 individuals with H1/H1 genotypes (defined using the intron 9 in/del [Baker et al. 1999]), in an attempt to discover H1-specific variability, herein denoted “H1-SNPs.” SNP and H1-SNP identification numbers, primers (5′→3′), and assay details are given in tables 1 and 2.
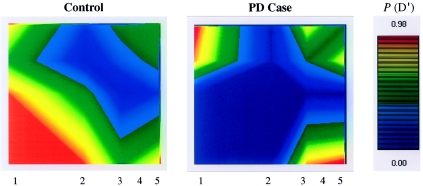
A matched subset of 81 probable PD cases and 81 controls, all homozygous for H1 (defined using the intron 9 in/del), were selected for H1-SNP genotyping, LD, and haplotype-association analyses. The median age was 76 ± 3.9 years (range 71–86 years) in the case group and 81 ± 5.1 years (range 72–93 years) in the control group. Initially, pairwise LD statistics were generated for five MAPT H1-SNPs (fig. 1).
Figure 1.
Graphical overview of LD within the MAPT locus. PD cases and controls each consist of matched groups of 81 individuals homozygous for H1/H1 genotypes (as defined by the intron 9 in/del [Baker et al. 1999]). The relative positions of H1-SNPs are numbered on both axes, and all pairwise measures of D′ are considered. The relative positions of H1-SNPs are the same on both axes, of both plots, and as numbered for each abscissa; all pairwise measures of D′ are considered.
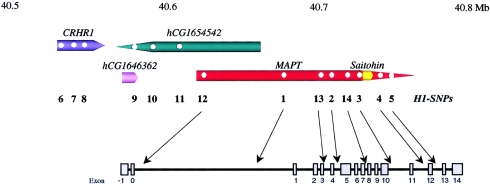
LD between H1-SNPs 1–4 was considerably greater in PD cases than in controls, and, in the former, the LD appears to extend further 5′ of MAPT. Genotyping additional SNPs in H1/H1, H1/H2, and H2/H2 control individuals (n=15) showed that the nonrecombining region between H1 and H2 haplotypes extends at least 100 kb 5′ of MAPT, including the corticotrophinreleasing hormone receptor locus (CRHR1). Hence,additional H1-SNPs were sought within the CRHR1-MAPT interval, and a further nine were identified and genotyped within matched Norwegian cases of PD (n=81) and controls (n=81). Relative H1-SNP positions are illustrated on an ideogram of genes in the region (fig. 2).
Figure 2.
Ideogram of the CRHR1-MAPT region. Physical distances are given in Mb; gene assignments are oriented 5′→3′ with respect to their promoters (top). H1-SNPs are indicated by circles within genes and by numbers beneath, and their relative positions within MAPT are indicated (center). The relative positions of MAPT exons are also shown but are not to scale (bottom). Note, 4 denotes exon 4 and exon 4A.
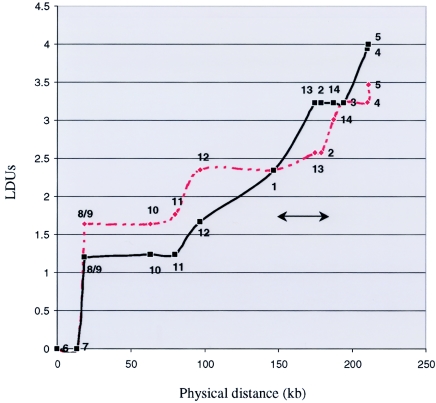
SNPtagger was used to identify the minimum set—9 of the 14 H1-SNPs—required to capture 100% of the haplotype diversity within the CRHR1-MAPT region (Ke and Cardon 2003). For these “haplotype-tagging” H1-SNPs, multilocus-haplotype frequencies were compared between PD cases and controls (Johnson et al. 2001). A total of 17 different multilocus haplotypes have frequencies of ⩾2% and account for >74% of the haplotypes estimated in the case and control groups combined (table 4). Of note, there are six H1 subhaplotypes in the PD case group that are not represented in the control group (XII–XVII), and seven haplotypes in the control group are not present in PD cases (V–XI). The comparison of multilocus haplotype counts between patients with PD and controls was highly significant: χ2=78.8; P<1.0e-6 (16 df). Thus, the MAPT H1 haplotype term is a misnomer; H1 actually represents a clade of haplotypes on the same H1 backbone (e.g., H1I, H1II, etc.) that are not H2. We postulated that one or more H1 subhaplotypes harbor sequence variability associated with disease. We attempted to use PHYLIP to model the evolutionary history of H1 subhaplotypes; however, multiple solutions were possible that fit the data equally well (data not shown). Nevertheless, results are indicative of ancestral recombination between H1 subhaplotypes. This is more formally demonstrated in terms of the LD map-unit profile across MAPT H1. Regions of low LD (indicative of recombination) appear as sloping “steps,” and regions of high LD (limited haplotypic diversity) appear as horizontal “plateaus” (Tapper et al. 2003) (fig. 3).
Table 4.
MAPTH1 Subhaplotype Frequencies in PD Cases and Controls
|
Frequency (n) among |
||
| H1 Subhaplotypea(Allelic Conformation) | Controls | PD Cases |
| I (GGTACGCGT) | .16 (26) | .22 (35) |
| II (GGCGTGTGT) | .09 (14) | .08 (13) |
| III (GGTACGTGT) | .06 (9) | .04 (7) |
| IV (AGTACGCGT) | .05 (8) | .06 (10) |
| V (AACACATGC) | .05 (8) | … |
| VI (AACATGCAT) | .04 (7) | … |
| VII (GGCACATGC) | .04 (6) | … |
| VIII (GGTATGCAC) | .03 (5) | … |
| IX (GGCACGCGT) | .03 (5) | … |
| X (GGTATGCAT) | .03 (5) | … |
| XI (GGCGTGCGT) | .02 (3) | … |
| XII (AACGTGTGT) | … | .08 (12) |
| XIII (GGCACACGC) | … | .04 (6) |
| XIV (AACACATGT) | … | .03 (5) |
| XV (GGCATGCGT) | … | .03 (4) |
| XVI (GGCACGTGT) | … | .02 (4) |
| XVII (GGCATATAC) | … | .02 (3) |
H1 subhaplotypes are comprised of “haplotype tagging” H1-SNPs 7-8-9-10-11-1-14-3-4.
Figure 3.
LD units are shown with respect to the physical position of H1-SNPs across the CRHR1-MAPT loci (Maniatis et al. 2002). H1-SNPs 1–14 are indicated on each curve. Probable PD cases (red dashed line) and controls (black solid line) each consist of matched groups of 81 individuals homozygous for H1/H1 (as defined by the intron 9 in/del [Baker et al. 1999]). The H1 subhaplotype overrepresented in patients with PD, as defined by H1-SNPs 1 and 2, is indicated (bar with arrows).
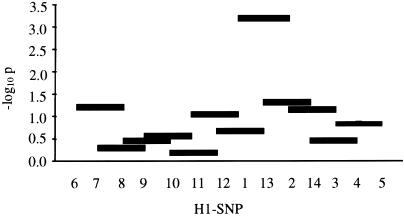
To refine the genomic region within CRHR1-MAPT that contributes the most to disease association, we compared haplotypes for smaller subsets of adjacent markers, sliding across all 14 H1-SNPs. χ2 analyses of 2-, 3-, and 4-marker haplotype counts in matched PD cases and controls (n=162) were performed using CLUMP (Sham and Curtis 1995). For illustration, the −log10P values for haplotype trios are shown (fig. 4).
Figure 4.
−log10 simulated P values are shown for counts of each haplotype trio compared between PD cases and controls. Empirical and simulated (10,000 Monte Carlo–Markov Chain iterations) P values were equivalent.
The number of H1 subhaplotypes containing markers 1, 13, and 2 is strikingly different between probable PD cases and controls (-logP<3.22; P<.0006). H1-SNPs 1, 13, and 2 were relatively informative, with minor allele frequencies of 0.40, 0.26, and 0.36 (within H1/H1 controls), respectively. Inspection of all possible pairwise combinations shows that the “A-A” haplotype, denoted by H1-SNPs 1 and 2, is the most significantly associated with disease (P<.003) (table 5). Significant LD exists between these markers in cases (PD′ < .00002) compared with in controls (PD′ > .9). These findings remain significant when other H1/H1 PD cases (possible and probable PD, n=201; probable PD, n=83) and H1/H1 controls are considered (n=278; P<.02); the distribution of haplotypes (possible and probable PD cases, probable PD cases, controls) is G-G (0.35, 0.38, 0.44), A-G (0.17, 0.18, 0.14), G-A (0.17, 0.17, 0.23), and A-A (0.31, 0.27, 0.19). Figure 4 highlights the position of H1-SNPs 1 and 2 within the MAPT gene.
Table 5.
Assessment of Bilocus Haplotypes (H1-SNP 1–2)
|
No. with Haplotype (Frequency)a |
||||
| Sample | G-G | A-G | G-A | A-A |
| PD cases | 63 (.39) | 22 (.13) | 31 (.19) | 46 (.29) |
| Controls | 61 (.38) | 42 (.26) | 36 (.22) | 23 (.14) |
Total number of PD cases and controls is 162.
Discussion
Our work has focused on the relatively homogeneous population of Trondheim, Norway, since isolated populations are generally more powerful for LD and disease-association mapping. In conclusion, we replicated and extended the association of MAPT H1 variability with PD in Norway (Farrer et al. 2002). The H1 haplotype extends 5′ of MAPT and includes the neighboring gene, CRHR1, a region in which LD is considerable in patients with PD. Given that PD typically affects individuals beyond their reproductive lifespan, this result was surprising. It was intriguing that the oldest and most extensively studied patients with probable PD and controls showed greatest evidence for MAPT H1 association, suggesting that the role of tau in PD is influenced by age. By genotyping H1-SNPs within the CRHR1-MAPT interval, we explored the hypothesis that genetic variability in a neighboring gene may be responsible for disease association (genetic “hitchhiking”); CRHR1 is a good candidate, since it is important in both nervous and immune systems (Webster et al. 1998). However, the H1 subhaplotype associated with PD maps to a genomic interval of ∼90 kb that contains MAPT exons 1–4.
Given the past literature on extended “nonrecombining” MAPT haplotypes, it is ironic that this assignment was facilitated by the genetic architecture of H1 subhaplotypes and the recombination between them. We focused on high-resolution analysis of H1 alleles, since they are associated with disease. One explanation for the lack of H1 and H2 recombination and for inconsistencies and/or marker duplication in the 17q21 physical map is that H2 represents a paracentric inversion. This hypothesis is not inconsistent with the evolution of syntenic primate chromosomes (Kehrer-Sawatzki et al. 2002) and requires that the ends of the nonrecombining H2 haplotype be defined.
Unifying hypotheses have been constructed to explain how genetic variability in MAPT H1 may be associated with PD, an α-synucleinopathy, and with 4R tauopathies (de Silva and Farrer 2002). Of note, aberrant splicing of MAPT exon 10 and its subsequent inclusion/translation are responsible for the overproduction of 4R tau in many familial cases of FTDP-17 (Hutton 2001). In contrast, isoforms of tau without exons 2–3 have less propensity to polymerize in vitro (King et al. 2000). Given our discovery that the MAPT association in PD includes exons 1–4, we propose that the PD-associated haplotype harbors genetic variability that influences splicing of exons 2–3. Of note, population genetics exploits ancestral recombination events and may facilitate high-resolution mapping of genes and sequence variability, but LD cannot be used to bound an interval associated with disease (this is in contrast to lowerresolution linkage mapping within families, in which obligate recombinants are used to define a candidate region). Hence, variability 5′ of exon 1, within the promoter, or 3′ of exon 4 should not be ruled out. In PSP, a similar mechanism may be conceived, albeit one owing to different H1-sequence variability. Both MAPT gene splicing and expression may be important.
A useful analogy is the genetics of amyloid precursor protein—mutations in the gene may lead to simple overexpression (Prasher et al. 1998) or may directly affect protease cleavage and the production of amyloidogenic Aβ40/42 peptides, and either mechanism may predispose to Alzheimer disease (Sambamurti et al. 2002). Functional studies of MAPT promoter function and splicing of exons 2–3 are now needed.
Acknowledgments
We thank the participants; Dr. Corrinne Aasly, for recruitment of control subjects; and Minnie Schreiber, for technical assistance. M.F. and the study were funded by the Society for Progressive Supranuclear Palsy, the Norwegian Parkinson’s Association, and Reberg’s Legacy. O.A.R. was a Visiting Fellow funded by the R&D Office, Health and Personal Social Services, Northern Ireland.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Celera, http://publication.celera.com/humanpub/index.jsp
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/ (for SNP IDs and rs numbers)
- Genbank, http://www.ncbi.nlm.nih.gov/Genbank/ (for MAPT mouse [accession number AC091629] and MAPT human [accession number AC091628])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FTDP-17, PSP, and PD)
- PHYLIP, http://evolution.genetics.washington.edu/phylip.html
- SNPtagger, http://www.well.ox.ac.uk/~xiayi/haplotype/
References
- Abecasis GR, Cookson WO (2000) GOLD: graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 10.1093/bioinformatics/16.2.182 [DOI] [PubMed] [Google Scholar]
- Baker M, Litvan I, Houlden H, Adamson J, Dickson D, Perez-Tur J, Hardy J, Lynch T, Bigio E, Hutton M (1999) Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet 8:711–715 10.1093/hmg/8.4.711 [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E (2003) Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol Aging 24:197–211 10.1016/S0197-4580(02)00065-9 [DOI] [PubMed] [Google Scholar]
- Conrad C, Andreadis A, Trojanowski JQ, Dickson DW, Kang D, Chen X, Wiederholt W, Hansen L, Masliah E, Thal LJ, Katzman R, Xia Y, Saitoh T (1997) Genetic evidence for the involvement of tau in progressive supranuclear palsy. Ann Neurol 41:277–281 [DOI] [PubMed] [Google Scholar]
- Cox DG, Canzian F (2001) Genotype transposer: automated genotype manipulation for linkage disequilibrium analysis. Bioinformatics 17:738–739 10.1093/bioinformatics/17.8.738 [DOI] [PubMed] [Google Scholar]
- de Silva R, Farrer M (2002) Tau neurotoxicity without the lesions: a fly challenges a tangled web. Trends Neurosci 25:327–329 10.1016/S0166-2236(02)02170-7 [DOI] [PubMed] [Google Scholar]
- Fahn S, Elton R, Members of the UPDRS Development Committee (1987) Recent developments in Parkinson’s disease. Macmillan, New York [Google Scholar]
- Fahn S, Tolosa E, Marin C (1993) Recent developments in Parkinson’s disease. Williams and Wilkins, Baltimore [Google Scholar]
- Farrer M, Skipper L, Berg M, Bisceglio G, Hanson M, Hardy J, Adam A, Gwinn-Hardy K, Aasly J (2002) The tau H1 haplotype is associated with Parkinson’s disease in the Norwegian population. Neurosci Lett 322:83–86 10.1016/S0304-3940(02)00106-4 [DOI] [PubMed] [Google Scholar]
- Gelb DJ, Oliver E, Gilman S (1999) Diagnostic criteria for Parkinson disease. Arch Neurol 56:33–39 10.1001/archneur.56.1.33 [DOI] [PubMed] [Google Scholar]
- Golbe LI, Lazzarini AM, Spychala JR, Johnson WG, Stenroos ES, Mark MH, Sage JI (2001) The tau A0 allele in Parkinson’s disease. Mov Disord 16:442–447 10.1002/mds.1087 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Lee VM, Trojanowski JQ (2002) Tau and axonopathy in neurodegenerative disorders. Neuromolecular Med 2:131–150 10.1385/NMM:2:2:131 [DOI] [PubMed] [Google Scholar]
- Houlden H, Baker M, Morris HR, MacDonald N, Pickering-Brown S, Adamson J, Lees AJ, Rossor MN, Quinn NP, Kertesz A, Khan MN, Hardy J, Lantos PL, St George-Hyslop P, Munoz DG, Mann D, Lang AE, Bergeron C, Bigio EH, Litvan I, Bhatia KP, Dickson D, Wood NW, Hutton M (2001) Corticobasal degeneration and progressive supranuclear palsy share a common tau haplotype. Neurology 56:1702–1706 [DOI] [PubMed] [Google Scholar]
- Hutton M (2001) Missense and splice site mutations in tau associated with FTDP-17: multiple pathogenic mechanisms. Neurology 56:S21–S25 [DOI] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29:233–237 10.1038/ng1001-233 [DOI] [PubMed] [Google Scholar]
- Ke X, Cardon LR (2003) Efficient selective screening of haplotype tag SNPs. Bioinformatics 19:287–288 10.1093/bioinformatics/19.2.287 [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Schreiner B, Tänzer S, Platzer M, Müller S, Hameister H (2002) Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am J Hum Genet 71:375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King ME, Gamblin TC, Kuret J, Binder LI (2000) Differential assembly of human tau isoforms in the presence of arachidonic acid. J Neurochem 74:1749–1757 10.1046/j.1471-4159.2000.0741749.x [DOI] [PubMed] [Google Scholar]
- Lang AE, Lozano AM (1998a) Parkinson’s disease: first of two parts. N Engl J Med 339:1044–1053 10.1056/NEJM199810083391506 [DOI] [PubMed] [Google Scholar]
- ——— (1998b) Parkinson’s disease: second of two parts. N Engl J Med 339:1130–1143 10.1056/NEJM199810153391607 [DOI] [PubMed] [Google Scholar]
- Maniatis N, Collins A, Xu CF, McCarthy LC, Hewett DR, Tapper W, Ennis S, Ke X, Morton NE (2002) The first linkage disequilibrium (LD) maps: delineation of hot and cold blocks by diplotype analysis. Proc Natl Acad Sci USA 99:2228–2233 10.1073/pnas.042680999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraganore DM, Hernandez DG, Singleton AB, Farrer MJ, McDonnell SK, Hutton ML, Hardy JA, Rocca WA (2001) Case-control study of the extended tau gene haplotype in Parkinson’s disease. Ann Neurol 50:658–661 10.1002/ana.1228 [DOI] [PubMed] [Google Scholar]
- Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, et al (2001) Association of single-nucleotide polymorphisms of the tau gene with late-onset Parkinson disease. Jama 286:2245–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047 10.1093/bioinformatics/16.11.1046 [DOI] [PubMed] [Google Scholar]
- Panda D, Samuel JC, Massie M, Feinstein SC, Wilson L (2003) Differential regulation of microtubule dynamics by three- and four-repeat tau: implications for the onset of neurodegenerative disease. Proc Natl Acad Sci USA 100:9548–9553 10.1073/pnas.1633508100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor P, Ezquerra M, Tolosa E, Munoz E, Marti MJ, Valldeoriola F, Molinuevo JL, Calopa M, Oliva R (2002) Further extension of the H1 haplotype associated with progressive supranuclear palsy. Mov Disord 17:550–556 10.1002/mds.10076 [DOI] [PubMed] [Google Scholar]
- Prasher VP, Farrer MJ, Kessling AM, Fisher EM, West RJ, Barber PC, Butler AC (1998) Molecular mapping of Alzheimer-type dementia in Down’s syndrome. Ann Neurol 43:380–383 [DOI] [PubMed] [Google Scholar]
- Sambamurti K, Greig NH, Lahiri DK (2002) Advances in the cellular and molecular biology of the beta-amyloid protein in Alzheimer’s disease. Neuromolecular Med 1:1–31 10.1385/NMM:1:1:1 [DOI] [PubMed] [Google Scholar]
- Schneider S, Kueffer J-M, Roessli D, Excoffier L (1997) Arlequin v.1.1: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland [Google Scholar]
- Schrag A, Ben-Shlomo Y, Quinn NP (1999) Prevalence of progressive supranuclear palsy and multiple system atrophy: a cross-sectional study. Lancet 354:1771–1775 10.1016/S0140-6736(99)04137-9 [DOI] [PubMed] [Google Scholar]
- Sham PC, Curtis D (1995) Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet 59:97–105 [DOI] [PubMed] [Google Scholar]
- Tapper WJ, Maniatis N, Morton NE, Collins A (2003) A metric linkage disequilibrium map of a human chromosome. Ann Hum Genet 67:487–494 10.1046/j.1469-1809.2003.00050.x [DOI] [PubMed] [Google Scholar]
- Togo T, Sahara N, Yen SH, Cookson N, Ishizawa T, Hutton M, de Silva R, Lees A, Dickson DW (2002) Argyrophilic grain disease is a sporadic 4-repeat tauopathy. J Neuropathol Exp Neurol 61:547–556 [DOI] [PubMed] [Google Scholar]
- Webster EL, Torpy DJ, Elenkov IJ, Chrousos GP (1998) Corticotropin-releasing hormone and inflammation. Ann NY Acad Sci 840:21–32 [DOI] [PubMed] [Google Scholar]
- West A, Lockhart PJ, O’Farrell C, Farrer MJ (2003) Identification of a novel gene linked to parkin via a bi-directional promoter. J Mol Biol 326:11–19 10.1016/S0022-2836(02)01376-1 [DOI] [PubMed] [Google Scholar]