Abstract
Miscarriage is a condition that affects 10%–15% of all clinically recognized pregnancies, most of which occur in the first trimester. Approximately 50% of first-trimester miscarriages result from fetal chromosome abnormalities. Currently, G-banded chromosome analysis is used to determine if large-scale genetic imbalances are the cause of these pregnancy losses. This technique relies on the culture of cells derived from the fetus, a technique that has many limitations, including a high rate of culture failure, maternal overgrowth of fetal cells, and poor chromosome morphology. Comparative genomic hybridization (CGH)–array analysis is a powerful new molecular cytogenetic technique that allows genomewide analysis of DNA copy number. By hybridizing patient DNA and normal reference DNA to arrays of genomic clones, unbalanced gains or losses of genetic material across the genome can be detected. In this study, 41 product-of-conception (POC) samples, which were previously analyzed by G-banding, were tested using CGH arrays to determine not only if the array could identify all reported abnormalities, but also whether any previously undetected genomic imbalances would be discovered. The array methodology detected all abnormalities as reported by G-banding analysis and revealed new abnormalities in 4/41 (9.8%) cases. Of those, one trisomy 21 POC was also mosaic for trisomy 20, one had a duplication of the 10q telomere region, one had an interstitial deletion of chromosome 9p, and the fourth had an interstitial duplication of the Prader-Willi/Angelman syndrome region on chromosome 15q, which, if maternally inherited, has been implicated in autism. This retrospective study demonstrates that the DNA-based CGH-array technology overcomes many of the limitations of routine cytogenetic analysis of POC samples while enhancing the detection of fetal chromosome aberrations.
Introduction
Approximately 10%–15% of all clinically recognized pregnancies end in miscarriage, most of which occur in the first trimester. Of these first-trimester miscarriages, ∼50% are due to fetal chromosome abnormalities (Hassold et al. 1980), the majority of which (86%) arise from numerical abnormalities, including trisomies, monosomies, and polyploidies (triploidy or tetraploidy). Structural abnormalities account for 6% of such losses, and other abnormalities, such as single-gene mutations and mosaicism, account for 8% (Goddijn and Leschot 2000).
Cytogenetic analysis of tissue from spontaneous abortions provides valuable insights into the cause of miscarriage, which can eliminate further costly testing. In addition, recurrence-risk estimates for subsequent pregnancies can also be determined. Routine cytogenetic analysis relies on the successful culture of fetal tissue and preparation of metaphase cells, a well-established methodology in clinical cytogenetics laboratories. However, diagnosis in product of conception (POC) samples is often hindered by a relatively high (10%–40%) rate of tissue-culture failure (Lomax et al. 2000) and the suboptimal quality of chromosome preparations. In addition, selective overgrowth of maternally derived cells can occur, thereby erroneously yielding a normal karyotype despite possible underlying fetal chromosomal abnormalities (Bell et al. 1999).
Comparative genomic hybridization (CGH) is a molecular cytogenetic technique in which fluorescently labeled patient and control whole-genomic DNA is hybridized to normal metaphase slides. Differential hybridization signals allow the detection of unbalanced gains and losses of chromosomal material across the whole genome at a resolution of ∼3–10 Mb (Kallioniemi et al. 1992; Kirchhoff et al. 2001). Recent studies have shown that CGH analysis of DNA extracted from uncultured or paraffin-embedded fetal tissue provides an effective alternative for the detection of fetal chromosome anomalies (Daniely et al. 1998; Bell et al. 2001; Fritz et al. 2001; Tabet et al. 2001). Many of the limitations of routine G-banding analysis, including cell-culture failure and poor chromosome morphology, are circumvented by the use of genomic DNA. However, this technique is limited by the inability to identify balanced translocations, triploidies, and tetraploidies. Since triploidies and tetraploidies are common findings in POC samples, one group overcame this limitation by subsequently using flow cytometry on samples for which CGH analysis yielded a normal result (Lomax et al. 2000).
A new molecular cytogenetic technique, CGH-array analysis, has recently been developed. Although based on the same principle as conventional CGH, array CGH differs in that genomic clones from selected regions of the genome replace the normal “control” metaphase cells as the target DNA (Pinkel et al. 1998; Snijders et al. 2001). Since genomic clones are used as the target DNA, the resolution of the technique is theoretically increased to the size of an individual clone, depending on its size and spacing. Therefore, rearrangements and deletions—that are not visible by either routine G-banding analysis or conventional CGH methods—can be detected.
CGH arrays have been successfully utilized to identify amplifications and deletions in multiple cancer types (Hodgson et al. 2001; Cai et al. 2002; Paris et al. 2003; Schwaenen et al. 2003), and their use in studying constitutional rearrangements is increasing (Veltman et al. 2002; Gunn et al. 2003; Ki et al. 2003; reviewed by Albertson and Pinkel [2003] and Vissers et al. [2003]). In this study, DNA arrays containing genomic clones for every telomere, as well as clones for all of the microdeletion syndromes and additional selected loci spanning the genome, have been used to analyze 41 previously karyotyped POC samples. The ability of array CGH to not only verify all abnormalities found by G-banding analysis but also to identify additional submicroscopic rearrangements was retrospectively assessed to validate the utility of array CGH in analyzing POC samples in the clinical cytogenetics setting.
Material and Methods
Sample Collection, DNA Isolation, and Cytogenetic Analysis
Fetal tissue samples were obtained from females who experienced spontaneous pregnancy losses (⩽20 wk), following approved institutional review board protocols. After removal of the pregnancy tissue from the uterus, the tissues were examined grossly, and a small portion of chorionic villi was placed in a sterile container with AmnioMAX media (GIBCO Invitrogen).
Chorionic villi were separated from maternal deciduae by use of a dissecting microscope, were dissociated using trypsin and collagenase, and were initiated in culture with AmnioMAX media. Cells were established in culture and grown on coverslips, as well as in T25 flasks. The cultures on coverslips were harvested for routine G-banding analysis following standard protocols. For each sample, 20 metaphase cells were counted, and 5 were fully analyzed. For DNA extraction, cells from two T25 flasks at 80% confluency were trypsinized and washed in PBS. DNA was isolated from these cells by use of the PureGene DNA isolation kit (Gentra Systems). Results from G-banding analysis were blinded to the experimenters before array analysis was initiated.
DNA Labeling, Array Hybridization, and Analysis
Test (POC) DNA was quantitated using a fluorometer and 100 ng each of test, and normal reference DNA (from an individual of the opposite sex of the fetal sample) was labeled with Cyanine 3- and Cyanine 5-dCTP fluorescent nucleotides (Perkin-Elmer), respectively, by use of the Microarray Random Priming Kit (Vysis/Abbott). In brief, DNA was denatured at 100°C for 10 min and was cooled to 4°C before the addition of Klenow fragments and nucleotide mix. After incubation at 37°C for 2 h, the samples were digested using a 1:20 DNAse dilution for 60 min at 15°C. Unincorporated nucleotides were then removed using Sephadex G-200 spin columns (Amersham-Biosciences). Probes were precipitated with 3 M sodium acetate and 100% ethanol and were resuspended in 10 mM Tris, pH 8.0.
Test and reference DNA was prepared for hybridization, according to manufacturer recommendations, with the GenoSensor Array 300 Kit (Vysis/Abbott). Equal aliquots of labeled test and reference DNA were combined in a tube with hybridization buffer, were denatured at 80°C for 10 min, and were incubated for 1 h at 37°C to allow for blocking of repetitive sequences. The solutions were then hybridized at 37°C for 72 h with a GenoSensor Array 300. This array contains 287 genomic clones, including those for each human telomere, as well as all of the known microdeletion syndromes and additional selected loci representing each chromosome arm (see fig. A1 [online only]).
After hybridization, the arrays were washed in 50% formamide/2× SSC at 40°C and 1× SSC at 25°C, and coverslips were mounted to the array with 4′,6-diamidino-2-phenylindole (DAPI). Finally, the arrays were imaged and data analyzed using the GenoSensor Reader and its accompanying software (Vysis/Abbott) (see table A [online only], a tab-delimited ASCII file that can be imported into a spreadsheet, for raw array data).
Targets with mean test-over-reference (T/R) ratios <0.8 were considered suggestive of losses, whereas those with ratios >1.2 were considered suggestive of gains. If questionable results were found on one array, the assay was repeated on a second array to confirm the results as a true gain or loss or to exclude such results as false positives. The data from one clone from the telomeric region of 4q (target 66, BAC clone CTB-31J3) was not included in the final analysis because of repeatedly poor performance, underscoring the importance of monitoring individual clone performance data (unpublished data). The G-banding diagnosis was revealed to the experimenters after final array results were interpreted.
FISH Analysis
After decoding the blinded samples, any losses or gains of a specific target clone(s) that did not correlate with the previous karyotype were verified using FISH. The clone on the array was used as the FISH probe in the analysis. The DNA was directly labeled with Spectrum Orange or Spectrum Green (Vysis/Abbott). The probes were hybridized to slides prepared from the sample in question. The probe and slide were codenatured on an automated hybridization chamber (HyBrite, Vysis/Abbott) for 2 min at 73°C. Slides were hybridized at 37°C for 24 h and were washed in 0.4× SSC/0.3% Tween 20 at 73°C for 2 min, 2× SSC/0.1% NP-40 at 25°C for 2 min, and 4× SSC/0.1% Tween 20 at 25°C for 4 min. Slides were counterstained with DAPI in antifade solution and were analyzed using a Zeiss Axiophot fluorescence microscope equipped with SmartCapture2 digital imaging software (Digital Scientific). Analysis was performed on a minimum of 50 interphase cells to verify array results.
Results
CGH-array analysis was utilized to examine 41 POC samples in a blind study. The results from this analysis were compared with results from G-banding analysis to ascertain whether CGH arrays could verify all abnormalities found previously by G-banding analysis and to discover whether additional, previously undetected submicroscopic rearrangements could be identified.
As shown in table 1, 37/41 POC cases analyzed by array were in exact concordance with the karyotype results. Of these matches, 24 cases showed normal karyotype and CGH-array results. Thirteen matches showed abnormal results for both tests, including 10 cases with trisomies, 2 cases with sex chromosome abnormalities (XYY and monosomy X), and 1 case with trisomy 17q and monosomy 20q, consistent with an unbalanced translocation.
Table 1.
Comparison of Conventional Cytogenetic Analysis with Array-CGH Analysis
|
Results of |
|||
| Sample | Cytogenetic Analysis | Array CGH | Matcha |
| 1 | 46,XX | Normal | Yes |
| 2 | 46,XX | Normal | Yes |
| 3 | 46,XX,der(20)t(17q;20q) | Trisomy 17q, monosomy 20q | Yes |
| 4 | 46,XX | Normal | Yes |
| 5 | 47,XY,+21 | Trisomy 21 | Yes |
| 6 | 47,XX,+16 | Trisomy 16 | Yes |
| 7 | 46,XY | Normal | Yes |
| 8 | 47,XYY | XYY | Yes |
| 9 | 46,XX | Normal | Yes |
| 10 | 46,XX | Normal | Yes |
| 11 | 46,XY | Normal | Yes |
| 12 | 46,XX | Normal | Yes |
| 13 | 46,XX | Normal | Yes |
| 14 | 47,XY,+14 | Trisomy 14 | Yes |
| 15 | 47,XX,+21 | Trisomy 21, trisomy 20 | Nob |
| 16 | 46,XX | Normal | Yes |
| 17 | 46,XX | Normal | Yes |
| 18 | 46,XY | Normal | Yes |
| 19 | 46,XX | Normal | Yes |
| 20 | 46,XY | Normal | Yes |
| 21 | 46,XY | Normal | Yes |
| 22 | 46,XY | Normal | Yes |
| 23 | 46,XX | Normal | Yes |
| 24 | 45,X | Monosomy X | Yes |
| 25 | 46,XY | Normal | Yes |
| 26 | 47,XX,+7 | Trisomy 7 | Yes |
| 27 | 46,XX | Normal | Yes |
| 28 | 47,XY,+10 | Trisomy 10 | Yes |
| 29 | 47,XY,+13 | Trisomy 13, del 9p21 | Noc |
| 30 | 46,XX | Normal | Yes |
| 31 | 46,XX | Normal | Yes |
| 32 | 46,XX | Normal | Yes |
| 33 | 46,XX | Normal | Yes |
| 34 | 47,XY,+13 | Trisomy 13 | Yes |
| 35 | 47,XX,+7 | Trisomy 7 | Yes |
| 36 | 47,XX,+21 | Trisomy 21 | Yes |
| 37 | 46,XX | Normal | Yes |
| 38 | 47,XY,+16 | Trisomy 16, dup 15q | Nod |
| 39 | 47,XX,+13 | Trisomy 13 | Yes |
| 40 | 46,XX | Dup 10qtel | Noe |
| 41 | 47,XX,+16 | Trisomy 16 | Yes |
Match compares array results with conventional cytogenetic results; for those that did not match, the additional abnormalities detected by array are described.
FISH analysis with probes for the centromeres of chromosomes 20 and 21 verified mosaic trisomy for chromosome 20.
Additional chromosome 9p21 deletion identified by array. Cells unavailable for FISH verification.
Chromosome 15q duplication verified by FISH analysis.
10q telomere duplication verified by FISH analysis.
As shown in table 1, the array identified additional abnormalities that were not detected by routine chromosome analysis in 4/41 (9.8%) cases. In case 15, the array identified the trisomy 21 observed by G-banding analysis but also suggested a trisomy for chromosome 20. The clones for chromosome 21 all showed ratios >1.2 with significant P values, clearly indicating a gain. For chromosome 20, the 20p telomere, 20q12, and 20q telomere clones, which are spaced on each end and at the middle of the chromosome, all showed increased ratios with significant P values, strongly suggesting a gain. Furthermore, all clones between these three clones showed slightly elevated T/R ratios. CGH-array analysis uses the total genomic DNA of a sample and sets its thresholds for gains (1.2) and losses (0.8) when 100% of the genetic material is affected. However, it is theoretically possible that the elevated signals in this region indicated that some, if not all, cells have additional material from chromosome 20. The array analysis was repeated, and the same results were obtained. As shown in figure 1A, subsequent FISH analysis verified the array results by revealing two cell populations, one with trisomy 21 and one with trisomy 20 and trisomy 21. Thirty percent of the cells exhibited trisomy 20 and trisomy 21, with the remainder showing trisomy 21 only.
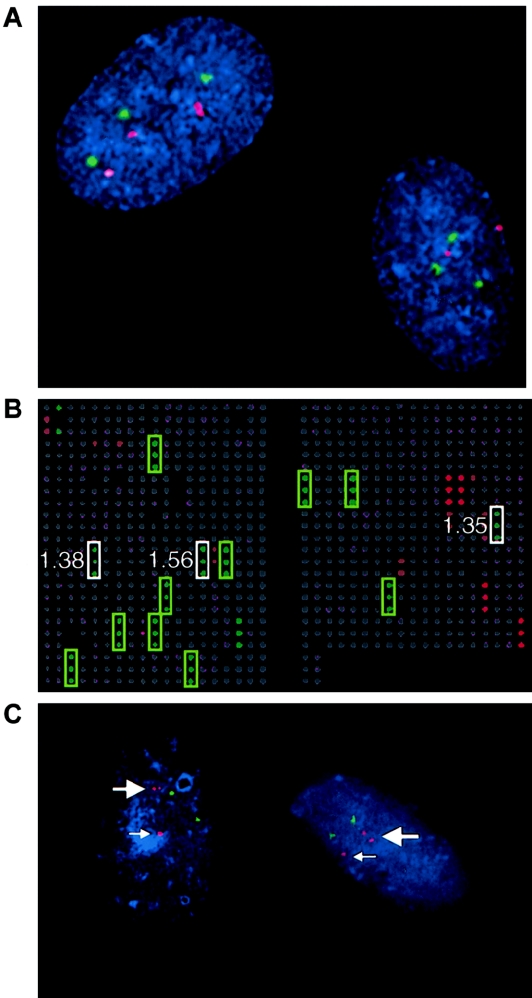
Figure 1.
Representative results from FISH and CGH-array analyses for cases 15 and 38. A, FISH verification of trisomy 21 and mosaic trisomy 20 array results for case 15. FISH probes corresponding to the centromeres of chromosome 20 (labeled with Spectrum Orange and shown in red) and chromosome 21 (labeled with Spectrum Green and shown in green) were hybridized to interphase cells from case 15. The cell on the left has trisomy for both chromosome 20 and chromosome 21, whereas the cell on the right shows only trisomy 21. There are only two hybridization signals for chromosome 20, verifying mosaicism for this trisomy. B, Representative pseudocolored image from CGH-array analysis for case 38, which has trisomy 16 and a duplication of the Prader-Willi/Angelman syndrome region on chromosome 15q. Male POC DNA (green) was hybridized with female reference DNA (red). Each clone is spotted in triplicate on the array, and clones with a gain in the POC sample are represented in green, loss in red, and normal copy number in gray. The green boxes mark the chromosome 16 clones that showed trisomy. The white boxes highlight the clones from the Prader-Willi/Angelman syndrome region that are duplicated; the corresponding ratios are shown next to each target. The other red and green signals correspond to clones from the X or Y chromosomes, respectively. C, Interphase FISH analysis of case 38 verifying the interstitial duplication of chromosome 15 that was identified by CGH array. A probe for the SNRPN gene (red) was used to test for the 15q11-q13 duplication, and a probe for the PML gene (green) in distal 15q was used as a control. In each cell, the small arrow points to the normal signal for SNRPN, and the large arrow indicates the duplicated chromosome 15, which shows two hybridization signals for SNRPN.
The trisomy 16 observed by G-banding analysis in case 38 was confirmed by CGH-array analysis. However, an interstitial duplication of 15q was also detected by array analysis. The array analysis showed four clones in the 15q11-q13 region (D15S11, SNRPN, UBE3A/D15S10, and GABRB3) with ratios of ⩾1.2. Figure 1B shows a representative image from CGH-array analysis of case 38. As shown in figure 1C, FISH analysis, with a probe for SNRPN, verified this interstitial duplication.
In case 29, two arrays detected not only a trisomy for chromosome 13 but also an interstitial deletion of chromosome 9. Two clones at 9p21 showed a ratio of 0.81 but without a significant P value. The clones on either side of these two clones showed normal ratios. We repeated this case on a second array and obtained similar results for the two clones at 9p21. Therefore, the data strongly suggest that case 29 exhibits a small interstitial deletion in addition to trisomy 13. FISH validation was not possible for this case, since additional cells were not available.
Finally, in case 40, two clones from the telomeric region of the long arm of chromosome 10 showed ratios of 1.25 and 1.39, suggestive of a duplication of this region. This result was verified by FISH analysis by use of one of the 10q clones from the array.
Discussion
In this study, array-based CGH was compared with routine G banding for the detection of fetal chromosomal abnormalities. Using array-based methods, we were able to detect all abnormalities previously identified by G-banding analysis. Additionally, four abnormalities were detected by the array that were not identified by chromosome analysis.
In case 15, the array data suggested mosaicism for trisomy 20 that was not identified by cytogenetic analysis. We pursued additional FISH analysis of the chromosome 20 clones and verified the mosaicism in this case, because multiple clones on the array showed an abnormal T/R ratio, demonstrating the usefulness of clone coverage redundancy on arrays. Several factors could account for the failure of cytogenetic analysis to detect the trisomy 20 cells. First, the cell line with trisomy 20 could be slow to grow and divide, thereby presenting very few mitotic cells of this type. It is interesting that this hypothesis is supported by the observation that trisomy for chromosome 20 was observed only in interphase cells and not in metaphase cells. It is also possible that a trisomy 20 cell could have been detected by cytogenetic analysis, but, since the diagnosis of trisomy 21 was already established, it was not further investigated. This example highlights two limitations of cytogenetic analysis, principally its reliance on cell culture and on subjective analysis of metaphase spreads, while illustrating the objective nature of DNA-based CGH-array analysis. It also demonstrates the potential for CGH-array results to suggest the presence of mosaicism.
A duplication of the proximal region of chromosome 15 was found in case 38, in addition to a previously identified trisomy. Although trisomy for chromosome 16 is the likely cause of the spontaneous abortion, the chromosome 15 duplication, if maternally inherited, has been associated with autistic disorder (Cook et al. 1997). Therefore, this finding could have important implications for genetic counseling and subsequent pregnancies. In addition to identifying the most likely cause of the miscarriage, case 38 shows the ability of array CGH to yield new, more complete diagnoses that may provide additional important information.
Through the detection of all chromosomal anomalies that were reported by routine G-banding analysis and the identification of four new abnormalities not detected by cytogenetic analysis, the efficacy of array-based CGH in analyzing POC samples has been established. In addition, this study demonstrates how several key shortcomings associated with routine cytogenetic methods of analysis of POCs could be avoided by use of this method. In array CGH, the use of genomic DNA circumvents the culture of fetal cells and the many problems (culture contamination, maternal overgrowth of cells, poor chromosome morphology, or culture failure) inherent in that technique. Moreover, the potential higher resolution of genomic microarrays allows for the detection of smaller unbalanced duplications and deletions of the genome. These factors, together with the objective nature of array CGH analysis, allows for a more refined and complete diagnosis.
The detection capabilities of array CGH are limited by the format of the array being used. In this study, the GenoSensor Array 300 was utilized over a higher-density array, such as an array with a 1-Mb resolution. Although a higher resolution could be obtained with a different array, the lower-density array was used, since it contains clinically relevant clones currently implicated in cytogenetic diagnoses, which is important for interpreting the significance of the results. In addition, both conventional and array-based CGH are limited, in being unable to detect polyploidies and balanced chromosomal rearrangements. Lomax et al. (2000) used flow cytometry as an adjunct to CGH to test for triploidy or tetraploidy. Alternatively, interphase FISH analysis of cells prepared from uncultured fetal tissue could be used to test the ploidy level or to validate results from array analysis.
Additional large-scale, prospective studies with uncultured POC samples—as well as peripheral blood, amniotic fluid, and other specimen types—need to be conducted to further assess the feasibility of utilizing array CGH in the clinical cytogenetics-laboratory setting. CGH-array analysis has the potential advantage of being automatable and of providing a shorter time frame for obtaining results. Future studies can test the benefits of the array technique in regard to higher detection parameters, more accurate and less failure-prone diagnoses, and a greater ease of use, as compared with conventional cytogenetic analysis.
Supplementary Material
Acknowledgments
This work was supported, in part, by March of Dimes grant 6-FY00-404 (to C.L.M. and D.H.L.) and a grant from the University of Chicago Pritzker School of Medicine Summer Research Program. The authors thank Kim Wilber and Teresa Ruffalo (Vysis/Abbott) and members of the University of Chicago Clinical Cytogenetics Laboratory for their technical expertise and assistance.
Appendix
Figure A1.
Schematic ideogram representation of the genomic clone coverage of the GenoSensor Array 300. Clones corresponding to each unique telomere region are represented by black circles, clones for all microdeletion syndromes are shown as gray circles, and additional selected loci representing each chromosome arm are depicted as white circles.
References
- Albertson DG, Pinkel D (2003) Genomic microarrays in human genetic disease and cancer. Hum Mol Genet Spec No 12:R145–R152 [DOI] [PubMed] [Google Scholar]
- Bell KA, Van Deerlin PG, Feinberg RF, du Manoir S, Haddad BR (2001) Diagnosis of aneuploidy in archival, paraffin-embedded pregnancy-loss tissues by comparative genomic hybridization. Fertil Steril 75:374–379 10.1016/S0015-0282(00)01703-9 [DOI] [PubMed] [Google Scholar]
- Bell KA, Van Deerlin PG, Haddad BR, Feinberg RF (1999) Cytogenetic diagnosis of “normal 46,XX” karyotypes in spontaneous abortions frequently may be misleading. Fertil Steril 71:334–341 10.1016/S0015-0282(98)00445-2 [DOI] [PubMed] [Google Scholar]
- Cai WW, Mao JH, Chow CW, Damani S, Balmain A, Bradley A (2002) Genome-wide detection of chromosomal imbalances in tumors using BAC microarrays. Nat Biotechnol 20:393–396 10.1038/nbt0402-393 [DOI] [PubMed] [Google Scholar]
- Cook EH Jr, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, Lord C, Courchesne E (1997) Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet 60:928–934 [PMC free article] [PubMed] [Google Scholar]
- Daniely M, Aviram-Goldring A, Barkai G, Goldman B (1998) Detection of chromosomal aberration in fetuses arising from recurrent spontaneous abortion by comparative genomic hybridization. Hum Reprod 13:805–809 10.1093/humrep/13.4.805 [DOI] [PubMed] [Google Scholar]
- Fritz B, Hallermann C, Olert J, Fuchs B, Bruns M, Aslan M, Schmidt S, Coerdt W, Muntefering H, Rehder H (2001) Cytogenetic analyses of culture failures by comparative genomic hybridisation (CGH): re-evaluation of chromosome aberration rates in early spontaneous abortions. Eur J Hum Genet 9:539–547 10.1038/sj.ejhg.5200669 [DOI] [PubMed] [Google Scholar]
- Goddijn M, Leschot NJ (2000) Genetic aspects of miscarriage. Baillieres Best Pract Res Clin Obstet Gynaecol 14:855–865 10.1053/beog.2000.0124 [DOI] [PubMed] [Google Scholar]
- Gunn SR, Mohammed M, Reveles XT, Viskochil DH, Palumbos JC, Johnson-Pais TL, Hale DE, Lancaster JL, Hardies LJ, Boespflug-Tanguy O, Cody JD, Leach RJ (2003) Molecular characterization of a patient with central nervous system dysmyelination and cryptic unbalanced translocation between chromosomes 4q and 18q. Am J Med Genet 120A:127–135 [DOI] [PubMed] [Google Scholar]
- Hassold T, Chen N, Funkhouser J, Jooss T, Manuel B, Matsuura J, Matsuyama A, Wilson C, Yamane JA, Jacobs PA (1980) A cytogenetic study of 1000 spontaneous abortions. Ann Hum Genet 44:151–178 [DOI] [PubMed] [Google Scholar]
- Hodgson G, Hager JH, Volik S, Hariono S, Wernick M, Moore D, Nowak N, Albertson DG, Pinkel D, Collins C, Hanahan D, Gray JW (2001) Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet 29:459–464 10.1038/ng771 [DOI] [PubMed] [Google Scholar]
- Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D (1992) Comparative genomic hybridization: a powerful new method for cytogenetic analysis of solid tumors. Science 258:818–821 [DOI] [PubMed] [Google Scholar]
- Ki A, Rauen KA, Black LD, Kostiner DR, Sandberg PL, Pinkel D, Albertson DG, Norton ME, Cotter PD (2003) Ring 21 chromosome and a satellited 1p in the same patient: novel origin for an ectopic NOR. Am J Med Genet 120A:365–369 [DOI] [PubMed] [Google Scholar]
- Kirchhoff M, Rose H, Lundsteen C (2001) High resolution comparative genomic hybridisation in clinical cytogenetics. J Med Genet 38:740–744 10.1136/jmg.38.11.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax B, Tang S, Separovic E, Phillips D, Hillard E, Thomson T, Kalousek DK (2000) Comparative genomic hybridization in combination with flow cytometry improves results of cytogenetic analysis of spontaneous abortions. Am J Hum Genet 66:1516–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris PL, Albertson DG, Alers JC, Andaya A, Carroll P, Fridlyand J, Jain AN, Kamkar S, Kowbel D, Krijtenburg PJ, Pinkel D, Schroder FH, Vissers KJ, Watson VJ, Wildhagen MF, Collins C, Van Dekken H (2003) High-resolution analysis of paraffin-embedded and formalin-fixed prostate tumors using comparative genomic hybridization to genomic microarrays. Am J Pathol 162:763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG (1998) High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet 20:207–211 10.1038/2524 [DOI] [PubMed] [Google Scholar]
- Schwaenen C, Wessendorf S, Kestler HA, Dohner H, Lichter P, Bentz M (2003) DNA microarray analysis in malignant lymphomas. Ann Hematol 82:323–332 10.1007/s00277-003-0649-6 [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG (2001) Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 29:263–264 10.1038/ng754 [DOI] [PubMed] [Google Scholar]
- Tabet AC, Aboura A, Dauge MC, Audibert F, Coulomb A, Batallan A, Couturier-Turpin MH, Feldmann G, Tachdjian G (2001) Cytogenetic analysis of trophoblasts by comparative genomic hybridization in embryo-fetal development anomalies. Prenat Diagn 21:613–618 10.1002/pd.115 [DOI] [PubMed] [Google Scholar]
- Veltman JA, Schoenmakers EFPM, Eussen BH, Janssen I, Merkx G, van Cleef B, van Ravenswaaij CM, Brunner HG, Smeets D, van Kessel AG (2002) High-throughput analysis of subtelomeric chromosome rearrangements by use of array-based comparative genomic hybridization. Am J Hum Genet 70:1269–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LELM, de Vries BBA, Osoegawa K, Janssen IM, Feuth T, Choy CO, Straatman H, van der Vliet W, Huys EHLPG, van Rijk A, Smeets D, van Ravenswaaij-Arts CMA, Knoers NV, van der Burgt I, de Jong PJ, Brunner HG, van Kessel AG, Schoenmakers EFPM, Veltman JA (2003) Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet 73:1261–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.