Abstract
Specific language impairment (SLI) is defined as an unexplained failure to acquire normal language skills despite adequate intelligence and opportunity. We have reported elsewhere a full-genome scan in 98 nuclear families affected by this disorder, with the use of three quantitative traits of language ability (the expressive and receptive tests of the Clinical Evaluation of Language Fundamentals and a test of nonsense word repetition). This screen implicated two quantitative trait loci, one on chromosome 16q (SLI1) and a second on chromosome 19q (SLI2). However, a second independent genome screen performed by another group, with the use of parametric linkage analyses in extended pedigrees, found little evidence for the involvement of either of these regions in SLI. To investigate these loci further, we have collected a second sample, consisting of 86 families (367 individuals, 174 independent sib pairs), all with probands whose language skills are ⩾1.5 SD below the mean for their age. Haseman-Elston linkage analysis resulted in a maximum LOD score (MLS) of 2.84 on chromosome 16 and an MLS of 2.31 on chromosome 19, both of which represent significant linkage at the 2% level. Amalgamation of the wave 2 sample with the cohort used for the genome screen generated a total of 184 families (840 individuals, 393 independent sib pairs). Analysis of linkage within this pooled group strengthened the evidence for linkage at SLI1 and yielded a highly significant LOD score (MLS = 7.46, interval empirical P<.0004). Furthermore, linkage at the same locus was also demonstrated to three reading-related measures (basic reading [MLS = 1.49], spelling [MLS = 2.67], and reading comprehension [MLS = 1.99] subtests of the Wechsler Objectives Reading Dimensions).
Introduction
Specific Language Impairment (SLI [MIM 602081]) is diagnosed as a disorder in the development of language despite adequate educational opportunity and normal intelligence. A diagnosis requires a significant discrepancy between the child’s verbal and nonverbal abilities in the absence of any additional disorders that might underlie the language problems (e.g., hearing loss, mental retardation, and autism).
The profile of deficits associated with SLI shows great variation between individuals, with differences both in the range of linguistic domains that are involved (phonology, morphology, syntax, semantics, and pragmatics) and in the language modality that is affected (expressive and/or receptive). Thus, whereas SLI is classified into three broad categories—phonological disorder, expressive-language disorder, and mixed expressive/receptive-language disorder—it has been suggested that a more sophisticated classification system, one capable of reflecting variations in the severity of the underlying impairment, is required (Bishop 1987).
It is estimated that 3%–7% of English-speaking preschool children are affected by SLI (Law et al. 1998), and, although a proportion of these children learn to compensate for their problems, a significant number experience severe and persistent language difficulties throughout their lives (Beitchman et al. 1994; Johnson et al. 1999).
The familial nature of language impairments has been well documented and, despite great variation in study design and disorder classification, many investigations have demonstrated an increased frequency of language problems in individuals with first-degree relatives affected by SLI, compared with that in controls (Bishop and Edmundson 1986; Neils and Aram 1986; Tallal et al. 1989; Stromswold 1998). Moreover, twin studies have indicated a consistent increase in concordance rates in MZ twins, compared with those in DZ twins, suggesting that much of the familial aggregation can be attributed to genetic influences (Lewis and Thompson 1992; Bishop et al. 1995; Tomblin and Buckwalter 1998). However, with the exception of one prominent pedigree (Lai et al. 2001), family studies have failed to find any clear segregation between genotype and phenotype, and it is generally accepted that the genetics underlying SLI are complex, involving several genes that interact—both with each other and with the linguistic environment—to produce an overall susceptibility to the development of the disorder.
To date, two genome screens have been performed for genetically complex forms of SLI. The first reported linkage to chromosomes 16q and 19q (SLIC 2002), and the second implicated loci on chromosomes 2p and 13q (Bartlett et al. 2002).
The SLIC study scanned 473 individuals from 98 nuclear families ascertained through epidemiological and clinical-based studies. All probands were selected to have language scores ⩾1.5 SD below the mean for their chronological age and a performance IQ score within the “normal” range (>80). Three quantitative measures of language abilities were employed for the genome screen: expressive and receptive language skills were assessed by the Clinical Evaluation of Language Fundamentals (CELF-R) test battery and a test of nonsense word repetition was used as a marker of phonological short-term memory. Systematic genomewide linkage analyses yielded two prominent regions of linkage, one on chromosome 16q24 (SLI1 [MIM 606711]) and a second on chromosome 19q13 (SLI2 [MIM 606712]). Although both of these loci generated a maximum LOD score (MLS) that bordered on genomewide significance (MLS = 3.55; pointwise empirical P=.0003 and P=.0004, respectively), the linkage to chromosome 16 was found only with the nonword repetition test, whereas the linkage on chromosome 19 appeared to be specific to the CELF-R expressive language score. No corresponding linkage was seen for other measures. Separation of the complete sample into its constituent clinical and epidemiological cohorts demonstrated that each group was contributing independently at both peaks of linkage.
In contrast to the SLIC study (2002), the second genome screen (Bartlett et al. 2002) employed large extended pedigrees, a selection of binary affection statuses, and parametric genetic analyses in their investigation of language impairment. This study scanned DNA from 86 individuals collected from several branches of five Canadian families. Although not interrelated or a population isolate, these families were selected from a relatively homogeneous sample of individuals with Celtic ancestry. All families were assessed with a comprehensive battery of tests covering expressive (Test of Language Development [TOAL:2, TOLDP:2, or TOLDI:2]) and receptive (token test) language skills, IQ (Wechsler Intelligence scales [WISC, WAIS, WPPSI, or WASI]), and reading development (nonword and single-word reading). The test scores were then used to derive three diagnostic categories. A statistical cutoff was applied to spoken language scores (Spoken Language Quotient of the Test of Language Development) and was used to define a group of “language impaired” individuals, a nonword reading/IQ discrepancy criterion was used to select a group of “reading impaired” individuals, and a set of more relaxed conditions (e.g., a history of speech therapy or reading intervention) were applied to specify a “clinically impaired” group. Each of these three diagnostic groups was then analyzed for linkage under dominant and recessive models of inheritance, both of which assumed a population prevalence of 7%. Genomewide analyses revealed two regions of linkage: one on chromosome 2p22 and one on chromosome 13q21 (SLI3 [MIM 607134]). The chromosome 13q peak was seen under a recessive model of reading impairment, with a four-point MLS of 3.92 (empirical genomewide adjusted P<.01), whereas linkage to chromosome 2 was found under a recessive model with the use of the “language impaired” group, with a four-point MLS of 2.79 (empirical genomewide adjusted P<.06).
In a recent abstract presented at the American Society of Human Genetics meeting, Bartlett et al. (2003) reported the completion of a third genome screen for SLI with the use of 22 multiplex nuclear and extended pedigrees ascertained in the United States. Using a methodology similar to their previous scan, Bartlett et al. reported four regions with LOD scores >2 on 18p11 (dominant clinical impairment), 1q31 (recessive clinical impairment), 7q11 (dominant reading impairment), and 6p21 (recessive reading impairment). Although none of these regions overlap with those reported elsewhere, Bartlett et al. did find LOD scores >1.0 on chromosomes 13q and 19q (2003).
The apparent lack of consensus between the results of SLIC and those of Bartlett et al. may be attributable, at least in part, to the differences in study design. Although both screens were related to SLI, they involved completely different approaches to subject selection, phenotype assessment, and genetic-linkage strategies. Both methods have their own strengths (the use of quantitative traits circumvents the need for the derivation of a consistent affection status, whereas the use of large pedigrees can increase the homogeneity of the sample) and weaknesses (parametric analyses make assumptions about the basis of the disorder, whereas the selection of phenotypes used for QTL analyses may not represent the full scope of disorder). It is possible that both screens have revealed loci that are of general importance to the SLI phenotype, but, as with other complex disorders, only the independent replication of these studies can evaluate the importance of individual chromosomal regions.
Interestingly, although none of the regions in the two published genome screens overlap, several of the identified loci have been associated with other neurodevelopmental disorders. It is a well-documented fact that the childhood conditions SLI, dyslexia, autism, and attention deficit hyperactivity disorder (ADHD [MIM 143465]) share a close, if intriguing, relationship. Individuals with autism have distinct problems with early language development (Lord et al. 1994), and children with language impairments are at an increased risk of developing dyslexia and ADHD (Cantwell and Baker 1987; Snowling et al. 2000, 2001; Kovac et al. 2001). Moreover, family and twin studies consistently demonstrate a marked clustering of these disorders (Gallagher et al. 2000; Bishop 2001; Brown et al. 2001; Tomblin et al. 2003) and have led to the proposal that there may exist shared genetic risk factors (Bishop 2001; Doyle et al. 2001; Tomblin et al. 2003; Stein et al. 2004). However, the exact relationship between these disorders remains unclear, and the importance of their relationship, in terms of diagnosis, continues to be a matter of debate. In a genomewide screen, the Collaborative Linkage Study of Autism (CLSA) found the strongest linkage result (MLS = 3.0, P=.008) to AUTS4 (MIM 608049) (CLSA 1999) with the same chromosome 13 marker (D13S800) as Bartlett et al. (2003). Furthermore, when a follow-up study stratified the CLSA cohort, by the language status of the proband, they found that the linkage on chromosome 13 arose predominantly from the language-abnormal (i.e., onset of speech >36 mo) proband subset (Bradford et al. 2001). In addition, a recent meta-analysis of four genome screens for autism indicated that the locus on chromosome 13 formed the second most significant region of linkage (after 7q31) across the entire genome (Badner and Gershon 2002). In a genomewide screen for dyslexia, Fisher et al. (2002) also found linkage between this region of 13q and a measure of orthographic choice (P=.001) within a sample of reading-impaired American families.
Dyslexia has also been linked to regions on chromosomes 2p (DYX3 [MIM 604254]) (Fagerheim et al. 1999; Fisher et al. 2002), 18p (DYX6 [MIM 606616]) (Fisher et al. 2002), and 6p (DYX2 [MIM 600202]) (Cardon et al. 1994; Grigorenko et al. 1997; Fisher et al. 1999; Gayán et al. 1999; Kaplan et al. 2002), all of which coincide with those identified by Bartlett et al. (2002, 2003) in their language impaired, clinically impaired, and reading impaired samples, respectively. In a study of families affected by speech-sound disorders (SSD) (i.e., a deficit that primarily involves articulation and/or the storage and processing of phonological information), Stein et al. (2004) found linkage to a region of chromosome 3 that elsewhere had been implicated in dyslexia in a Finnish family (Nopola-Hemmi et al. 2001). Linkage was found to quantitative measures relating to aspects of reading ability (e.g., Woodcock word identification), short-term memory (e.g., nonsense word repetition), and articulation (e.g., the Goldman-Fristoe Test of Articulation). Furthermore, Stein et al. demonstrated that, in this region, those families who showed a strong level of linkage to language-related traits (nonsense word repetition and multisyllabic word test) also contributed to the linkage to reading-related measures (Woodcock word identification and word attack). Therefore, they concluded that this locus is likely to act in a pleiotropic fashion, influencing both SSD and developmental dyslexia (Stein et al. 2004).
The current study focuses on the SLI1 and SLI2 loci and aims to analyze QTL linkage at these loci within an extended sample of families affected by SLI. In addition, we aim to investigate the possibility of a shared genetic etiology between SLI and dyslexia, by the analysis of three reading-related measures at these loci.
Subjects and Methods
Subjects
A total of 840 subjects (393 sib pairs) were recruited from 184 families by four separate centers (table 1)—the Newcomen Centre at Guy’s Hospital, London; the Cambridge Language and Speech Project (CLASP); the Child Life and Health Department at the University of Edinburgh; and the Department of Child Health at the University of Aberdeen.
Table 1.
Sample Groups Divided by Ascertainment Cohorts and by Waves[Note]
|
No. in Sample |
|||||||||||
| Cambridge |
Guys |
||||||||||
| Sample Group | Aberdeen (Wave 2 Only) | Wave 1 | Wave 2 | Total | Edinburgh (Wave 2 Only) | Wave 1 | Wave 2 | Total | All Wave 1 | All Wave 2 | All |
| Families | 10 | 55 | 10 | 65 | 48 | 43 | 18 | 61 | 98 | 86 | 184 |
| Individuals | 40 | 266 | 42 | 310 | 206 | 207 | 79 | 287 | 473 | 367 | 840 |
| Sib pairs (all possible) | 11 | 113 | 19 | 132 | 111 | 106 | 33 | 139 | 219 | 174 | 393 |
Note.— Wave 1 consists of those samples included in the SLIC genome screen (2002) and originates from the Cambridge and Guys groups. Wave 2 consists of all samples collected since the SLIC genome screen (2002) and originates from Aberdeen, Cambridge, Edinburgh, and Guys groups. The total sample, which includes all possible samples, was used for the extension study.
The cases recruited by the University of Aberdeen were selected from specialist language units within the Aberdeen and Aberdeenshire areas. These individuals represent a self-referred sample of children with severe and persistent language difficulties. Ethical approval was granted by the Grampian local research ethic committee. The Aberdeen sample was small (10 families, 11 sib pairs) and, as such, was amalgamated with the Guys sample for all analyses. Both of these groups were selected from clinical samples of severely affected children and, therefore, were considered to be the most similar of the samples in terms of ascertainment and clinical presentation. This assumption was supported by the distributions of the phenotypes within the two cohorts (table 2).
Table 2.
Results of Mann-Whitney U-Test of Phenotypic Means[Note]
|
Similarity of Distributions (U) |
||||||||||||
| Aberdeen |
Cambridge |
Edinburgh |
Probands |
|||||||||
| Group | ELS | RLS | NWR | ELS | RLS | NWR | ELS | RLS | NWR | ELS | RLS | NWR |
| Cambridge | .087 | .230 | .003 | |||||||||
| Edinburgh | .407 | .234 | .005 | .000 | .000 | .000 | ||||||
| Guys | .449 | .872 | .410 | .035 | .008 | .000 | .015 | .062 | .000 | |||
| Sibs | .000 | .000 | .000 | |||||||||
Note.— The Mann-Whitney U-test was used to ascertain the level of difference between the phenotypic means of each cohort, prior to their amalgamation for analyses. In many cases, the means differed significantly between the groups. Significant differences are shown in italics. Note that, for all phenotypes (ELS, RLS, and NWR), the probands were found to differ significantly from the sibs.
The cases referred by the University of Edinburgh were selected originally to participate in a study of children with severe receptive-language impairments. These individuals were recruited from eastern and central Scotland on a prospective basis through consultant pediatricians and speech and language therapists. All probands required specialist educational support and were selected to have a historical language comprehension score >2 SD below that expected for their age. Families were initially approached by letter, and those who expressed an interest in the study were visited at home by a trained speech and language therapist. A detailed family history was taken by this professional, and all children who did not meet the SLIC study criteria (see below) were excluded. The final sample consisted of 48 families (111 sib pairs). The research program at Edinburgh was approved by local ethic committees in East and Central Scotland.
Both the Guys and the Cambridge cohorts have been described in detail elsewhere (Burden et al. 1996; SLIC 2002). In brief, the cases referred by Guy's Hospital represent a clinical sample selected through specialist language schools and through Afasic, a support organization for people with developmental and language impairments. The Cambridge cases were identified through epidemiological screening and from a subset of a sample collected for a community-based longitudinal investigation of speech and language difficulties.
The Guys and Cambridge samples consist of 98 families, included in the original SLIC genome screen (wave 1), plus an additional 28 families (18 from Guy's Hospital and 10 from Cambridge) recruited since that point. Thus, the entire cohort can be considered in terms of two waves: wave 1, consisting of the original genome screen sample of 98 families (from Guys and Cambridge), and wave 2, consisting of a replication set of 86 families collected after the completion of the genome screen (from Guys, Cambridge, Edinburgh, and Aberdeen). Alternatively, each cohort can be studied as an independent sample, resulting in three separate groups: Cambridge, Edinburgh, and Guys/Aberdeen (table 1).
As with the genome-screen study, all families were selected to include a proband with, either currently or in the past, language skills ⩾1.5 SD below the normative mean for their chronological age on the expressive and/or receptive scales of the CELF-R battery (Semel et al. 1992). Any child reported to have a nonverbal IQ <80 was excluded from the study. Additional exclusion criteria included MZ twinning, chronic illness requiring multiple hospital visits or admissions, deafness, a clinical diagnosis of autism, English as a second language, children with known neurological disorders, and children under local authority care.
Whole blood (Guys and Edinburgh) or buccal swab (Cambridge and Aberdeen) samples were collected from all available family members, regardless of language ability. DNA was extracted using standard protocols, and all buccal swab DNAs were preamplified using a primer extension preamplification technique (Zhang et al. 1992).
Phenotypic Measures
Six phenotypic scores, measuring different aspects of language and reading abilities, were analyzed for linkage across chromosomes 16 and 19. Expressive language scores (ELS) and receptive language scores (RLS) were determined with the use of CELF-R (Semel et al. 1992), a test of nonword repetition (NWR) was used as a marker of phonological short-term memory (SE Gathercole, personal communication), and the Wechsler Objective Reading Dimensions (WORD) (basic reading [BR], reading comprehension [RC], and spelling [SP]) were analyzed as gross measures of individuals’ reading abilities (Rust et al. 1993). No parental phenotype data was collected, as the linkage analysis utilizes only information from sib pair phenotype data. A summary of all the phenotypic measures can be found in tables 3 and 4.
Table 3.
Descriptive Statistics for Analyzed Phenotypes[Note]
|
Result for Phenotype Test |
||||||
| CELF-R |
WORD |
|||||
| Statistic andSample Group | ELS | RLS | NWR | BR | SP | RC |
| Mean: | ||||||
| All | 79.03 | 88.34 | 91.28 | 92.33 | 92.32 | 90.48 |
| Wave 1 | 81.68 | 91.11 | 97.29 | 93.20 | 93.14 | 92.09 |
| Wave 2 | 75.42 | 84.59 | 83.95 | 89.50 | 89.66 | 89.24 |
| Aberdeen | 75.35 | 87.88 | 89.35 | NA | NA | NA |
| Cambridge | 83.28 | 93.40 | 101.76 | 95.90 | 95.68 | 96.24 |
| Edinburgh | 73.90 | 82.67 | 77.33 | NA | NA | NA |
| Guys | 78.41 | 86.87 | 91.22 | 88.28 | 88.51 | 82.00 |
| SD: | ||||||
| All | 16.09 | 18.07 | 19.58 | 15.32 | 16.29 | 15.48 |
| Wave 1 | 16.05 | 18.23 | 17.92 | 15.56 | 16.54 | 15.60 |
| Wave 2 | 15.46 | 17.21 | 19.07 | 14.29 | 15.27 | 14.01 |
| Aberdeen | 11.58 | 13.17 | 14.27 | NA | NA | NA |
| Cambridge | 15.72 | 18.14 | 16.99 | 14.71 | 16.08 | 14.48 |
| Edinburgh | 15.49 | 16.91 | 18.10 | NA | NA | NA |
| Guys | 16.23 | 17.99 | 16.79 | 15.05 | 15.73 | 13.99 |
| Range: | ||||||
| All | 50–131 | 50–141 | 55–136 | 55–127 | 52–134 | 48–136 |
| Wave 1 | 50–131 | 50–141 | 55–136 | 55–127 | 52–134 | 48–136 |
| Wave 2 | 50–124 | 50–128 | 55–130 | 57–114 | 57–124 | 49–107 |
| Aberdeen | 50–95 | 70–107 | 58–116 | NA | NA | NA |
| Cambridge | 54–131 | 50–141 | 55–136 | 66–127 | 64–134 | 62–136 |
| Edinburgh | 50–124 | 50–128 | 55–130 | NA | NA | NA |
| Guys | 50–115 | 50–131 | 55–123 | 55–123 | 55–124 | 55–122 |
| Count: | ||||||
| All | 424 | 424 | 444 | 271 | 271 | 260 |
| Wave 1 | 244 | 244 | 244 | 207 | 207 | 202 |
| Wave 2 | 180 | 180 | 200 | 64 | 64 | 58 |
| Aberdeen | 17 | 17 | 17 | 0 | 0 | 0 |
| Cambridge | 158 | 158 | 161 | 144 | 134 | 140 |
| Edinburgh | 101 | 101 | 118 | 0 | 0 | 0 |
| Guys | 148 | 148 | 148 | 127 | 127 | 120 |
Note.— WORD scores were not collected for the Aberdeen and Edinburgh cohorts and were not analyzed with wave 1 and wave 2 as separate groups. NA = not available.
Table 4.
Age Statistics[Note]
|
Age by Phenotype(years/mo) |
||||
| WORD |
||||
| Statistic andSample Group | CELF-R(ELS and RLS) | NWR | BR and SP | RC |
| Age range: | ||||
| All | 4/7–17/0 | 3/4–29/1 | 6/1–18/5 | 6/1–17/0 |
| Wave 1 | 4/7–17/0 | 4/8–23/11 | NA | NA |
| Wave 2 | 4/9–16/10 | 3/4–29/1 | NA | NA |
| Aberdeen | 4/9–15/3 | 8/8–18/5 | NA | NA |
| Cambridge | 8/0–16/4 | 8/0–18/6 | 6/11–18/5 | 6/11–16/4 |
| Edinburgh | 4/11–16/10 | 3/4–29/2 | NA | NA |
| Guys | 4/7–17/0 | 4/8–23/11 | 6/1–17/0 | 6/1–17/0 |
| Mean age: | ||||
| All | 10/2 | 10/9 | 10/5 | 10/4 |
| Wave 1 | 10/2 | 10/6 | NA | NA |
| Wave 2 | 10/1 | 10/11 | NA | NA |
| Aberdeen | 9/2 | 13/2 | NA | NA |
| Cambridge | 10/3 | 10/6 | 10/5 | 10/2 |
| Edinburgh | 9/11 | 11/0 | NA | NA |
| Guys | 10/2 | 10/8 | 10/4 | 10/6 |
Note.— WORD scores were not collected for the Aberdeen and Edinburgh cohorts and were not analyzed with wave 1 and wave 2 as separate groups. NA = not available.
CELF-R
The CELF-R is a clinical tool widely used for the identification, diagnosis, and follow-up evaluation of language disorders in school-age children. The battery is split into receptive and expressive scales, which can be combined to provide a composite language score. Each scale consists of three subtests, designated to be primarily receptive or expressive in nature. The exact combination of individual tests that are used is dependent on the subject's age. Additive raw scores from each segment are then transformed to derive a standardized RLS and ELS, each with a mean of 100 and a SD of 15 in the general population calibration sample (Semel et al. 1992).
NWR
In the NWR test, subjects are required to repeat tape-recorded nonsensical words of increasing length and complexity (e.g., “brufid” and “contramponist”). Studies show that individuals with current language impairments, as well as those who had language difficulties in early childhood that later resolved, perform poorly on this test (Gathercole et al. 1994; Bishop et al. 1996). All available children were tested using the NWR test (S. E. Gathercole, personal communication). Note that this test version is prepublication, since we found that the published standardization (Gathercole et al. 1994) introduced flooring effects in our more severely affected samples.
Children selected by the Edinburgh team were assessed with a Scottish version of the NWR test, in which all words were presented in a Scottish accent.
WORD
This test consists of three separate components (BR, SP, and RC), each of which assesses a different literacy skill. The BR tasks are designed to assess the subject's ability to identify letter-sound correspondences and to read single words, the SP tests involve writing individual letters and words to dictation, and the RC tasks require the child to read short passages in silence and to answer questions about them (Rust et al. 1993).
Each WORD scale produces an age-standardized score with a mean of 100 and a SD of 15. These measures can be used independently or can be combined with the Wechsler IQ to produce IQ-discrepancy information. Owing to logistical constraints, WORD data was collected only for the Guys and Cambridge groups.
Data Transformation
The Mann Whitney U-test was performed to ascertain whether the phenotypic means differed between cohorts prior to amalgamation for analysis. In many cases, the means were found to vary significantly between groups (table 2). These differences are attributable to the fact that, although the selection criteria applied to each group were identical, the ascertainment criteria and severity of disorder differed between cohorts. The Guys and Aberdeen collections were chosen from severely affected clinical distributions; the Edinburgh cohort was ascertained to have mainly receptive language impairments; and the Cambridge group represents a mainstream, epidemiologically selected sample. To combine the groups for variance-components analysis, which creates a model around a single mean, all phenotypes, therefore, were standardized to a z score (z=(x-μ)/σ, where x = attained score, μ = mean, and σ = SD; note that the mean and SD are taken from each group separately). This standardization creates a distribution centered around a single mean (0), while preserving the variance of the original samples, and is in accordance with the standardization performed for the original genome screen (SLIC 2002). The standardized scores are represented by the terms “RLStrans,” “ELStrans,” “NWRtrans,” “BRtrans,” “SPtrans,” and “RCtrans” and were used for all analyses performed.
Genotyping and Data Handling
All 871 individuals were genotyped for 40 polymorphic microsatellite markers spanning chromosome 16 (table B1 [online only]), in addition to 21 microsatellite markers across chromosome 19 (table B2 [online only]). The genotyping methods have been described in detail elsewhere (SLIC 2002). In brief, marker regions were amplified within a 10-μl PCR reaction using fluorescently labeled primers (Applied Biosystems). PCR products were pooled, allowing concurrent detection by ABI 3700 sequencers (Applied Biosystems). Data were extracted using Genescan (version 3.1) and Genotyper (version 2.0) (Applied Biosystems) and were checked using Genetic Analysis Software (GAS version 2.0) (A. Young). Marker haplotypes were generated within GENEHUNTER2.0 (Kruglyak et al. 1996), and all chromosomes showing an excessive number of recombination events were re-examined at the genotype level. Sex-averaged marker maps were taken from the deCODE map (Kong et al. 2002) and were supplemented with data from the human genome map (UCSC, April 2003 assembly). Final marker density was estimated at ∼10 cM across the entire length of both chromosomes (tables B1 and B2 [online only]).
Linkage Analysis
Haseman-Elston (HE) (Haseman and Elston 1972) and variance-components (VC) (Amos 1994; Pratt et al. 2000) methods were used with GENEHUNTER2.0 (Kruglyak et al. 1996) to calculate both single-point and multipoint LOD scores for both chromosomes, with the use of the ELStrans, RLStrans, NWRtrans, BRtrans, SPtrans, and RCtrans scores as quantitative measures of language ability.
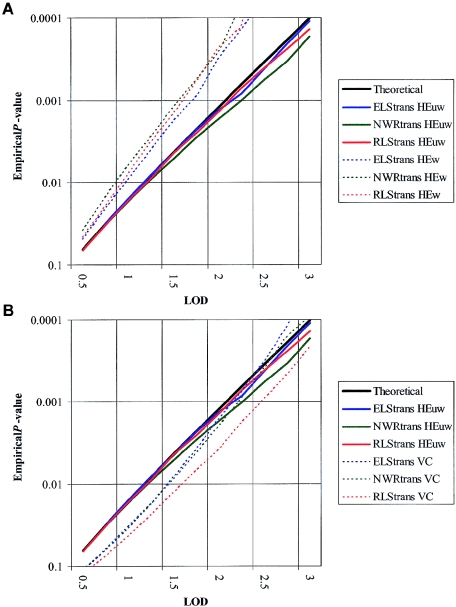
Simulations indicated that, of the three analyses performed (HE, with and without weighting, and VC), the HE method with no weighting for multiple sib pairs provided the best possible fit to theoretical statistics within our data set (fig. 1). Results hereafter, therefore, are presented for unweighted HE analyses only. In general, the results obtained by VC analyses supported those arising from HE. Full VC results can be found on the Wellcome Trust Center for Human Genetics (WTCHG) Web site.
Figure 1.
LOD-score significance distributions for different linkage methods. Simulations demonstrate that the unweighted HE method (HEuw; solid lines) gives a better fit to the theoretical distribution (black line) than either the weighted HE (HEw; graph A) or the VC (graph B) methods (dotted lines). Data is shown for the total data set.
The analyses were performed in three distinct stages. The chief aim of this study was to replicate the findings of the SLIC genome screen on chromosomes 16 and 19. Primary analyses, therefore, involved the wave 2 replication set (table 1) and the three language-related traits used in the original investigation (ELStrans, RLStrans, and NWRtrans).
Following this initial analysis, the wave 1 and wave 2 data sets were pooled to allow an extension study of linkage within the regions of interest. This strategy follows the guidelines suggested by Lander and Kruglyak (1995) for cases in which the original linkage was suggestive (MLS < 3.6). Linkage was assessed also for each of the constituent cohorts (Guys/Aberdeen, Cambridge, and Edinburgh) across chromosomes 16 and 19. Finally, three measures of reading ability (BRtrans, SPtrans, and RCtrans) were analyzed for linkage to chromosomes 16 and 19, both in the combined sample and in the various subgroups.
Simulations
Simulations were used to assess the suitability of the three analysis options available (HE, with and without weighting for multiple sib ships, and VC) and to calculate significance values for the LOD scores obtained.
Pedigree structure and phenotype data were maintained for each family within the sample, and SIMULATE (J. Terwilliger) was used to generate random genotypes for a single marker with four equally frequent alleles (75% heterozygosity) within this framework. A total of 100,000 replications were run, and linkage was assessed for each with the use of VC, weighted HE, and unweighted HE approaches.
The LOD significance distributions arising from such simulations are applicable generally for estimating the pointwise significance of linkage peaks (Fisher et al. 2002). As recommended by Lander and Kruglyak (1995), intervalwide significance was derived from these pointwise P values by performance of a Bonferonni correction for the appropriate number of markers tested within each region (i.e., P/40 for chromosome 16 and P/21 for chromosome 19). Note that such an adjustment is considered generally to be overly conservative, since the marker data are not truly independent.
Results
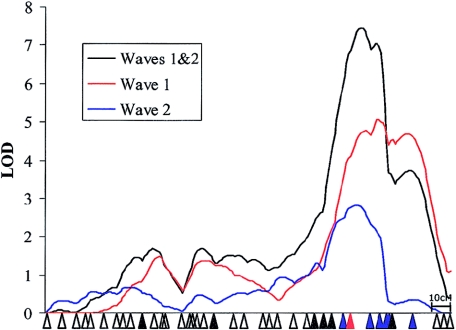
Analysis of the three language-related measures within the wave 2 cohort demonstrated independent linkages at both the SLI1 and SLI2 loci (figs. 2 and 3, respectively). Linkage to chromosome 16 (MLS = 2.84) was found with the same trait (NWRtrans) and in the same region as that described in the original study (SLIC 2002) (fig. 2). Linkage to chromosome 19 (MLS = 2.31) also coincided with that reported in the original genome screen (SLIC 2002) but was found with an alternative language trait. Whereas the SLIC study (2002) reported that the chromosome 19 linkage appeared to be specific to ELStrans, the wave 2 analysis found that all the chromosome 19 linkage came from the NWRtrans trait, with only a nominal level of linkage to ELStrans (MLS = 0.27) (fig. 3).
Figure 2.
HE linkage to chromosome 16 within the wave 1 and wave 2 samples. Linkage to chromosome 16 within the replication sample (wave 2) was found, with the NWRtrans trait in a similar region to that seen in the genome screen (wave 1). The positions of all markers are shown by triangles. Markers that gave single-point LOD scores >1.0 are shown in black, markers that gave single-point LOD scores >2.0 are shown in blue, and the marker that gave the maximum single-point LOD score (D16S3040; HE MLS = 4.41, VC MLS = 2.73) is shown in red. Single-point data refers to the total data set. Plots are shown only for HE analyses and the NWRtrans trait. Note that the linkage in wave 1 differs from that reported in SLIC 2002, owing to the addition of missing data points and to the use of different genetic maps.
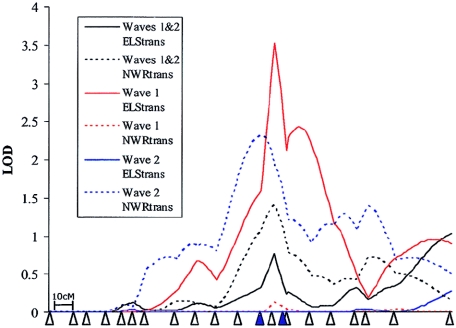
Figure 3.
Linkage to chromosome 19 within the wave 1 and wave 2 samples. Linkage to chromosome 19 within the replication sample (wave 2) was found with the NWRtrans trait in a similar region to that of ELStrans seen in the genome screen (wave 1). The positions of all markers are shown by triangles. Markers that gave single-point LOD scores >1.0 (waves 1 and 2) are shown in black. Single-point data refers to the total data set. Plots are shown only for HE analyses and the NWRtrans and ELStrans traits. Note that the linkage in wave 1 differs from that reported in SLIC 2002, owing to the addition of missing data points and to the use of different genetic maps.
Simulations of the wave 2 data set indicated that both linkages were significant at the 2% level (chromosome 16, HE empirical intervalwide P=.0108; chromosome 19, HE empirical intervalwide P=.019).
Amalgamation of the wave 1 and wave 2 data sets resulted in a strengthening of the linkage to chromosome 16 beyond the accepted threshold indicative of “highly significant linkage” (i.e., LOD >5.4) (Lander and Kruglyak 1995) (fig. 2). This locus was supported by both multipoint (MLS = 7.46) and single-point (MLS = 4.41) linkage analyses. D16S3040 yielded the most significant single-point LOD and was found to be flanked by a cluster of 11 markers, all with single-point LOD >1.0 (fig. 2). A multipoint LOD score of this magnitude was never observed in 100,000 simulations, resulting in an interval-corrected significance of P<.0004. Within this region, only a negligible level of linkage was seen to either the ELStrans (MLS = 0.39) or RLStrans (MLS = 0.22) traits.
In contrast, although linkage was observed to chromosome 19 within the wave 2 sample, the fact that this was with a phenotype alternative to that seen in the genome screen led to a weakening of both results within the amalgamated sample. In the combined sample, a similar level of linkage was found to both the NWRtrans (MLS = 1.40) and ELStrans (MLS = 1.02) traits, with two markers under the peak generating a single-point LOD >1.0 (D19S420 and D19S606) (fig. 3). None of these LODs were found to be significant at the 5% level.
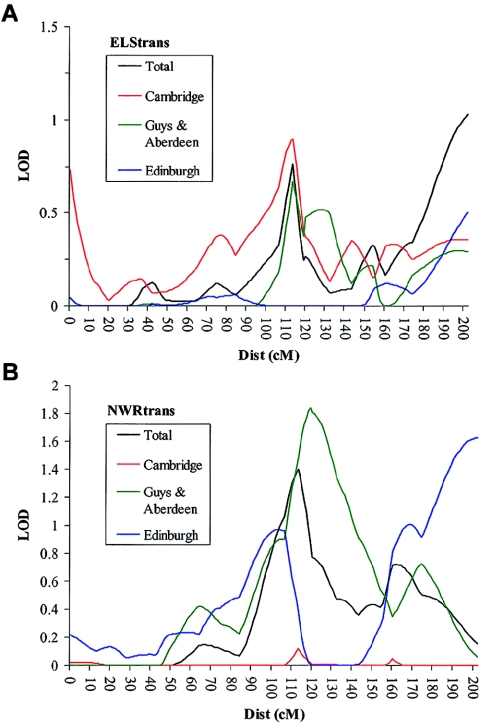
To clarify the contribution that each cohort made to each of the linkage peaks, we divided the entire sample into its constituent groupings (Guys/Aberdeen, Cambridge, and Edinburgh) and reanalyzed both regions. These analyses indicated that each group contributes to the peak of linkage on both chromosomes 16 and 19 (figs. 4 and 5). On chromosome 19, however, the Cambridge sample shows linkage to ELStrans, the Edinburgh cohort appears to be linked primarily to the NWRtrans trait, and the Guys/Aberdeen group is linked to both measures (fig. 5).
Figure 4.
Linkage to chromosome 16 within the constituent groups. Linkage is demonstrated independently within each independent cohort. Traces are shown only for HE analyses and the NWRtrans trait.
Figure 5.
Linkage to chromosome 19 within the constituent groups. Linkage is demonstrated independently within each independent cohort. A, Linkage to ELStrans on chromosome 19. B, Linkage to NWRtrans on chromosome 19. The Cambridge cohort shows linkage to ELStrans only, the Edinburgh cohort shows linkage to NWRtrans only, and the Guys/Aberdeen group shows linkage to both ELStrans and NWRtrans. In A and B, traces are shown for HE analyses and the ELStrans or NWRtrans trait, respectively.
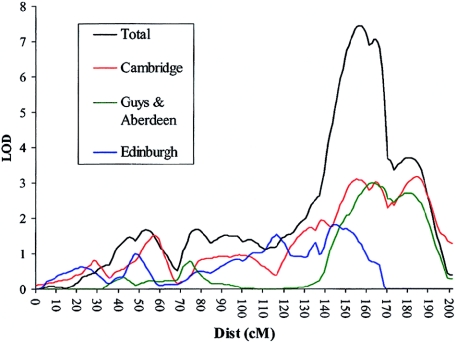
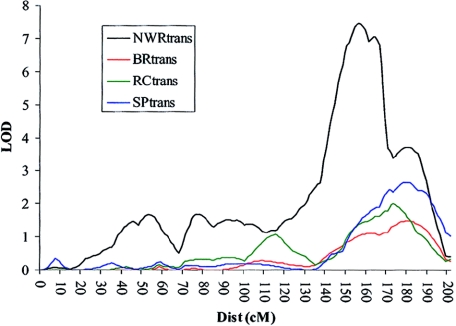
Analyses of the additional WORD phenotypes (BRtrans, RCtrans, and SPtrans) within the entire sample provided further support for the linkage on chromosome 16, with all three reading measures showing linkage to this region (fig. 6). SPtrans provided the highest LOD in this region, with a MLS of 2.67. The remaining two reading scores (BRtrans and RCtrans) yielded MLS of 1.49 and 1.99, respectively. None of the reading measures were linked to chromosome 19 (SPtrans MLS = 0.46, RCtrans MLS = 0.16, and BRtrans MLS = 0.07). A summary of all the LOD scores can be found in table 5.
Figure 6.
Linkage of reading measures to chromosome 16. Linkage was also found between three measures of reading ability on chromosome 16q. Traces are shown for HE analyses only.
Table 5.
MLS for Chromosomes 16 and 19 in All Sample Groups
|
Linkage |
||||||
| Multipoint |
Single-Point |
|||||
| Chromosome and Sample Group | MLS | Trait | Position(cM) | MLS | Trait | Marker |
| 16: | ||||||
| Waves 1 and 2 | 7.46 | NWRtrans | 156 | 4.41 | NWRtrans | D16S3040 |
| Wave 1 | 5.07 | NWRtrans | 165 | 3.52 | NWRtrans | D16S3091 |
| Wave 2 | 2.84 | NWRtrans | 153 | 1.45 | NWRtrans | D16S534 |
| Cambridge | 3.17 | NWRtrans | 184 | 2.75 | NWRtrans | D16S3040 |
| Guys/Aberdeen | 2.99 | NWRtrans | 163 | 1.60 | NWRtrans | D16S505 |
| Edinburgh | 1.81 | NWRtrans | 144 | 1.61 | NWRtrans | D16S3040 |
| 19: | ||||||
| Waves 1 and 2 | 1.40 | NWRtrans | 113 | 1.45 | NWRtrans | D19S606 |
| Wave 1 | 3.52 | ELStrans | 113 | 2.51 | ELStrans | D19S908 |
| Wave 2 | 2.31 | NWRtrans | 107 | 2.59 | NWRtrans | D19S420 |
| Cambridge | .90 | ELStrans | 113 | .46 | ELStrans | D19S908 |
| Guys/Aberdeen | 1.84 | NWRtrans | 119 | 2.18 | NWRtrans | D19S606 |
| Edinburgh | .97 | NWRtrans | 103 | 2.23 | NWRtrans | D19S572 |
Discussion
This report describes the further investigation of two QTLs previously implicated in the complex disorder of Specific Language Impairment. A significant level of linkage (P<.02) was demonstrated to both chromosomes 16 and 19 within an independent wave of families affected by SLI (figs. 2 and 3), and a highly significant level of linkage (P<.0004) was found at the chromosome 16 locus within the total data set, consisting of both the wave 2 sample and the original SLIC genome-screen cohort (fig. 6). Separation of the sample into its constituent groupings demonstrated that each area of linkage was contributed to by all cohorts (figs. 4 and 5).
The most striking outcome of this study is the chromosome 16 replication. This locus was found to be linked to NWR, BR, RC, and SP within both the genome screen and the wave 2 samples, resulting in an MLS of 7.46 to the NWR trait within the total data set. However, no linkage was observed between this region and the two CELF measures studied (ELS and RLS).
The NWR test was developed to test the capacity of the phonological short-term memory for novel speech sounds. It has been proposed that a deficit in this memory may underlie many cases of language impairment. Under such a model, children with SLI are unable to hold unfamiliar phonological forms in their short-term memory for a sufficient amount of time to allow in-depth processing and transfer to the long-term memory (Baddeley and Wilson 1993; Baddeley et al. 1998).
In a computer-based segregation analysis of two short-term memory tasks (digit span and NWR) in dyslexic subjects, Wijsman et al. (2000) indicated that different specific aspects of working memory may be under the control of alternative QTLs. They estimated that ∼2–3 QTLs affect NWR performance. When they tried to fit a genetic model to the NWR task, the most parsimonious model involved a major gene pattern of inheritance, with a few modifying loci with smaller effects. Furthermore, they suggested that at least one of these QTLs may influence a memory component that is unique to the NWR task and does not play a role even in digit span, which is usually conceived of as a highly related test. Given the proposed specificity of such QTLs, it may be not surprising that the CELF measures show no linkage to chromosome 16. The CELF scores were selected for the genome screen because they are derived from an established test battery for assessing language and, therefore, are able to characterize an individual’s performance across a whole range of cognitive and linguistic processes, while maintaining a minimum number of phenotypes for analysis. However, such an approach has the potential disadvantage that the phenotypes measured by the CELF-R scales may be rather distant from the underlying neurocognitive processes. Conversely, specific measures, such as NWR, could be construed as neurolinguistic “endophenotypes.” Although such measures still rely upon an array of neural processes, they are likely to be more closely related to the underlying genetic determinants. From this view, distinct as well as overlapping linkage signals for the two quantitative measures would be expected. Other studies indicate that there may exist shared genetic influences between performance on NWR tests and measures of literacy (Bishop 2001; Hsu et al. 2002). Hsu et al. (2002) used reciprocal aggregation analyses to investigate a series of measures, including nonword memory and spelling (WRAT3 spelling subtest), in dyslexic individuals. Their results indicated that phonological short-term memory, as assessed by NWR, may account for much of the familial aggregation pattern of the spelling measure. They concluded that these phonological processes may exert an influence on orthographic processes, which are important in reading tasks. Interestingly, the present study found that the area of linkage to NWR was also linked to three measures of reading ability, with the highest LOD coming from the SP measure. This is the first study to demonstrate linkage between measures of reading ability and chromosome 16q. However, it should be noted that only two full-genome screens have been completed with reading-disabled sib pairs (Fisher et al. 2002) and that neither of these used the WORD battery. It is widely reported that the deficits that underlie speech and language impairments may also render an individual at an increased risk for the onset of dyslexia. However, since the overlap between dyslexia and SLI is far from absolute, the existence of distinct etiological mechanisms is also expected (Snowling et al. 2000). Thus, one may postulate that the selection of a sample on the basis of language abilities may lead inadvertently to the creation of a more homogeneous group of individuals, in terms of reading impairment, than a strategy on the basis of reading ability alone. Such a sample, therefore, would have an increased power to detect the genetic loci underlying those specific forms of dyslexia. Recent studies suggest that those individuals with speech disorders and phonological impairments may be at the greatest risk for developing reading problems (Snowling et al. 2000). Thus, given that the chromosome 16q locus was detected with the use of a measure of phonological short-term memory, it may be not surprising that this region also was found to be linked to reading ability.
The region implicated on chromosome 16 is large in size and spans from D16S515 to D16S413. However, although this region extends over 60 cM, it measures only 11.3 Mb on the physical map. This area contains ∼75 genes, a number of which represent good candidates for involvement in SLI. These include CDH13 (MIM 601364), which encodes Cadherin 13, a calcium-dependent glycoprotein; KCNG4 (MIM 607603), which encodes a voltage-gated potassium channel with strong expression in the brain; and JPH3 (MIM 605268), which encodes Junctophilin3, a component of junctional complexes, and various zinc finger proteins.
In contrast with chromosome 16, chromosome 19 was found to be linked to two separate language-related measures (NWR and ELS). In the original screen, linkage to chromosome 19 appeared to be specific to the ELS trait (SLIC 2002). Within wave 2, however, this region appeared to be linked only to NWR. Dissection of the sample into its constituent groupings demonstrated that, although the Cambridge sample showed linkage specific to the ELS trait, Edinburgh, which formed the majority of the wave 2 sample, was linked only to the NWR measure, whereas Guys/Aberdeen showed linkage to both phenotypes. This divergence resulted in a minimal level of linkage to both ELS and NWR within the combined sample.
In the CELF battery, the specific combination of subtests that are applied depends primarily on the age of the subject. For children <8 years of age, one of the three subtests (sentence assembly) on the expressive scale is substituted for a more suitable test (word structure). In the Cambridge group, which shows the most consistent linkage to the ELS trait, all individuals were age ⩾8 years when tested. In the Edinburgh and Guys/Aberdeen cohorts, a significant proportion of children (∼30% and ∼20%, respectively) were age <8 years and would have completed the alternative subtest. Thus, it would appear that as the proportion of children age <8 years in each subgroup increases, the degree of linkage to the ELS trait decreases (fig. 5).
In the study of polygenic traits, discrepancies between results are not uncommon (Grigorenko et al. 1997; Fisher et al. 1999; Fisher and DeFries 2002) and may be due to chance uncontrolled factors and low power caused by small sample sizes. It should be noted that, in univariate linkage analyses, the magnitude of linkage does not necessarily provide a reliable reflection of the effect of a genetic locus on a given phenotype, as the extent of linkage may be also affected by a number of variables that are independent of the genetic effects (e.g., the sensitivity of the psychometric test within the given sample and the distribution, ascertainment, and size of the sample) (Fisher and DeFries 2002). Thus, given the diversity in the origins of each of our samples, a certain amount of divergence between the results may be expected. As discussed above, these results demonstrate the difficulty of delineating a complex syndrome, such as SLI, into a small number of relevant quantitative scores.
Concerns regarding the variation in results between and within studies may be further evaluated with the use of multivariate linkage analyses. Such methods apply a VC model, as described in “Subjects and Methods,” to detect loci underlying QTL measures. However, in addition to the trait variances, multivariate techniques also consider the covariances between traits. Thus, such techniques not only enable the investigation of multiple traits relating to a given phenotype, they also allow the exploration of the relationships between those measures at a specific QTL (Marlow et al. 2003). It is hoped that such multivariate analyses will allow a more thorough investigation of the subtests comprising the CELF scales and will aid in the clarification of the relationships between the factors underlying both the chromosome 16 and the chromosome 19 loci.
In summary, the present study has demonstrated a replication of the elsewhere reported linkage between NWR and chromosome 16q and has extended this finding to three reading-related measures within the same area. Furthermore, analysis of this region within a pooled sample of 184 language-impaired families resulted in a LOD of 7.46, representing one of the most significant findings to date for a complex behavioral trait.
Investigation of chromosome 19q implied that this locus is also likely to play a role in the genetic determination of susceptibility to language impairment, although perhaps in a more complex manner than that underlying SLI1. It is hoped that the further characterization of both of these loci may allow the identification of genes underlying cases of language impairment and, thus, promote a better understanding of the processes involved in language acquisition and aid in the diagnosis and treatment of individuals affected by this disorder.
Acknowledgments
First, we would like to thank all the families who have participated in the study and the professionals who continue to make this study possible. We thank everyone at the Newcomen, CLASP, Edinburgh, and Aberdeen centers for their involvement in the project and Leila Jannoun, Jane Addison, Claire Craven, Deborah Jones, Tilly Storr, and Til Utting-Brown for their assistance with data collection and management. All laboratory work and the collection of families from Guy's Hospital were funded by the Wellcome Trust. CLASP is funded by the Wellcome Trust, British Telecom, the Isaac Newton Trust, an NHS Anglia & Oxford Regional R&D Strategic Investment Award, and an NHS Eastern Region R&D Training Fellowship Award. The Edinburgh group was supported by the Chief Scientist’s Office, Scotland. The Aberdeen group was supported by grants from Grampian Healthcare Trust and Grampian Primary Care NHS Trust. A.P.M. and D.V.M.B. are Wellcome Trust Principal Research Fellows. S.E.F. is a Royal Society Research Fellow.
Appendix A: Members of the SLIC
The Wellcome Trust Centre for Human Genetics, Oxford: D. F. Newbury, J. D. Cleak, E. Banfield, A. J. Marlow, S. E. Fisher, and A. P. Monaco; Cambridge Language and Speech Project (CLASP): C. M. Stott, M. J. Merricks, and I. M. Goodyer; Child and Adolescent Psychiatry Department and Medical Research Council Centre for Social, Developmental, and Genetic Psychiatry, Institute of Psychiatry: P. F. Bolton; Newcomen Centre, Guy’s Hospital: V. Slonims and G. Baird; Department of Child Health, the University of Aberdeen: A. Everitt, E. Hennessy, M. Main, and P. Helms; The Raeden Centre and Grampian University Hospitals Trust: A. D. Kindley; Speech and Language Sciences, Queen Margaret University College: A. Hodson and J. Watson; Department of Reproductive and Developmental Sciences, University of Edinburgh: A. O’Hare; Molecular Medicine Centre, University of Edinburgh: W. Cohen, H. Cowie, J. Steel, A. MacLean, and J. Seckl; Academic Unit of Neurology, University of Sheffield Medical School: J. Nasir; Department of Experimental Psychology, University of Oxford: D. V. M. Bishop; Human Communication and Deafness, School of Education, the University of Manchester: Z. Simkin and G. Conti-Ramsden; Biostatistics Group, School of Epidemiology and Health Science, the University of Manchester: A. Pickles.
Appendix B
Table B1.
Markers Used for Analysis of Chromosome 16
| Marker | Position(cM)a |
| D16S423 | 16.0 |
| D16S418 | 23.3 |
| D16S404 | 30.8 |
| D16S519 | 35.9 |
| D16S3102 | 37.9 |
| D16S500 | 44.3 |
| D16S3103 | 51.1 |
| D16S3056 | 54.5 |
| D16S3036 | 58.0 |
| D16S403 | 64.0 |
| D16S3068 | 70.8 |
| D16S3100 | 74.6 |
| D16S3093 | 75.3 |
| D16S753 | 83.9 |
| D16S409 | 86.8 |
| D16S3080 | 88.0 |
| D16S3136 | 90.3 |
| D16S416 | 94.1 |
| D16S3034 | 100.5 |
| D16S3112 | 110.2 |
| D16S3057 | 115.8 |
| D16S3132 | 124.7 |
| D16S3089 | 126.4 |
| D16S503 | 131.6 |
| D16S3095 | 139.2 |
| D16S512 | 146.6 |
| D16S515 | 150.7 |
| D16S3138 | 153.4 |
| D16S3049 | 158.7 |
| D16S504 | 164.3 |
| D16S3040 | 167.3 |
| D16S505 | 177.3 |
| D16S534 | 182.3 |
| D16S422 | 186.0 |
| D16S3091 | 186.4 |
| D16S402 | 188.8 |
| D16S3061 | 200.9 |
| D16S520 | 211.4 |
| D16S3123 | 215.3 |
| D16S413 | 217.9 |
Positions are given in Haldane centimorgans.
Table B2.
Markers Used for Analysis of Chromosome 19
| Marker | Position(cM)a |
| D19S886 | 0.0 |
| D19S209 | 12.7 |
| D19S216 | 19.5 |
| D19S884 | 28.1 |
| D19S865 | 36.7 |
| D19S221 | 42.2 |
| D19S226 | 48.5 |
| D19S566 | 64.2 |
| D19S882 | 74.3 |
| D19S225 | 83.9 |
| D19S224 | 95.3 |
| D19S420 | 106.3 |
| D19S908 | 112.9 |
| D19S606 | 118.9 |
| D19S902 | 120.0 |
| D19S904 | 131.8 |
| D19S553 | 142.9 |
| D19S888 | 154.5 |
| D19S572 | 160.0 |
| D19S418 | 174.1 |
| D19S210 | 202.5 |
Positions are given in Haldane centimorgans.
Electronic-Database Information
URLs for data presented herein are as follows:
- Applied Biosystems, http://www.appliedbiosystems.com
- GENEHUNTER2.0, http://www.broad.mit.edu/ftp/distribution/software/genehunter/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- SIMULATE, ftp://linkage.cpmc.columbia.edu/software/simulate
- UCSC, http://genome.ucsc.edu/
- WTCHG, http://www.well.ox.ac.uk/~dianne/wave2results (for full results of all analyses performed)
References
- Amos CI (1994) Robust variance components approach for assessing genetic linkage in pedigrees. Am J Hum Genet 54:535–543 [PMC free article] [PubMed] [Google Scholar]
- Baddeley A, Wilson BA (1993) A developmental deficit in short-term phonological memory: implications for language and reading. Memory 1:65–78 [DOI] [PubMed] [Google Scholar]
- Baddeley AD, Gathercole SE, Papagno C (1998) The phonological loop as a language learning device. Psychol Rev 105:158–173 [DOI] [PubMed] [Google Scholar]
- Badner JA, Gershon ES (2002) Regional meta-analysis of published data supports linkage of autism with markers on chromosome 7. Mol Psychiatry 7:56–66 10.1038/sj.mp.4001922 [DOI] [PubMed] [Google Scholar]
- Bartlett CW, Flax JF, Li W, Reaple-Bonilla T, Hayter J, Logue MW, Zimmerman R, Tallal P, Brzustowicz LM (2003) A genome scan of specific language impairment loci in families from the United States. Am J Hum Genet Suppl 73:A1884 [Google Scholar]
- Bartlett CW, Flax JF, Logue MW, Vieland VJ, Bassett AS, Tallal P, Brzustowicz LM (2002) A major susceptibility locus for specific language impairment is located on chromosome 13q21. Am J Hum Genet 71:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitchman JH, Browlie EB, Inglis A, Wild J, Matthews R, Schachter D, Kroll R, Martin S, Ferguson B, Lancee W (1994) Seven-year follow-up of speech/language-impaired and control children: speech/language stability and outcome. J Am Acad Child Adolesc Psychiatry 33:1322–1330 [DOI] [PubMed] [Google Scholar]
- Bishop DVM (1987) The causes of specific developmental language disorder. J Child Psychol Psychiatry 28:1–8 [DOI] [PubMed] [Google Scholar]
- ——— (2001) Genetic influences on language impairment and literacy problems in children: same or different? J Child Psychol Psychiatry 42:189–198 10.1017/S002196300100662X [DOI] [PubMed] [Google Scholar]
- Bishop DVM, Edmundson A (1986) Is otitis media a major cause of specific developmental language disorders? Br J Disord Comm 21:321–338 [DOI] [PubMed] [Google Scholar]
- Bishop DVM, North T, Donlan C (1995) Genetic basis of specific language impairment: evidence from a twin study. Dev Med Child Neurol 37:56–71 [DOI] [PubMed] [Google Scholar]
- ——— (1996) Nonword repetition as a behavioural marker for inherited language impairment: evidence from a twin study. J Child Psychol Psychiatry 37:391–403 [DOI] [PubMed] [Google Scholar]
- Bradford Y, Haines, J, Hutcheson H, Gardiner M, Braun T, Sheffield V, Cassavant T, Huang W, Wang K, Vieland V, Folstein S, Santangelo S, Piven J (2001) Incorporating language phenotypes strengthens evidence of linkage to autism. Am J Med Genet 105:539–547 10.1002/ajmg.1497 [DOI] [PubMed] [Google Scholar]
- Brown RT, Freeman WS, Perrin JM, Stein MT, Amler RW, Feldman HM, Pierce K, Wolraich ML (2001) Prevalence and assessment of attention-deficit/hyperactivity disorder in primary care settings. Pediatrics 107:E43 [DOI] [PubMed] [Google Scholar]
- Burden V, Stott CM, Forge J, Goodyer I (1996) The Cambridge Language and Speech Project (CLASP). I. Detection of language difficulties at 36 to 39 months. Dev Med Child Neurol 38:613–631 [DOI] [PubMed] [Google Scholar]
- Cantwell DP, Baker L (1987) Prevalence and type of psychiatric disorder and developmental disorders in three speech and language groups. J Commun Disord 20:151–160 10.1016/0021-9924(87)90006-2 [DOI] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC (1994) Quantitative trait locus for reading disability on chromosome 6. Science 266:276–279 (correction 268:1553) [DOI] [PubMed] [Google Scholar]
- Collaborative Linkage Study of Autism (CLSA): Barrett S, Beck JC, Bernier R, Bisson E, Braun TA, Casavant TL, Childress D, et al (1999) An autosomal genome screen for autism. Am J Med Genet 88:609–615 [DOI] [PubMed] [Google Scholar]
- Doyle AE, Faraone SV, DuPre EP, Biederman J (2001) Separating attention deficit hyperactivity disorder and learning disabilities in girls: a familial risk analysis. Am J Psychiatry 158:1666–1672 10.1176/appi.ajp.158.10.1666 [DOI] [PubMed] [Google Scholar]
- Fagerheim T, Raeymaekers P, Tønnessen FE, Pedersen M, Tranebjærg L, Lubs HA (1999) A new gene (DYX3) for dyslexia is located on chromosome 2. J Med Genet 36:664–669 [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, DeFries JC (2002) Developmental dyslexia: genetic dissection of a complex trait. Nat Rev Neurosci 3:767–780 10.1038/nrn936 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Francks C, Marlow AJ, MacPhie L, Newbury DF, Cardon LR, Ishikawa-Brush Y, Richardson AJ, Talcott JB, Gayán J, Olson RK, Pennington BF, Smith SD, DeFries JC, Stein JF, Monaco AP (2002) Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat Genet 30:86–91 10.1038/ng792 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP (1999) A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet 64:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher A, Frith U, Snowling MJ (2000) Precursors of literacy delay among children at genetic risk of dyslexia. J Child Psychol Psychiatry 41:203–213 10.1017/S0021963099005284 [DOI] [PubMed] [Google Scholar]
- Gathercole SE, Willis C, Baddeley AD, Emslie H (1994) The children’s test of nonword repetition: a test of phonological working memory. Memory 2:103–127 [DOI] [PubMed] [Google Scholar]
- Gayán J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, Pennington BF, DeFries JC (1999) Quantitative trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet 64:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Hart LA, Speed WC, Shuster A, Pauls DL (1997) Susceptibility loci for distinct components of developmental dyslexia on chromosomes 6 and 15. Am J Hum Genet 60:27–39 [PMC free article] [PubMed] [Google Scholar]
- Haseman JK, Elston RC (1972) The investigation of linkage between a quantitative trait and a marker locus. Behav Genet 2:3–19 [DOI] [PubMed] [Google Scholar]
- Hsu L, Wijsman EM, Berninger VW, Thomson JB, Raskind WH (2002) Familial aggregation of dyslexia phenotypes. II. Paired correlated measures. Am J Med Genet 114:471–478 10.1002/ajmg.10523 [DOI] [PubMed] [Google Scholar]
- Johnson CJ, Beitchman JH, Young A, Escobar M, Atkinson L, Wilson B, Browlie EB, Douglas L, Taback N, Lam I, Wang M (1999) Fourteen-year follow-up of children with and without speech/language impairments: speech/language stability and outcomes. J Speech Lang Hear Res 42:744–760 [DOI] [PubMed] [Google Scholar]
- Kaplan DE, Gayán J, Ahn J, Won T-W, Pauls D, Olson RK, DeFries JC, Wood F, Pennington BF, Page GP, Smith SD, Gruen JR (2002) Evidence for linkage and association with reading disability on 6p21.3–22. Am J Hum Genet 70:1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kovac I, Gavabedian B, Du Souich C, Palmour RM (2001) Attention deficit/hyperactivity in SLI children increases risk of speech/language disorders in first-degree relatives: a preliminary report. J Commun Disord 34:339–354 10.1016/S0021-9924(01)00054-5 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lai CSL, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP (2001) A novel forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413:519–523 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- Lander ES, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Law J, Boyle J, Harris F, Harkness A, Nye C (1998) Screening for speech and language delay: a systematic review of the literature. Health Technol Assess 2:1–184 [PubMed] [Google Scholar]
- Lewis BA, Thompson LA (1992) A study of developmental speech and language disorders in twins. J Speech Hear Res 35:1086–1094 [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A (1994) Autism diagnostic interview—revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24:659–685 [DOI] [PubMed] [Google Scholar]
- Marlow AJ, Fisher SE, Francks C, Macphie IL, Cherny SS, Richardson AJ, Talcott JB, Stein JF, Monaco AP, Cardon LR (2003) Use of multivariate linkage analysis for dissection of a complex cognitive trait. Am J Hum Genet 72:561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neils J, Aram DM (1986) Family history of children with developmental language disorders. Percept Mot Skills 63:655–658 [DOI] [PubMed] [Google Scholar]
- Nopola-Hemmi J, Myllyluoma B, Haltia T, Taipale M, Ollikainen V, Ahonen T, Voutilainen A, Kere J, Widen E (2001) A dominant gene for developmental dyslexia on chromosome 3. J Med Genet 38:658–664 10.1136/jmg.38.10.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt SC, Daly MJ, Kruglyak L (2000) Exact multipoint quantitative-trait linkage analysis in pedigrees by variance components. Am J Hum Genet 66:1153–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust J, Golombok S, Trickey G (1993) Wechsler objective reading dimensions. Psychological Corporation, Sidcup, Kent, United Kingdom [Google Scholar]
- Semel EM, Wiig EH, Secord W (1992) Clinical evaluation of language fundamentals, revised. Psychological Corporation, San Antonio [Google Scholar]
- The SLI Consortium (2002) A genomewide scan identifies two novel loci involved in specific language impairment. Am J Hum Genet 70:384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowling M, Bishop DVM, Stothard SE (2000) Is preschool language impairment a risk factor for dyslexia in adolescence? J Child Psychol Psychiatry 41:587–600 10.1017/S0021963099005752 [DOI] [PubMed] [Google Scholar]
- Snowling MJ, Adams JW, Bishop DVM, Stothard SE (2001) Educational attainments of school leavers with a preschool history of speech-language impairments. Int J Lang Commun Disord 36:173–183 10.1080/13682820010019892 [DOI] [PubMed] [Google Scholar]
- Stein CM, Schick JH, Taylor HG, Shriberg LD, Millard C, Kundtz-Kluge A, Russo K, Minich N, Hansen A, Freebairn LA, Elston RC, Lewis BA, Iyengar SK (2004) Pleiotropic effects of a chromosome 3 locus on speech-sound disorder and reading. Am J Hum Genet 74:283–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromswold K (1998) Genetics of spoken language disorders. Hum Biol 70:297–324 [PubMed] [Google Scholar]
- Tallal P, Ross R, Curtiss S (1989) Familial aggregation in specific language impairment. J Speech Hear Disord 54:167–173 [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Buckwalter PR (1998) Heritability of poor language achievement among twins. J Speech Lang Hear Res 41:188–199 [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Hafeman LL, O’Brien M (2003) Autism and autism risk in siblings of children with specific language impairment. Int J Lang Commun Disord 38:235–250 10.1080/1368282031000086363 [DOI] [PubMed] [Google Scholar]
- Wijsman, EM, Petersen D, Leutenegger, AL, Thomson JB, Goddard KAB, Hsu L, Beringer VW, Raskind WH (2000) Segregation analysis of phenotypic components of learning disabilities. I. Nonword memory and digit span. Am J Hum Genet 67:631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N (1992) Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci USA 89:5847–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]