Abstract
Small patella syndrome (SPS) is an autosomal-dominant skeletal dysplasia characterized by patellar aplasia or hypoplasia and by anomalies of the pelvis and feet, including disrupted ossification of the ischia and inferior pubic rami. We identified an SPS critical region of 5.6 cM on chromosome 17q22 by haplotype analysis. Putative loss-of-function mutations were found in a positional gene encoding T-box protein 4 (TBX4) in six families with SPS. TBX4 encodes a transcription factor with a strongly conserved DNA-binding T-box domain that is known to play a crucial role in lower limb development in chickens and mice. The present identification of heterozygous TBX4 mutations in SPS patients, together with the similar skeletal phenotype of animals lacking Tbx4, establish the importance of TBX4 in the developmental pathways of the lower limbs and the pelvis in humans.
Small patella syndrome (SPS [MIM 147891]), also referred to as “Scott-Taor syndrome,” “ischio-pubic-patellar syndrome,” “coxo-podo patellar syndrome,” or “ischiopatellar dysplasia,” is a rare autosomal-dominant disorder affecting skeletal structures of the lower limb and the pelvis (Scott and Taor 1979; Vaněk 1981; Morin et al. 1985; Dellestable et al. 1996; Poznanski 1997). Essential features for the clinical diagnosis of SPS are patellar aplasia or hypoplasia, associated with absent, delayed, or irregular ossification of the ischiopubic junctions and/or the infra-acetabular axe-cut notches (fig. 1A, 1B, and 1C) (Bongers et al. 2001). In addition, femur and foot anomalies—including a wide space between the first and second toes, short fourth and fifth rays of the feet (fig. 1D), and pes planus—may accompany SPS. Craniofacial dysmorphisms have been described in reports of four sporadic individuals with SPS and one familial case of SPS including micrognathia and/or cleft palate (Sandhaus 1987; Kozlowski and Nelson 1995; Azouz and Kozlowski 1997; Habboub and Thneibet 1997; Bongers et al. 2001). Offiah et al. (2002) proposed that “ischio-pubic-patellar syndrome” and SPS should be distinguished clinically, because the former condition presents a more severe skeletal phenotype and facial features that are not seen in SPS. SPS is clearly clinically different from autosomal-dominant familial patella aplasia hypoplasia (PTLAH [MIM 168860]) (Bernhang and Levine 1973; Mangino et al. 1999). In PTLAH, congenital aplasia or hypoplasia of the patellae are the only clinical features. PTLAH is genetically heterogeneous with one locus mapped to an ∼12-cM region on chromosome 17q22 (Mangino et al. 1999; Bongers et al. 2002). Elsewhere, we found possible linkage of SPS with this PTLAH locus on chromosome 17q22 in two SPS families, suggesting that one single gene may be involved in both disorders (families A and B in figs. 2 and 3A) (Bongers et al. 2001). Recently, patellar and pelvic anomalies similar to those of SPS and facial features as micrognathia/retrognathia and high or cleft palate were recognized in individuals with surviving campomelic dysplasia (MIM 114290) caused by mutations in the SOX9 gene, located at chromosome 17q24.3 (Mansour et al. 2002). Here, we demonstrate that mutations in the TBX4 gene underlie SPS.
Figure 1.
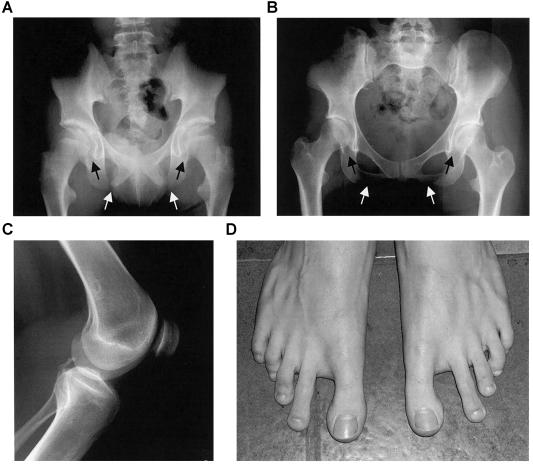
Characteristic features of the pelvis and the lower limb in individuals with SPS. A, Radiograph of the pelvis of family A's proband at the age of 12 years and 11 mo, showing bilaterally absent ossification of the ischiopubic junction (unblackened arrows), infra-acetabular axe-cut notches (blackened arrows), and elongated femoral necks. B, Radiograph of the pelvis of family C's proband at the age of 19 years and 7 mo. Note that the irregular ossification of the ischiopubic junction (unblackened arrows) and infra-acetabular axe-cut notches (blackened arrows) are less severe, as compared to the pelvic anomalies seen in the proband of family A. C, Radiograph of the knee of family C's proband at the age of 19 years and 7 mo, demonstrating a small patella. D, Feet of an affected male from family C at age 14 years and 8 mo, showing short fourth and fifth rays and an increased space between the first and second toes. (Radiograph A is reproduced from the work of Bongers et al. [2001] with permission from the BMJ Publishing Group.)
Figure 2.
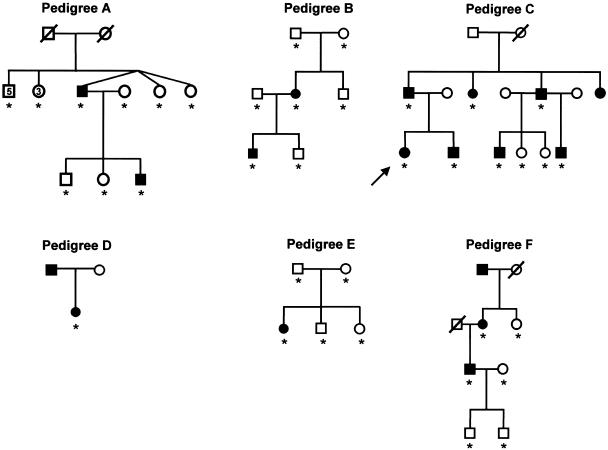
Pedigrees of six unrelated kindreds with SPS. Blackened symbols denote individuals with SPS, and unblackened symbols denote unaffected individuals. TBX4 mutation analysis was performed in all individuals indicated by an asterisk (*). The proband of the newly identified Dutch family is indicated by an arrow.
Figure 3.
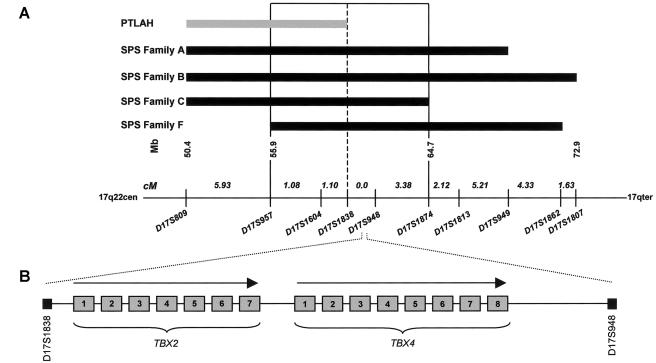
Summary of the linkage analysis and identification of positional candidate genes. A, Linkage analysis of four families with SPS and one family with PTLAH. Microsatellite markers (below the line) span an interval of 24.8 cM at chromosome 17q22. The SPS critical region (D17S957 and D17S1874) is boxed. Linkage analysis of families A and B and the clinical phenotype of families A, B, and F were described elsewhere (Vaněk 1981; Bongers et al. 2001). Previous linkage studies revealed a PTLAH candidate region at chromosome 17q22 (Mangino et al. 1999). B, Localization and schematic genomic structure of candidate genes TBX2 (exons 1–7) and TBX4 (exons 1–8) (not drawn to scale) (UCSC Genome Bioinformatics).
After informed consent and approval were obtained from the human research ethics committees of the corresponding institutions, blood samples were obtained from 15 patients who were part of five families with SPS, comprising two Dutch families reported elsewhere (families A and B) (Bongers et al. 2001), one newly identified Dutch family (family C), one Belgian family (family D) (Bongers et al. 2001), and one Czech family (family F) (Vaněk 1981); from 2 sporadic Swiss patients (families E and G) (Burckhardt 1988) (fig. 2 [family G not shown]); and from unaffected family members. The proband of the newly identified Dutch family (pedigree C in fig. 2), age 29 years at the time of examination, was referred because of dislocation of the patellae and familial occurrence of knee complaints. At physical examination, small patellae, an increased space between the first and second toes, and short fourth and fifth rays were found bilaterally. She had no facial dysmorphisms. Radiographic examination of her knees confirmed patellar hypoplasia, and radiographs of the pelvis showed irregular ossification of the ischio-pubic junction. Physical and radiographic examination of her affected family members demonstrated small patellae associated with pelvic and feet anomalies characteristic of SPS in all cases (figs. 1B–1D and 2). All patients included in this study had the classical SPS phenotype and fulfilled the essential diagnostic criteria for SPS, comprised of patellar and characteristic pelvic anomalies (Bongers et al. 2001). The phenotype of the individuals with SPS in this study ranged from small patellae, associated with irregular ossification of the ischiopubic junctions or infra-acetabular axe-cut notches, to absent patellae, severe pelvic and femur anomalies, and more obvious foot anomalies. The skeletal findings varied both within and between SPS families, except for one family (C), in which all affected individuals had patellar hypoplasia and only minor skeletal anomalies of the pelvis. A distinctive facial appearance, including a high nasal bridge, micrognathia, and a high-arched palate were found in one female patient with SPS (family D) (Bongers et al. 2001). None of the other patients showed facial dysmorphisms. Extensive clinical details and photographs of SPS families A, B, and D-G have been published elsewhere (Vaněk 1981; Burckhardt 1988; Bongers et al. 2001). Control DNA samples were obtained from 50 anonymous, unrelated Dutch individuals. Genomic DNA was extracted from peripheral lymphocytes by use of standard techniques (Miller et al. 1988).
Previous studies in families A and B showed possible linkage of SPS with a locus on chromosome 17q22 (fig. 3A) (Bongers et al. 2001). The SOX9 gene is located in this wider SPS linkage interval (data not shown). To investigate whether SOX9 mutations could be responsible for SPS, we first performed sequence analysis of the SOX9 gene (exons 1–3). No pathogenic SOX9 aberrations were found in these two families with SPS (data not shown; primers and PCR conditions are available on request).
Subsequently, two additional SPS families became available for molecular investigations. We directed our study to find evidence for linkage of SPS with the elsewhere reported candidate locus on chromosome 17q22, in a new family (family C) and in a previously described family (family F) (Vaněk 1981). Manual genotyping of microsatellite markers from chromosome 17q22 was performed as described elsewhere (Kremer et al. 1994). Haplotype analysis demonstrated linkage of SPS with the chromosome 17q22 disease locus in both families with SPS (families C and F in fig. 3A). We refined the 18.8-cM candidate-gene interval to a critical region of 5.6 cM between microsatellite marker D17S957, centromerically, and D17S1874, telomerically, with a 2.2-cM overlap with the PTLAH candidate region (fig. 3A). The SOX9 gene is located 6 Mb distal from the telomeric flanking marker, D17S1874, of this critical interval.
Inspection of genes in the SPS critical region revealed the TBX2 and TBX4 genes as positional candidates (fig. 3A and 3B). TBX2 and TBX4 belong to the T-box gene family, encoding transcription factors characterized by a strongly conserved DNA-binding motif (T-box domain) (Yi et al. 2000). Tbx2 is involved in forelimb and hindlimb development and in the specification of posterior digit identity in chickens (Gibson-Brown et al. 1998; Suzuki et al. 2004). Studies in mice and chickens have revealed a crucial role for Tbx4 in the development of the lower limbs (Rodriguez-Esteban et al. 1999; Naiche and Papaioannou 2003; Takeuchi et al. 2003). In chickens, it has been shown that misexpression of dominant-negative Tbx4 results in legless phenotypes and in a deformed and hypoplastic pelvis, including absent pubis (Takeuchi et al. 2003). In Tbx4 null mice, the initial patterning of the hindlimb bud appears normal, but further hindlimb development and outgrowth does not occur (Naiche and Papaioannou 2003). These Tbx4 animal models clearly resemble the phenotype of individuals with SPS. On the basis of these findings, we hypothesized that mutations in the human homologue of Tbx4 result in SPS. We sequenced TBX4 (exons 1–8) in the four families mapping to 17q22, in one familial case, and in the two unrelated sporadic individuals with SPS (primers and PCR conditions are available on request). We identified six different heterozygous mutations in the TBX4 gene in five families and in one sporadic individual with SPS (fig. 4A, 4D). These include one nonsense, two missense, one frameshift, and one splice-site mutation and the skipping of exon 7 of the TBX4 gene; all of these mutations were absent in 100 control chromosomes. In family A, a G→T transversion at nucleotide 743 that predicts a G248V substitution in the T-box domain was found (fig. 4A). Glycine 248 is evolutionarily strongly conserved among the T-box genes of many species (fig. 4B). A structural model of the T-box predicts that substitution of the small glycine 248 by a valine will create steric hindrance that impairs DNA binding by the neighboring lysine 247 (fig. 5). The G248V mutation is also present in the affected son but not in any of the healthy relatives (fig. 4C). Haplotype analysis revealed that G248V is a de novo mutation (pedigree A in fig. 4C), which is compelling evidence for the G248V mutation being causative for SPS in this family. In family B, a C→T transition at nucleotide 184 was found. This mutation is predicted to result in premature termination of the protein (Q62X) with abolition of the T-box domain (fig. 4A). This mutation occurred de novo in the germline of one of the healthy grandparents and cosegregates with the disease in the affected offspring. In one new family with SPS, a 1592A→G transition in exon 8 that predicts a missense mutation (Q531R), which cosegregates with the disease, was identified (family C in fig. 4A). Glutamine 531 is strictly conserved between T-box genes of various species (fig. 4B), strongly suggesting its causative role in the disease. In a familial case with SPS, an insertion-deletion (Indel) consisting of an 18-nt deletion (13 nt from intron 2 and 5 nt from exon 3) and an AGC triplet insertion was found at the 3′ splice site of intron 2 (family D in fig. 4A). Finally, one of the two sporadic individuals with SPS carried an insertion 1112_1113C in intron 8, which is predicted to result in a truncated protein lacking 75 amino acids (family E in fig. 4A). Seven different polymorphisms were identified in seven SPS kindreds and 100 control chromosomes (table 1). These sequence variants were considered nonpathogenic for SPS, since they were identified in both affected and unaffected individuals from multiple families, in 50 control individuals, and in addition to clear pathogenic mutations in SPS patients. Moreover, some of the sequence variants are within introns or result in synonymous substitutions. In family F, in which SPS is linked to chromosome 17q22 (fig. 3A), and in one sporadic individual with SPS (a member of family G), sequence analysis of the TBX4 exons and intron-exon boundaries did not reveal any pathogenic mutation. The possibility of a causative deletion of the entire TBX4 gene was excluded in family F by the identification of heterozygous polymorphisms within this gene (table 1). To identify small deletions or alternatively spliced exons, we performed an RT-PCR of TBX4 in families F and G. Total RNA was prepared from EBV-transformed lymphoblastoid cell lines of families F and G by use of the Rneasy Midi kit (Qiagen) according to the manufacturers' conditions. In the examined affected individuals of family F, RT-PCR analysis of the fragment encompassing the coding sequences of exons 5–8 showed an aberrantly sized fragment of 172 bp, which was absent in the unaffected members of this family (fig. 4D). Sequencing of the latter fragment demonstrated skipping of exon 7, resulting in the absence of 230 nt. This leads to a shift in the reading frame that is predicted to create a truncated protein of 307 amino acids, of which the 44 C-terminal residues are generated from the incorrect reading frame. It is possible that a small deletion encompassing exon 7 is responsible for this abnormal TBX4 transcript, as sequencing of genomic DNA showed no mutations in the splice sites and introns flanking exon 7. Alternatively, disruption of splice-enhancer sequences, buried further into an intron of the TBX4 gene, could be causative for the exon 7 skipping. In the sporadic individual in which no mutation was identified, mutations in regulatory elements, introns, alternatively spliced exons, or small deletions (exons 1–4) within TBX4 may underlie SPS. A more tempting explanation is that mutations in the positional candidate gene TBX2 may be responsible for the SPS phenotype, as TBX2 expression seems to be partly regulated by TBX4 (Naiche and Papaioannou 2003). However, TBX2 sequence analysis revealed no pathogenic mutations in family G (primers and PCR conditions are available on request).
Figure 4.
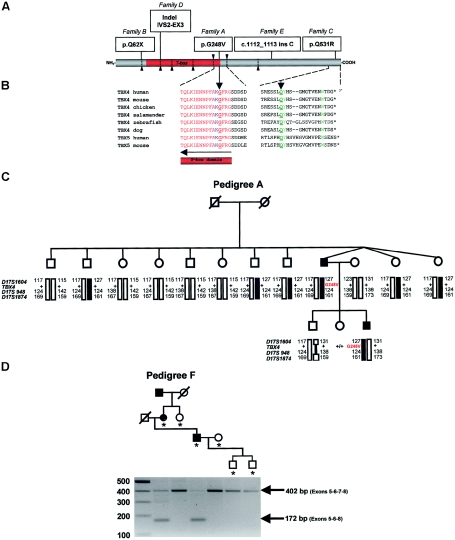
Molecular analysis of TBX4. A, Distribution of TBX4 gene mutations identified by sequencing. Exon boundaries are identified by arrowheads and dotted lines, and the DNA-binding motif (T-box domain) is colored red. B, Sequence homology analysis of amino acids 248 and 531 and flanking residues (NCBI BLAST). The strongly conserved amino acids of the T-box domain and the C-terminal domain are indicated in red and green colors, respectively. The amino acids that directly contact the DNA are depicted in blue. Glycine 248 and glutamine 531 (amino acids underlined with an arrow) are strongly conserved. C, Segregation of G248V mutation in SPS pedigree A (Bongers et al. 2001). The mutations are present in affected individuals (blackened symbols, G248V) and absent in their healthy siblings (unblackened symbols, +/+). Haplotypes are defined by markers D17S1604, D17S948, and D17S1874 and are encoded by numbers. Blackened bars indicate the risk allele. Note from the mutation and haplotype analysis that the mutation has occurred de novo in the germline of one of the grandparents. D, Segregation of exon 7 skipping in SPS pedigree F (Vaněk, 1981) detected by RT-PCR analysis of the fragment encompassing exons 5–8 of the TBX4 coding region, showing an aberrant sized transcript of 172 bp in the affected individuals on agarose gel. The RT-PCR experiment was performed in all individuals indicated by an asterisk (*).
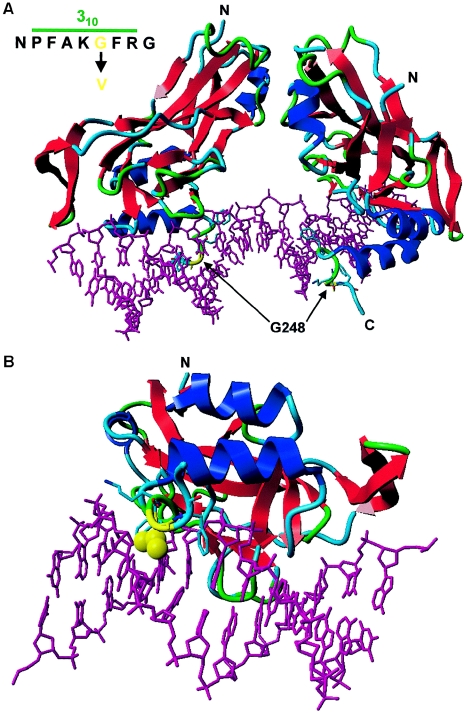
Figure 5.
Protein model of the T-box domain of TBX4, created with the help of the modelling methods described by Chinea et al. (1995) and the resolved structure of TBX3 as template (Coll et al. 2002) (Protein Data Bank). The T-box domains of TBX3 and TBX4 show a high degree of homology (80%) and identity (64%), which ensures a faithful prediction of the TBX4 structure. The 310 helical axis of the T-box domains of TBX3 and TBX4 are 100% identical. The residues of the DNA are represented as a purple stick model. Protein is shown as a ribbon model with red strands, blue helices, and green/turquoise turns and loops. The protein-DNA complex is viewed along the T-box. A, T-box dimer binding to a DNA consensus sequence. Residue G248 (yellow) is located in the C-terminal 310 helix that interacts with the DNA in the minor groove. B, View along the 310 helical axis of the T-box domain. The G248V mutation leads to the introduction of a hydrophobic side chain (yellow atoms in the ball display) that is normally absent. This likely displaces water molecules that stabilize the protein-DNA interaction through hydrogen bonding. Therefore, the G248V mutation is predicted to destabilize the TBX4-DNA interaction. Other highlighted amino acids include F245, K247, and F249 (stick representations).
Table 1.
Sequence Anomalies and Small Deletions Found in the TBX4 Gene in SPS Families and Control Chromosomes
|
Presence of Change |
||||
| DNA Changea | Predicted Effect | Location | SPS Familyb | Control Chromosomesc |
| Likely pathogenic mutations: | ||||
| 184C→T | Q62X | Exon 1 | B | |
| IVS2-13_284del18insAGC | Abnormal splicing | Intron 2, Exon3 | D | |
| 743G→T | G248V | Exon 6 | A | |
| Not detectedd | Skipping exon 7 | Exon 7 | F | |
| 1112_1113insC | Frameshift | Exon 8 | E | |
| 1592A→G | Q531R | Exon 8 | C | |
| Likely nonpathogenic polymorphisms: | ||||
| 17G→C | G6A | Exon 1 | A, E, F | + |
| 104C→T | A35V | Exon 1 | + | |
| 276T→G | A92A | Exon 2 | A, E, F | |
| IVS3-8G→A | None | Intron 4 | D | |
| IVS6-18T→C | None | Intron 7 | E | |
| 1224C→T | D408D | Exon 8 | + | |
| 1446C→T | V482V | Exon 8 | A, B, D, F | + |
No mutation had been identified in family G.
A plus sign (+) indicates that the polymorphism was found in the control chromosomes.
The genomic basis of exon 7 skipping remains undefined because the splice sites and intronic sequences flanking exon 7 were normal.
The precise pathogenic mechanism by which TBX4 mutations cause dominant SPS remains to be elucidated. Haploinsufficiency appears to be responsible for dominantly inherited SPS in at least some of the families. It is highly likely that the Q62X and the Indel mutation will create null alleles. Furthermore, protein modeling predicts that the G248V mutation partially or completely disrupts DNA binding activity. Mice embryos homozygous for the Tbx4 null allele die because of failure of chorioallantoic fusion, whereas heterozygous mutants appear to have a normal phenotype (Naiche and Papaioannou 2003). The observation that the human heterozygous phenotype resembles the phenotype of Tbx4 null mice may reflect differences in dosage sensitivity for this gene in the two species. On the other hand, dominant-negative effects for some of the other TBX4 mutations cannot be excluded.
The identification of TBX4 mutations and exclusion of SOX9 mutations in the present SPS families indicate that SPS and surviving campomelic dysplasia are genetically heterogeneous disorders. We found that TBX4 mutations account for both an SPS familial case with a distinctive facial appearance and SPS families without facial features. The sporadic reports on minor facial features in individuals with skeletal features of SPS do not allow any conclusions on the previous suggestion that “ischio-pubic-patellar syndrome” and SPS are clinically heterogeneous disorders (Offiah et al. 2002). Intrafamilial variability of patellar, pelvic, and foot anomalies was found in all SPS families investigated in this study. At present, evidence for a genotype-phenotype correlation is lacking. Molecular analysis of additional individuals with SPS will facilitate further delineation of the phenotypic spectrum of SPS and, finally, will contribute to the question as to whether craniofacial anomalies are part of the syndrome.
Our studies render a role for TBX4 in PTLAH highly unlikely. First, the TBX4 gene is located outside the PTLAH critical region: D17S1838 is >80 kb proximal to TBX4. Second, we could not identify a causative TBX4 mutation in one PTLAH family (data not shown). However, disruption of long-range TBX4 regulatory elements in PTLAH cannot be formally excluded as yet, and we have to keep in mind that PTLAH is genetically heterogeneous (Bongers et al. 2002).
Mutations in several T-box genes are associated with human developmental disorders, including TBX1 with DiGeorge syndrome, TBX22 with isolated cleft palate, TBX3 with ulnar-mammary syndrome (UMS [MIM 181450]), and TBX5 with Holt-Oram syndrome (HOS [MIM 142900]) (Bamshad et al. 1997; Basson et al. 1997; Packham and Brook 2003). In both UMS and HOS, mutated TBX genes cause disrupted upper limb development (Khan et al. 2002; Agarwal et al. 2003). Critical stage-dependent roles of Tbx4 in lower-limb initiation and outgrowth were shown by misexpression studies of dominant-negative Tbx4 in chickens. A legless phenotype was seen after early disruption of development, and truncated legs and hypomorphic distal structures were seen during Tbx4 misexpression at later developmental stages (Takeuchi et al. 2003). Pelvic anomalies were also observed upon late misexpression of Tbx4. The SPS phenotype suggests that human TBX4 mutations do not affect early steps in limb development, such as limb-bud initiation, but do have a profound effect at later stages. The late effect of TBX4 mutations is manifested by patellar aplasia/hypoplasia and anomalies of the posterior structures of the foot, such as short fourth and fifth rays. Completion of ischiopubic formation is another very late process in human development that is disrupted in individuals with SPS. The type and location of TBX4 mutations reported here are comparable to those of heterozygous mutations in TBX3 and TBX5, found in UMS and HOS, respectively. UMS and HOS are characterized by severe upper-limb anomalies, including phocomelia, as well as several other developmental defects. The relatively mild limb phenotype and the absence of other developmental anomalies in SPS suggest a higher degree of redundancy for TBX4 in comparison to TBX3 and TBX5.
Our data provide the first evidence for a crucial role of human TBX4 in the skeletal development of the patella, pelvis, and feet. Genes underlying syndromes characterized by patellar aplasia or hypoplasia appear to be involved in a common regulatory pathway of the patterning of dorsal structures of the lower limb. Heterozygous mutations in the LIM-homeodomain transcription factor LMX1B are responsible for nail-patella syndrome (MIM 161200). LMX1B cooperates with specific downstream PTX genes (Dreyer et al. 1998; Smidt et al. 2000). Pitx1, in turn, encodes a transcription factor that acts upstream of Tbx4 in hindlimb development (Logan and Tabin 1999). Mutations in two genes from this signaling cascade, LMX1B and TBX4, lead to abnormal patella formation. Therefore, it is very possible that other genetic disorders with patellar anomalies, such as PTLAH and Meier-Gorlin syndrome (also named “ear, patella, short stature syndrome” [MIM 224690]) are caused by a molecular defect in the same PTX/TBX signaling cascade.
Acknowledgments
We thank the patients and their families for their cooperation, B. van den Helm for cell culturing, E. van Beusekom for help with the linkage studies, W. Nillesen and A. Rikken for SOX9 mutation analysis, and J. van den Ende for providing material from one affected individual. This work was supported by the Council for Medical and Health Research of the Netherlands Organization for Scientific Research (ZON-MW), through grant 920-03-131.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/GenBank/ (for TBX4 [H. sapiens] mRNA [accession number AF188703] and for TBX2 [H. sapiens] mRNA [accession number U28049])
- NCBI BLAST, http://www.ncbi.nlm.gov/BLAST/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- Protein Data Bank, http://www.rcsb.org/pdb/ (for TBX3 [accession number 1H6F])
- UCSC Genome Bioinformatics, http://genome.cse.ucsc.edu/ (for Human Genome Browser)
References
- Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG (2003) Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130:623–633 10.1242/dev.00191 [DOI] [PubMed] [Google Scholar]
- Azouz EM, Kozlowski K (1997) Small patella syndrome: a bone dysplasia to recognize and differentiate from the nail-patella syndrome. Pediatr Radiol 27:432–435 10.1007/s002470050163 [DOI] [PubMed] [Google Scholar]
- Bernhang AM, Levine SA (1973) Familial absence of the patella. J Bone Joint Surg Am 55:1088–1090 [PubMed] [Google Scholar]
- Bamshad M, Lin R, Law Dj, Watkins WC, Krakowiak PA, Moore ME, Fransceschini P, Lala R, Holmes LB, Gebuhr TC, Bruneau GB, Schinzel A, Seidman JG, Seidman CE, Jorde LB (1997) Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet 16:311–315 [DOI] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill T, Leblanc-Straceski J, Renault B, Kucherlapati R, Seidman JG, Seidman CE (1997) Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15:30–35 [DOI] [PubMed] [Google Scholar]
- Bongers EMHF, van Bokhoven H, van Thienen M-N, Kooyman MAP, van Beersum SEC, Boetes C, Knoers NAVM, Hamel BCJ (2001) The small patella syndrome: description of five cases from three families and examination of possible allelism with familial patella aplasia-hypoplasia and nail patella syndrome. J Med Genet 38:209–213 10.1136/jmg.38.3.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers EMHF, van Bokhoven H, Knoers NVAM, Hamel BCJ, Woods CG (2002) Evidence for genetic heterogeneity in familial Isolated Patella Aplasia-Hypoplasia. Am J Med Genet 108:78–79 10.1002/ajmg.10198 [DOI] [PubMed] [Google Scholar]
- Burckhardt A (1988) “The small patella syndrome”: eine kombination von knie- und beckendysplasie. Z Orthop 126:22–29 [DOI] [PubMed] [Google Scholar]
- Chinea G, Padron G, Hooft RWW, Sander C, Vriend G (1995) The use of position specific rotamers in model building by homology. Proteins 23:415–421 [DOI] [PubMed] [Google Scholar]
- Coll M, Seidman JG, Muller CW (2002) Structure of the DNA-bound T-box domain of human TBX3, a transcription factor responsible for ulnar-mammary syndrome. Structure (Camb) 10:343–356 [DOI] [PubMed] [Google Scholar]
- Dellestable F, Péré P, Blum A, Gaucher A (1996) The “small-patella” syndrome, hereditary osteodysplasia of the knee, pelvis and foot. J Bone Joint Surg Br 78:63–65 [PubMed] [Google Scholar]
- Dreyer SD, Zhou G, Baldini A, Winterpacht A, Zabel B, Cole W, Johnson RL, Lee B (1998) Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat Genet 19:47–50 [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Silver LM, Niswander L, Papaioannou VE (1998) Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development 125:2499–2509 [DOI] [PubMed] [Google Scholar]
- Habboub HK, Thneibet WA (1997) Ischio-pubic-patellar hypoplasia: is it a new syndrome? Pediatr Radiol 27:430–431 10.1007/s002470050162 [DOI] [PubMed] [Google Scholar]
- Khan P, Linkhart B, Simon H-G (2002) Different regulation of T-box genes Tbx4 and Tbx5 during limb development and limb regeneration. Dev Biol 250:383–392 10.1016/S0012-1606(02)90801-8 [DOI] [PubMed] [Google Scholar]
- Kozlowski K, Nelson J (1995) Small patella syndrome. Am J Med Genet 57:558–561 [DOI] [PubMed] [Google Scholar]
- Kremer H, Pinckers A, van den Helm B, Deutman AF, Ropers HH, Mariman ECM (1994) Localization of the gene for dominant cystoid macular dystrophy on chromosome 7p. Hum Mol Genet 3:299–302 [DOI] [PubMed] [Google Scholar]
- Logan M, Tabin CJ (1999) Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science 283:1736–1739 10.1126/science.283.5408.1736 [DOI] [PubMed] [Google Scholar]
- Mangino M, Sanchez O, Torrente I, De Luca A, Capon F, Novelli G, Dallapiccola B (1999) Localization of a gene for familial patella aplasia-hypoplasia (PTLAH) to chromosome 17q21–22. Am J Hum Genet 65:441–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S, Offiah AC, McDowall S, Sim P, Tolmie J, Hall C (2002) The phenotype of survivors of campomelic dysplasia. J Med Genet 39:597–602 10.1136/jmg.39.8.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin Ph, Vielpeau C, Fournier L, Denizet D (1985) Le syndrome coxo-podo-patellaire. J Radiol 66:441–446 [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE (2003) Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development 130:2681–2693 10.1242/dev.00504 [DOI] [PubMed] [Google Scholar]
- Packham EA, Brook JD (2003) T-box genes in human disorders. Hum Mol Genet 12:R37–R44 10.1093/hmg/ddg077 [DOI] [PubMed] [Google Scholar]
- Offiah AC, Mansour S, McDowall S, Tolmie J, Sim P, Hall CM (2002) Surviving campomelic dysplasia has the radiological features of the previously reported ischio-pubic-patella syndrome. J Med Genet 39:e50 10.1136/jmg.39.9.e50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poznanski AK (1997) Editorial comments on the ischio-pubic-patellar syndrome. Pediatr Radiol 27:428–429 10.1007/s002470050161 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Esteban C, Tsukui T, Yonei S, Magallon J, Tamura K, Belmonte JCI (1999) The T-box genes Tbx4 and Tbx5regulate limb outgrowth and identity. Nature 398:814–818 10.1038/19769 [DOI] [PubMed] [Google Scholar]
- Sandhaus YS, Ben-Ami T, Chechick A, Goodman RM (1987) A new patella syndrome. Clin Genet 31:143–147 [DOI] [PubMed] [Google Scholar]
- Scott JE, Taor WS (1979) The “small patella” syndrome. J Bone Joint Surg Br 61:172–175 [DOI] [PubMed] [Google Scholar]
- Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP (2000) A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci 3:337–341 10.1038/73902 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Takeuchi J, Koshiba-Takeuchi K, Ogura T (2004) Tbx genes specify posterior digit identity through Shhand BMP signaling. Dev Cell 6:43–53 10.1016/S1534-5807(03)00401-5 [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Koshiba-Takeuchi K, Suzuki T, Kamimura M, Ogura K, Ogura T (2003) Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signalling cascade. Development 130:2729–2739 10.1242/dev.00474 [DOI] [PubMed] [Google Scholar]
- Vaněk J (1981) Ischiopatellare dysplasie (Syndrom der “kleinen patella” von Scott und Taor). Fortschr Röntgenstr 135:354–356 [PubMed] [Google Scholar]
- Yi CH, Russ, A, Brook, JD (2000) Virtual cloning and physical mapping of a human T-box gene, TBX4. Genomics 67:92–95 10.1006/geno.2000.6222 [DOI] [PubMed] [Google Scholar]