Abstract
Spinocerebellar ataxia type 10 (SCA10) is an autosomal dominant disorder characterized by ataxia, seizures, and anticipation. It is caused by an expanded ATTCT pentanucleotide repeat in intron 9 of a novel gene, designated “SCA10.” The ATTCT expansion in SCA10 represents a novel class of microsatellite repeat and is one of the largest found to cause human diseases. The expanded ATTCT repeat is unstably transmitted from generation to generation, and an inverse correlation has been observed between size of repeat and age at onset. In this multifamily study, we investigated the intergenerational instability, somatic and germline mosaicism, and age-dependent repeat-size changes of the expanded ATTCT repeat. Our results showed that (1) the expanded ATTCT repeats are highly unstable when paternally transmitted, whereas maternal transmission resulted in significantly smaller changes in repeat size; (2) blood leukocytes, lymphoblastoid cells, buccal cells, and sperm have a variable degree of mosaicism in ATTCT expansion; (3) the length of the expanded repeat was not observed to change in individuals over a 5-year period; and (4) clinically determined anticipation is sometimes associated with intergenerational contraction rather than expansion of the ATTCT repeat.
Introduction
Expanded nucleotide repeats have been identified as the genetic mutation in an increasing number of hereditary neurological diseases, such as fragile-X syndrome (FMR1 [MIM 309550]), Friedreich ataxia (FRDA [MIM 229300]), Huntington disease (HD [MIM 143100]), many spinocerebellar ataxias, and myotonic dystrophies. The expanded nucleotide repeats are unstable in soma and germline, causing somatic mosaicism and intergenerational instability. This dynamic property of nucleotide-repeat expansion is thought to contribute to variable manifestation of the diseases. Understanding repeat instability in various neurological disorders will shed light on the underlying basic mechanism for maintaining genomic stability.
Spinocerebellar ataxia type 10 (SCA10 [MIM 603516]) is an autosomal dominant neurodegenerative disorder characterized by ataxia, seizures, and anticipation (Grewal et al. 1998, 2002; Matsuura et al. 1999; Rasmussen et al. 2001). It belongs to a group of diseases known as “autosomal dominant cerebellar ataxias” (ADCAs) (Harding 1981). To date, 18 ADCA loci—SCA1 (MIM 164400), SCA2 (MIM 183090), SCA3 (MIM 109150), SCA4 (MIM 600223), SCA5 (MIM 600224), SCA6 (MIM 183086), SCA7 (MIM 164050), SCA8 (MIM 603680), SCA10, SCA11 (MIM 604432), SCA12 (MIM 604326), SCA13 (MIM 605259), SCA14 (MIM 605361), SCA15 (MIM 606658), SCA16 (MIM 606364), SCA17 (MIM 607136), SCA19 (MIM 607346), and SCA21 (MIM 607454)—and dentatorubral-pallidoluysian atrophy (DRPLA [MIM 125370]) have been mapped to specific chromosomal regions (Rosa and Ashizawa 2002) (see also Genome Database). Although the specific mutations have yet to be identified in SCA4, SCA5, SCA11, SCA13, SCA15, SCA16, SCA19, and SCA21, individuals with one of seven ADCAs—including SCA1, SCA2, SCA3 (also known as “Machado-Joseph disease” [MJD]), SCA6, SCA7, SCA17, and DRPLA—are known to have expanded CAG trinucleotide repeats, which encodes polyglutamine tracts within various contexts of different proteins. In most of these diseases, the age at onset is inversely correlated with the number of CAG repeats, and the repeat size tends to increase in successive generations, providing the molecular basis for anticipation. In addition to these ADCAs, HD (Huntington’s Disease Collaborative Research Group 1993) and Kennedy disease (SBMA [MIM 313200]) (La Spada et al. 1991) are also caused by polyglutamine-coding CAG repeat expansions, and HD clearly shows anticipation due to progressive repeat expansion in successive generations.
Different nucleotide-repeat expansions have also been found in other diseases. SCA8 has been associated with an expanded CTG repeat in the 3′ UTR of the SCA8 gene (Koob et al. 1999). In SCA12, a CAG repeat is expanded in the 5′ UTR of a protein phosphatase subunit gene, PPP2RB (Holmes et al. 1999). In FRDA, an autosomal recessive neurological disease, homozygous expansion of a GAA repeat located in the first intron of the FRDA gene is the principal disease-causing mutation (Campuzano et al. 1996). It is interesting that, although the expanded repeats are unstable, neither clinical anticipation nor progressive intergenerational expansion of repeats has been documented in SCA8, SCA12, and FRDA. Other diseases—such as myotonic dystrophy type 1 (DM1 [MIM 602668]), caused by CTG expansion in the 3′ UTR of the DMPK gene (Fu et al. 1992), and FMR1, caused by CGG expansion in the 5′ UTR of the FMR1 gene (Verkerk et al. 1991)—show anticipation and intergenerational expansion of the repeats. An expanded CCTG tetranucleotide repeat in intron 1 of ZNF9 gene causes myotonic dystrophy type 2 (DM2 [MIM 602668]) (Liquori et al. 2001). This repeat shows a high level of instability, but the intergenerational changes in repeat size result in either gain or loss of the repeats, which cannot fully explain the reported anticipation (Day et al. 2003).
We localized the SCA10 mutation to 22q13.3 (Matsuura et al. 1999; Zu et al. 1999) and demonstrated that the mutation is a large expansion of ATTCT pentanucleotide repeats with a gain of up to 4,500 repeats (Matsuura et al. 2000). The expanded ATTCT repeat is unstable and is located in intron 9 of a novel gene, designated “SCA10” (also known as “E46L”), which has no sequence homology to any known genes. We have reported a weak inverse correlation between the size of expansion and the age at onset and showed a considerable instability during intergenerational transmission. But little is known about this novel class of disease-causing microsatellite repeats. In the present study, we investigated the intergenerational instability of the expanded ATTCT repeats, focusing on its relationships to the sex of the transmitting parent, somatic and germline instability, and repeat-size changes during aging.
Material and Methods
Patients and DNA Samples
Under a consent procedure approved by the local internal review board, blood samples were drawn from affected and at-risk individuals of pedigrees with SCA10, and genomic DNA was extracted from peripheral blood leukocytes (PBL). Some patients provided two consecutive DNA samples, after 5-year intervals. We also obtained a buccal sample from six patients and semen samples from two affected males. Genomic DNA was prepared from buccal samples by use of MasterPure Complete DNA & RNA Purification Kit (Epicentre Technologies). To prepare sperm DNA, we followed the protocol described by Jeffreys et al. (1994).
Southern-Blot Analysis
Southern-blot analysis was performed using EcoRI restriction digestion of the genomic DNA and the probe generated by PCR amplification of the repeat-free intron 9 region flanked by primers DanL and DanR, as described elsewhere (Matsuura et al. 2000).
Serial Passages of Lymphoblastoid Cell Lines (LBCLs)
PBL were isolated from blood samples obtained from four patients with SCA10, by use of the Ficoll-Hypaque gradient, and were transformed into LBCLs by use of Epstein-Barr virus (EBV). The LBCLs were cultured, in a 75-cm2 flask, in RPMI-1640 containing 10% heat-inactivated fetal-bovine serum and antimycotics (GIBCO-BRL) in 5% CO2 at 37°C (Khajavi et al. 2001). After reaching ∼3×107 cells in culture, an aliquot of the cells (∼107 cells) was used for isolation of genomic DNA. Another aliquot underwent a passage, and the remaining cells were frozen in the culture medium containing 10% DMSO. Viability of cells was assessed by trypan blue exclusion. This passage procedure was repeated over 12 generations. The ATTCT-repeat size was monitored by Southern-blot analysis. The sizes of expanded ATTCT repeats in the original PBL of four patients were as follows: 800 repeats (patient 6), 1,300 repeats (patient 8), 2,780 repeats (patient 5), and 3,760 repeats (patient 9).
Cloning of LBCLs
By use of a hemocytometer, each cell line was single-cell cloned by limitation of dilutions at the concentration of 0.5 cell per well of a 96-well plate. The single-cell origin of each cell line was assured, as described elsewhere (Khajavi et al. 2001). Each clone was transferred to a 25-cm2 flask, once the cell number reached ∼106 cells. Clones were then transferred to 75-cm2 flasks and were allowed to grow until the total cell number reached 3×107 cells. An aliquot was harvested for DNA analysis, and another aliquot of 107 cells was passed. The remaining cells were frozen in the culture medium containing 10% DMSO. We analyzed three clones from patient 9 and four clones from patient 6. Passage step was repeated as long as the cells were viable (3–20 passages). The ATTCT repeat size was monitored by Southern-blot analysis.
Results
Intergenerational Changes of the Size of ATTCT Repeats
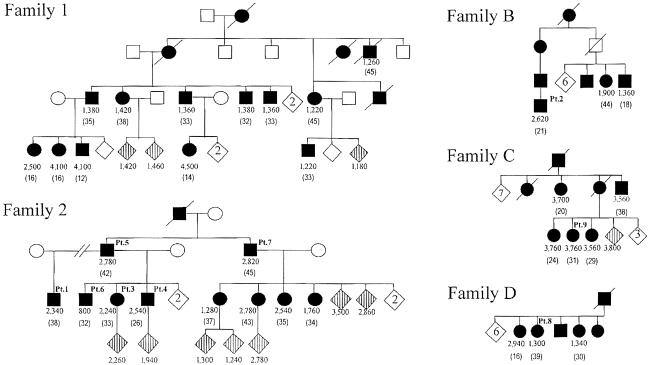
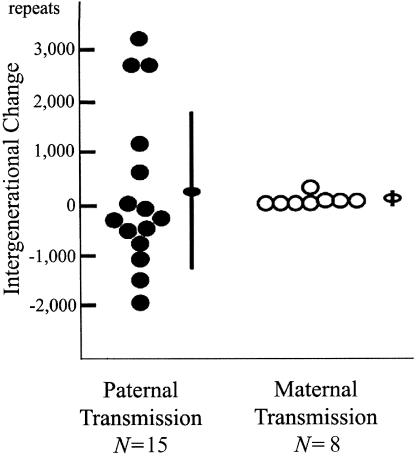
We analyzed the genomic DNA from five SCA10-affected families of Mexican descent (fig. 1) by PCR and subsequent Southern-blot analysis (see the “Material and Methods” section). We recently reported the genotype-phenotype correlation in the affected members of families 1 and 2 (Grewal et al. 2002). Here, we extended our analysis of the stability of expanded ATTCT repeats and provided additional data on these two families as well as three more families from Mexico (Rasmussen et al. 2001). The normal size of the EcoRI fragment containing ATTCT repeats is 2.5 kb, whereas the average size of the 39 affected and at-risk individuals is 13.1 kb (2,120 repeats), with a range of 6.5–25.0 kb (800–4,500 repeats). The expansion size (mean±SD) in 22 individuals with the disease allele transmitted from the affected father was 15.2±4.8 kb (2,540±960 repeats), whereas the expansion size of the remaining 17 individuals who received the disease allele from the affected mother was 10.5±4.8 kb (1,600±960 repeats). Among them, we were able to examine 23 parent-offspring pairs in families 1 and 2 (15 father-offspring pairs and 8 mother-offspring pairs) for repeat-size changes during the intergenerational transmissions. Because two affected males from family 2 had a large number of affected offspring, paternal transmission was overrepresented in that series. However, we found that paternal transmissions of the expanded alleles were highly unstable, with large expansions or contractions, whereas maternal transmissions showed few or smaller changes. The mean±SD of the absolute repeat-size difference between parent and child was 1,143±1,042 in the 15 paternal transmissions and 20±19 in the 8 maternal transmissions (P<.01). The mean±SD of the repeat-size change (i.e., the repeat size of the child minus the repeat size of the parent) was 247±1,555 in the paternal transmissions and 0±28 in the maternal transmissions, suggesting that there is a mild expansion bias with the paternal transmission, whereas neither expansion nor contraction bias is present with the maternal transmission (P<.01) (fig. 2). It should be noted that all paternal transmissions (n=4) of the expanded alleles in family 1 resulted in large expansions, whereas most paternal transmissions (n=11) in family 2 showed smaller expansions or contractions. It is interesting that all transmitting males in family 2 had alleles of >2,500 repeats, whereas those in family 1 had alleles of <1,500 repeats.
Figure 1.
Five families with SCA10. Unblackened symbols indicate unaffected individuals, and blackened symbols indicate individuals affected with SCA10. Circles indicate females, and squares indicate males. A diagonal line over a symbol indicates a deceased individual. Diamonds with vertical lines indicate asymptomatic mutation carriers. Unblackened diamonds indicate noncarrier unaffected sibling(s), and the number within the diamond shows the number of unaffected siblings. The number of ATTCT-repeat units in the expanded allele is shown under each symbol. Patients 1–9 (discussed in the text) are shown at the right upper corner of their respective symbols as “Pt. 1–9.” Patients 10–12 are from three additional families, which are not shown here. The expanded allele was paternally transmitted in patients 10 and 11 and was maternally transmitted in patient 12. The age at onset (in years) is shown in parentheses below the number of ATTCT-repeat units.
Figure 2.
Paternal transmission of the expanded allele (blackened circle) (n=15) results in a greater degree of intergenerational changes than does maternal transmission (unblackened circle) (n=8) (paternal change: 247±1,555 repeats; maternal: 0±28 repeats P<.01). Paternal transmissions gave rise to both elongations and contractions of the expanded alleles. Ovals and error bars indicate means and SDs, respectively.
Somatic and Germline Instability of the ATTCT Expansion
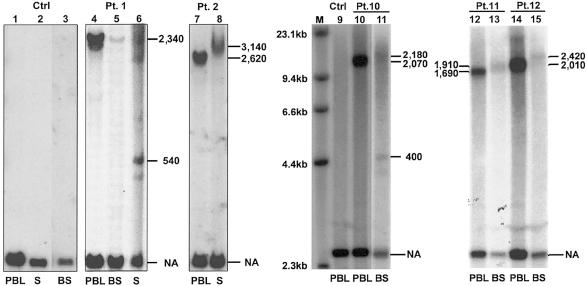
To study instability of ATTCT expansion in somatic tissues and germline, we extracted genomic DNA from sperm and PBL from two male patients; we also obtained DNA from buccal cells of six patients. It is unfortunate that DNA was degraded in buccal samples from two of the six patients. All normal alleles of these samples and normal control subjects showed the same 2.5-kb EcoRI fragment. PCR analysis confirmed the absence of intertissue variability of normal alleles in the samples (data not shown). In patient 1, the expanded allele in the buccal sample showed the same expansion size as that of PBL. Although the band of the buccal sample is fainter than other samples, probably due to degraded DNA, no major mosaicism was detected. In patient 10, however, the buccal sample showed clear repeat-size mosaicism with a fuzzy main band of slightly larger size accompanied by two or more smaller bands, in contrast to the single expanded allele in PBL. Since the undigested genomic DNA of this buccal sample showed no evidence of degradation, the observed mosaicism is unlikely to be degradation artifacts. Expanded alleles in buccal samples of patients 11 and 12 showed larger size than those in PBL. Thus, expanded SCA10 ATTCT repeats show evidence of somatic instability with intratissue mosaicism and intertissue repeat-size variability (fig. 3).
Figure 3.
Repeat instability in somatic tissues and sperm. A normal male control subject showed the same 2.5-kb EcoRI fragment. In patient 1, the sperm showed a smearlike pattern of multiple bands of expansion, whereas the other tissues shared the same size (2,340 repeats). The expanded band of the buccal sample is substantially fainter; this is probably due to degraded DNA. In patient 2, the sperm showed a smearlike band larger than the expanded allele of PBL. In patient 10, the buccal sample showed expanded alleles of heterogeneous sizes, presented as a fuzzy band slightly larger than the expanded allele of PBL. The buccal sample of this patient also showed smaller additional bands. Buccal samples from patients 11 and 12 showed patterns of expanded alleles similar to that of patient 10, although distinct mosaic alleles are lacking. NA = normal allele; S = sperm; BS = buccal sample; Ctrl = normal control subject; Pt. = patient; M = λ HindIII marker. Numbers on both sides of each panel indicate the number of ATTCT repeat units.
The expanded allele in sperm from patient 1 consisted of a heterogeneous smear with additional distinct bands, suggesting mosaicism in the male germline. The most evident mosaic band in sperm was ∼540 repeats, much smaller than that of PBL (∼2,340 repeats). Patient 2 showed a larger expanded allele of broader size range in sperm (∼3,140 repeats) than PBL (∼2,620 repeats) (fig. 3). Although the sample size is small, these results suggest that the expanded alleles in male germline are highly unstable.
Stability Over Time of ATTCT Expansion in PBL from Patients with SCA10
To determine whether the expanded ATTCT repeats in PBL change their size as patients age, we obtained consecutive samples of DNA from six patients (patients 1, 3, 4, 5, 6, and 7) collected over a 5-year interval. The size of the largest alleles did not show detectable changes (fig. 4). However, four of these patients (patients 1, 5, 6, and 7) showed additional bands, which are smaller than the main band, suggesting potential mosaicism. We believe that they are not artifacts, because the size of the additional band is variable from sample to sample. It is interesting that the pattern of this mosaicism did not change in each patient after the 5-year interval. These data suggest that the expanded alleles in PBL are typically relatively stable, although rare instability events do occur, as evidenced by the somatic mosaicism.
Figure 4.
ATTCT expansion is stable in PBL during a 5-year interval. The odd-numbered lanes represent initial PBL DNA, and the even-numbered lanes represent PBL DNA 5 years later. M = λ HindIII marker; NA = normal allele; nl = normal control individual.
Changes of ATTCT Expansion after Serial Passaging of SCA10 LBCLs in Culture
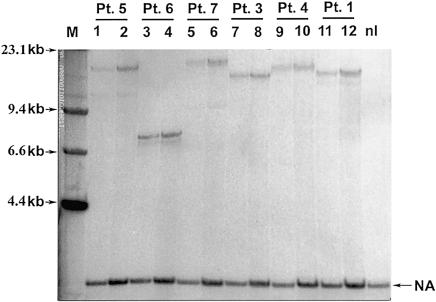
To study instability in cell culture, we compared the sizes of the ATTCT expansion between PBL and the LBCLs in nine affected individuals. Following EBV transformation, three of the nine LBCLs showed small contractions, when compared with the original alleles in PBL (100 to 780 fewer repeats). The remaining six LBCLs did not show any change in repeat size. The observed changes could be explained by the fact that EBV preferentially transforms B-lymphocytes, which represent only a minor subpopulation of PBL. Therefore, this observation might have revealed the mosaicism of repeat expansion in PBL. Alternatively, this shift in repeat size could be due to mutational events in culture, resulting in changes to the ATTCT expansion in LBCL.
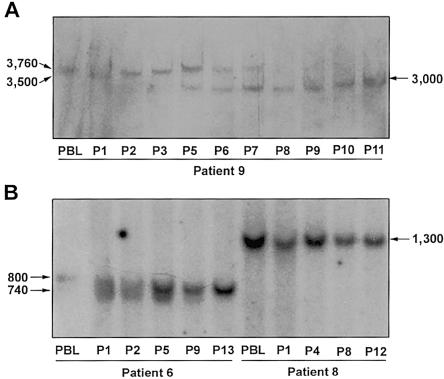
We also looked at the stability of expanded ATTCT repeats in these LBCLs during serial passaging (fig. 5) and found that the LBCLs showed variable degrees of instability. In two of the three LBCLs with contracted repeats after transformation, the expanded alleles showed further contractions after different passages. In the first LBCL (derived from patient 9 [fig. 5A]), a contraction from 3,760 to 3,000 repeats was detected at passage 5, and it gradually replaced the progenitor allele by passage 11. In the other LBCL, the expanded allele of 2,780 repeats changed its size in early passages and was replaced by an allele with 2,500 repeats, which seemed stable, with no further changes (data not shown). The third LBCL (derived from patient 6 [fig. 5B]) showed heterogeneous expansion of ∼800 repeats, as indicated by a smear immediately after the EBV transformation. The size heterogeneity gradually dissipated, and a sharp, narrow band eventually took over in the subsequent passages. These changes probably revealed a shift of cell population due to clonal expansion, although repeat-size mutations during culture cannot be excluded. It is noteworthy, however, that the normal alleles of all LBCLs had no changes. Moreover, in most of the patients, the size of expansion remained stable after EBV transformation and serial passages, as demonstrated by cells from patient 8 in figure 5B.
Figure 5.
Repeat instability in LBCL. Only expanded alleles are shown, to highlight the instability. A, In patient 9, the 3,760 repeat-expansion in PBL contracted to 3,500 through the EBV transformation (P1). The expanded allele gradually increased in size to 3,760 again (P1–P5). Moreover, a 3,000-repeat allele appeared in P5 and gradually replaced the 3,760 repeats. B, Patient 6 showed repeat-size heterogeneity through the transformation. Following the passages, the broad band converted to a tight one of 740 repeats. In patient 8, the allele with 1,300 repeats was stable throughout the passages and the transformation. P1–P12 = passages 1–12.
Instability of the Expanded ATTCT Repeat in Serially Passed Clonal LBCLs
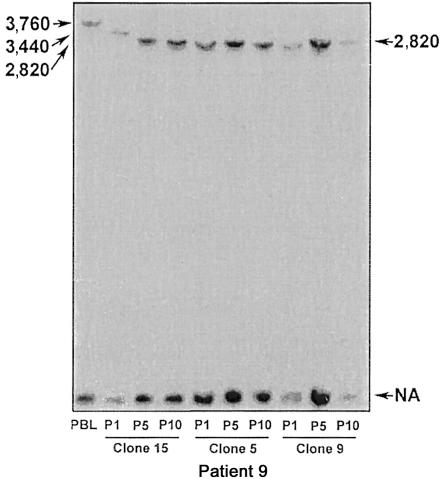
To explore mutation versus clonal expansion mechanisms for repeat instability in the heterogeneous bulk LBCLs, we established clonal LBCLs. Since clonal lines were derived from a single cell, changes in repeat size should represent mutation events occurring after cloning. Figure 6 shows the repeat-size changes in three clonal LBCLs (clones 5, 9, and 15) derived from patient 9. Clone 15 had a clear contraction of the expanded allele, from 3,760 to 2,820 repeats, during the first five passages, suggesting that a repeat-size mutation has occurred. In contrast, the expanded allele showed no detectable changes in clones 5 and 9. Similarly, the expanded ATTCT repeat was stable in four clonal LBCLs derived from patient 6 (data not shown).
Figure 6.
Repeat instability in clonal LBCLs (clones 5, 9, and 15) from patient 9, with a 3,760-repeat allele in PBL. Contraction was noted between passages 1 and 5 of clone 15. P1 = passage 1; P5 = passage 5; P10 = passage 10.
Discussion
Intergenerational Changes of the Size of ATTCT Repeats
Our data showed that the size of the expanded ATTCT repeat is unstably transmitted from generation to generation in our families with SCA10. The pattern of the intergenerational instability was dependent on the sex of the transmitting parent. The repeat was highly unstable during paternal transmission, whereas maternal transmission showed few changes and of a smaller magnitude. Unstable transmissions of expanded repeats have been observed in almost all diseases with expanded trinucleotide repeats. The parental sex is an important determinant of the intergenerational instability in many of these disorders. Most autosomal dominant diseases caused by an expanded CAG repeat coding for a polyglutamine tract—such as SCA1 (Chung et al. 1993), SCA2 (Schols et al. 1997), SCA3 (Maruyama et al. 1995), SCA7 (David et al. 1998), HD (Goldberg et al. 1993; Telenius et al. 1993), and DRPLA (Nagafuchi et al. 1994)—show greater instability in paternal transmission than in maternal transmission. The intergenerational instability is biased toward further expansion, giving rise to paternally driven anticipation in these disorders. In DM1, the expanded CTG repeat in the premutation range (37–50 CTGs) is comparable to the fully expanded alleles of polyglutamine diseases; further expansion of the CTG repeat is mostly associated with paternal transmission (Martorell et al. 2001). It is interesting that, when the CTG repeat size exceeds ∼1,000 CTGs in the transmitting father, the expanded alleles tend to contract in the offspring with increasing frequency, whereas maternally transmitted full-expansion alleles seldom contract, regardless of the repeat size (Ashizawa et al. 1994). In FMR1, the premutation CGG-repeat alleles expand to full-mutation alleles in the offspring when transmitted maternally, and the probability of expansion into the full-mutation range increases when the repeat size of the maternal premutation allele is larger (Fu et al. 1991; Nolin et al. 2003). Conversely, paternal transmission of fully expanded CGG repeat often gives rise to a contracted allele in the offspring (Malter et al. 1997). The GAA repeat of FRDA (De Michele et al. 1998) and the CTG repeat of SCA8 (Moseley et al. 2000) also contract during paternal transmission. In DM1, FMR1, FRDA, and SCA8, the expanded repeat is located in noncoding regions of the affected gene, and the size of full-expansion alleles is substantially larger than in those of polyglutamineexpansion diseases. Conversely, repeat expansion is stable with either paternal or maternal transmission in SCA6 and oculopharyngeal muscular dystrophy (MIM 164300), in which expansion of CAG and GCG repeats coding for polyglutamine and polyalanine, respectively, is much shorter (Zhuchenko et al. 1997; Brais et al. 1998). Thus, the sex of the transmitting parent, the motif of the repeat unit, the length of the repeat, the location of the repeat in the gene, and the surrounding sequences (i.e., cis elements) may influence the characteristics of instability.
The ATTCT repeat is a unique motif located in an intron, and the size of expansion in SCA10 is one of the largest among diseases caused by repeat expansion. Therefore, the unique characteristics of ATTCT-repeat instability shown in our study are attributable to these variables. The CCTG-repeat expansion found in DM2, like in SCA10, is located in an intron, and the expansion size is also comparable to the ATTCT-repeat expansion in SCA10 (Liquori et al. 2001). In DM2, repeat size did not correlate with age at disease onset, and affected offspring had markedly shorter expansions than did their affected parents, with a mean size difference of −17 kb (−4,250 CCTGs) (Day et al. 2003). However, expanded CCTG repeats in PBL are highly unstable in DM2-affected patients with conspicuous age-dependent repeat-size instability, unlike the expanded ATTCT repeats in SCA10.
Somatic and Germline Instability of Expanded ATTCT Repeats
In most of the repeat-expansion diseases, the expansion is unstable in both somatic and germline tissues. In the present study, we presented several lines of evidence that suggest instability of expanded ATTCT repeats in somatic tissues. First, the size of repeat expansion in some LBCLs is different from that of the original PBLs. This may be due to selection of B-lymphocytes from the PBL pool by EBV transformation, indicating that the PBL population is heterogeneous in repeat size because of somatic instability. Second, some single-cell cloned LBCLs showed changes in expansion size after passages in culture. Third, repeat mosaicism appears to exist in blood and buccal DNA of patients with SCA10. Most PBLs and LBCLs derived from patients with SCA10, however, had stable expansion, which is remarkable considering the enormous expansion size. This is quite different from other large repeats causing human diseases—such as CTG repeats in DM1 (Khajavi et al. 2001), CGG repeats in FMR1 (Verkerk et al. 1991), GAA repeats in FRDA (Bidichandani et al. 1999), and CCTG repeats in DM2 (Day et al. 2003)—where extensive instability is typical.
The frequency and magnitude of instability in somatic tissues, especially in blood, were much lower in comparison with sperm DNA in SCA10. It thus appears that the expanded ATTCT repeat is very unstable during spermatogenesis, which contributes to intergenerational instability through paternal transmission. But it is interesting that the expanded ATTCT repeat was remarkably more stable during maternal transmission, suggesting relatively stable repeat expansion during oogenesis. Thus, we believe that repeat stability is distinct between male and female germlines. The difference in repeat stability between male and female germlines probably underlies the parental sex effects observed in our study.
We also examined intertissue variability of the ATTCT repeat expansion in four patients by comparing the expansion size in PBLs and buccal cells. In three of these four patients, expanded alleles in the buccal-cell samples showed slightly larger sizes and greater degrees of repeat-size heterogeneity compared with those of PBLs. Furthermore, one sample showed additional smaller alleles. These findings suggest that expanded ATTCT repeats in buccal cells may have a greater degree of instability than those in PBL. It is obvious that the number of samples and tissue types studied here is too limited for general conclusions regarding ATTCT-repeat instability in other tissues, such as the brain. Nevertheless, our data are compatible with the idea of variable instability of the SCA10 ATTCT repeat in different somatic tissues.
The Size of Expanded ATTCT Repeat and the Age at Onset
It is puzzling that, in family 2, the expansion appears to have contracted in the offspring of patients 5 and 7, in spite of clinically observed anticipation. To exclude ascertainment bias, we have interviewed the patients and family members again and have cross-referenced the age at onset with additional information sources for each subject. Although determination of precise age at onset was often difficult, even with careful interviews, the history strongly suggested that the ages at onset of patients 5 and 7 were later than those of their offspring (fig. 1). This apparent discrepancy between the observed anticipation and shorter repeat lengths in the offspring could be explained by a difference in the sampling age, since affected parents are usually sampled at an older age than their affected offspring. Under this hypothesis, PBL DNA samples in the parents result from many more rounds of DNA replication. If the repeats tend to expand during mitosis, it is likely that repeat size in the parents might catch up with the offspring who inherited an expansion. Alternatively, repeat instability might be variable in different tissues; the size of repeat expansion in the affected tissues (such as the cerebellum) might be larger in the offspring, distinct from what is observed in PBL. Our data of somatic mosaicism and repeat instability in a cloned cell line suggest that somatic instability could occur during the individual’s life, although we were unable to demonstrate age-dependent repeat-length changes in blood DNA. We believe that family-specific modifying factors play an important role in this genotype-phenotype discrepancy (Grewal et al. 2002). To test these possibilities, further studies involving additional families will be required to examine the PBL alleles from long-term longitudinal sampling, to compare alleles in a variety of tissues, including cerebellum, and to identify the family-specific factors. Meanwhile, it should be noted that additional genetic and environmental factors can outweigh the mild correlation (r2=0.34, P=.018) between repeat length and age at onset. Thus, the use of repeat length in genetic counseling needs careful consideration.
Acknowledgments
This study was supported by National Institutes of Health grant NS041547 (T.A.) and grants from National Ataxia Foundation, National Organization for Rare Disorders, the Ichiro Kanehara Foundation (Tokyo), and Brain Science Foundation (Tokyo) (T.M.).
Electronic-Database Information
The URLs for data presented herein are as follows:
- Genome Database, http://www.gdb.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FMR1, FRDA, HD, SCA10, SCA1, SCA2, SCA3, SCA4, SCA5, SCA6, SCA7, SCA8, SCA11, SCA12, SCA13, SCA14, SCA15, SCA16, SCA17, SCA19, SCA21, DRPLA, SBMA, DM1, DM2, and oculopharyngeal muscular dystrophy)
References
- Ashizawa T, Dunne PW, Ward PA, Seltzer WK, Richards CS (1994) Effects of the sex of myotonic dystrophy patients on the unstable triplet repeat in their affected offspring. Neurology 44:120–122 [DOI] [PubMed] [Google Scholar]
- Bidichandani SI, Purandare SM, Taylor EE, Gumin G, Machkhas H, Harati Y, Gibbs RA, Ashizawa T, Patel PI (1999) Somatic sequence variation at the Friedreich ataxia locus includes complete contraction of the expanded GAA triplet repeat, significant length variation in serially passaged lymphoblasts and enhanced mutagenesis in the flanking sequence. Hum Mol Genet 8:2425–2436 10.1093/hmg/8.13.2425 [DOI] [PubMed] [Google Scholar]
- Brais B, Bouchard JP, Xie YG, Rochefort DL, Chretien N, Tome FM, Lafreniere RG, Rommens JM, Uyama E, Nohira O, Blumen S, Korczyn AD, Heutink P, Mathieu J, Duranceau A, Codere F, Fardeau M, Rouleau GA, Korcyn AD (1998) Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet 18:164–167 [DOI] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, et al (1996) Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423–1427 [DOI] [PubMed] [Google Scholar]
- Chung MY, Ranum LP, Duvick LA, Servadio A, Zoghbi HY, Orr HT (1993) Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet 5:254–258 [DOI] [PubMed] [Google Scholar]
- David G, Durr A, Stevanin G, Cancel G, Abbas N, Benomar A, Belal S, Lebre AS, Abada-Bendib M, Grid D, Holmberg M, Yahyaoui M, Hentati F, Chkili T, Agid Y, Brice A (1998) Molecular and clinical correlations in autosomal dominant cerebellar ataxia with progressive macular dystrophy (SCA7). Hum Mol Genet 7:165–170 10.1093/hmg/7.2.165 [DOI] [PubMed] [Google Scholar]
- Day JW, Ricker K, Jacobsen JF, Rasmussen LJ, Dick KA, Kress W, Schneider C, Koch MC, Beilman GJ, Harrison AR, Dalton JC, Ranum LPW (2003) Myotonic dystrophy type 2: molecular, diagnostic and clinical spectrum. Neurology 60:657–664 [DOI] [PubMed] [Google Scholar]
- De Michele G, Cavalcanti F, Criscuolo C, Pianese L, Monticelli A, Filla A, Cocozza S (1998) Parental gender, age at birth and expansion length influence GAA repeat intergenerational instability in the X25 gene: pedigree studies and analysis of sperm from patients with Friedreich’s ataxia. Muscle Nerve 21:390–393 [DOI] [PubMed] [Google Scholar]
- Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG Jr, Warren ST, Oostra BA, Nelson DL, Caskey CT (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67:1047–1058 10.1016/0092-8674(91)90283-5 [DOI] [PubMed] [Google Scholar]
- Fu YH, Pizzuti A, Fenwick RG Jr, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, de Jong P, Wieringa B, Korneluk R, Perryman MB, Epstein HF, Caskey CT (1992) An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science 255:1256–1258 [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Kremer B, Andrew SE, Theilmann J, Graham RK, Squitieri F, Telenius H, Adam S, Sajoo A, Starr E, Heiberg A, Wolff G, Hayden MR (1993) Molecular analysis of new mutations for Huntington’s disease: intermediate alleles and sex of origin effects. Nat Genet 5:174–179 [DOI] [PubMed] [Google Scholar]
- Grewal RP, Achari M, Matsuura T, Durazo A, Tayag E, Zu L, Pulst SM, Ashizawa T (2002) Clinical features and ATTCT repeat expansion in spinocerebellar ataxia type 10. Arch Neurol 59:1285–1290 10.1001/archneur.59.8.1285 [DOI] [PubMed] [Google Scholar]
- Grewal RP, Tayag E, Figueroa KP, Zu L, Durazo A, Nunez C, Pulst SM (1998) Clinical and genetic analysis of a distinct autosomal dominant spinocerebellar. Neurology 51:1423–1426 [DOI] [PubMed] [Google Scholar]
- Harding A (1981) Genetic aspects of autosomal dominant late onset cerebellar ataxia. J Med Genet 18:436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes SE, O’Hearn EE, McInnis MG, Gorelick-Feldman DA, Kleiderlein JJ, Callahan C, Kwak NG, Ingersoll-Ashworth RG, Sherr M, Sumner AJ, Sharp AH, Ananth U, Seltzer WK, Boss MA, Vieria-Saecker AM, Epplen JT, Riess O, Ross CA, Margolis RL (1999) Expansion of a novel CAG trinucleotide repeat in the 5′ region of PPP2R2B is associated with SCA12. Nat Genet 23:391–392 10.1038/70493 [DOI] [PubMed] [Google Scholar]
- Huntington’s Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–983 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Tamaki K, MacLeod A, Monckton DG, Neil DL, Armour JA (1994) Complex gene conversion events in germline mutation at human minisatellites. Nat Genet 6:136–145 [DOI] [PubMed] [Google Scholar]
- Khajavi M, Tari AM, Patel NB, Tsuji K, Siwak DR, Meistrich ML, Terry NH, Ashizawa T (2001) “Mitotic drive” of expanded CTG repeats in myotonic dystrophy type 1 (DM1). Hum Mol Genet 10:855–863 10.1093/hmg/10.8.855 [DOI] [PubMed] [Google Scholar]
- Koob MD, Moseley ML, Schut LJ, Benzow KA, Bird TD, Day JW, Ranum LP (1999) An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat Genet 21:379–384 10.1038/7710 [DOI] [PubMed] [Google Scholar]
- La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH (1991) Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352:77–79 10.1038/352077a0 [DOI] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LPW (2001) Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 293:864–867 10.1126/science.1062125 [DOI] [PubMed] [Google Scholar]
- Malter HE, Iber JC, Willemsen R, de Graaff E, Tarleton JC, Leisti J, Warren ST, Oostra BA (1997) Characterization of the full fragile X syndrome mutation in fetal gametes. Nat Genet 15:165–169 [DOI] [PubMed] [Google Scholar]
- Martorell L, Monckton DG, Sanchez A, Lopez De Munain A, Baiget, M (2001) Frequency and stability of the myotonic dystrophy type 1 premutation. Neurology 56:328–335 [DOI] [PubMed] [Google Scholar]
- Maruyama H, Nakamura S, Matsuyama Z, Sakai T, Doyu M, Sobue G, Seto M, Tsujihata M, Oh-I T, Nishio T, Sunohara N, Takahashi R, Hayashi M, Nishino I, Ohtake T, Oda T, Nishimura M, Saida T, Matsumoto H, Baba M, Kawaguchi Y, Kakizuka A, Kawakami H (1995) Molecular features of the CAG repeats and clinical manifestation of Machado-Joseph disease. Hum Mol Genet 4:807–812 [DOI] [PubMed] [Google Scholar]
- Matsuura T, Achari M, Khajavi K, Bachinski L, Zoghbi HY, Ashizawa T (1999) Mapping of the gene for a novel spinocerebellar ataxia with pure cerebellar signs and epilepsy. Ann Neurol 45:407–411 [DOI] [PubMed] [Google Scholar]
- Matsuura T, Yamagata T, Burgess DL, Rasmussen A, Grewal RP, Watase K, Khajavi M, McCall A, Davis CF, Zu L, Achari M, Pulst SM, Alonso E, Noebels JL, Nelson DL, Zoghbi HY, Ashizawa T (2000) Large expansion of ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet 26:191–194 10.1038/79911 [DOI] [PubMed] [Google Scholar]
- Moseley ML, Schut LJ, Bird TD, Koob MD, Day JW, Ranum LP (2000) SCA8 CTG repeat: en masse contractions in sperm and intergenerational sequence changes may play a role in reduced penetrance. Hum Mol Genet 9:2125–2130 10.1093/hmg/9.14.2125 [DOI] [PubMed] [Google Scholar]
- Nagafuchi S, Yanagisawa H, Sato K, Shirayama T, Ohsaki E, Bundo M, Takeda T, Tadokoro K, Kondo I, Murayama N, Tanaka Y, Kikushima H, Umino K, Kurosawa H, Furukawa T, Nihei K, Inoue T, Sano A, Komure O, Takahashi M, Yoshizawa T, Kanazawa I, Yamada M (1994) Dentatorubral and pallidoluysian atrophy expansion of an unstable CAG trinucleotide on chromosome 12p. Nat Genet 6:14–18 [DOI] [PubMed] [Google Scholar]
- Nolin SL, Brown WT, Glicksman A, Houck GE Jr, Gargano AD, Sullivan A, Biancalana V, Bröndum-Nielsen K, Hjalgrim H, Holinski-Feder E, Kooy F, Longshore J, Macpherson J, Mandel J-L, Matthijs G, Rousseau F, Steinbach P, Väisänen M-L, von Koskull H, Sherman SL (2003) Expansion of the fragile X CGG repeat in females with premutation or intermediate alleles. Am J Hum Genet 72:454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A, Matsuura T, Ruano L, Yescas P, Ochoa A, Ashizawa T, Alonso E (2001) Clinical and genetic analysis of four Mexican families with spinocerebellar ataxia type 10. Ann Neurol 50:234–239 10.1002/ana.1081 [DOI] [PubMed] [Google Scholar]
- Rosa AL, Ashizawa T (2002) Genetic ataxia. Neurol Clin 20:727–757 [DOI] [PubMed] [Google Scholar]
- Schols L, Gispert S, Vorgerd M, Menezes Vieira-Saecker AM, Blanke P, Auburger G, Amoiridis G, Meves S, Epplen JT, Przuntek H, Pulst SM, Riess O (1997) Spinocerebellar ataxia type 2: genotype and phenotype in German kindreds. Arch Neurol 54:1073–1080 [DOI] [PubMed] [Google Scholar]
- Telenius H, Kremer HP, Theilmann J, Andrew SE, Almqvist E, Anvret M, Greenberg C, Greenberg J, Lucotte G, Squitieri F (1993) Molecular analysis of juvenile Huntington disease: the major influence on (CAG)n repeat length is the sex of the affected parent. Hum Mol Genet 2:1535–1540 [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, Eussen BE, Van Ommen GJB, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Caskey CT, Nelson DL, Oostra BA, Warren ST (1991) Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell 65:905–914 [DOI] [PubMed] [Google Scholar]
- Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton DW, Amos C, Dobyns WB, Subramony SH, Zoghbi HY, Lee CC (1997) Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet 15:62–69 [DOI] [PubMed] [Google Scholar]
- Zu L, Figueroa KP, Grewal R, Pulst SM (1999) Mapping of a new autosomal dominant spinocerebellar ataxia to chromosome 22. Am J Hum Genet 64:594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]