Abstract
Spondylocostal dysostosis (SCD) is a term given to a heterogeneous group of disorders characterized by abnormal vertebral segmentation (AVS). We have previously identified mutations in the Delta-like 3 (DLL3) gene as a major cause of autosomal recessive spondylocostal dysostosis. DLL3 encodes a ligand for the Notch receptor and, when mutated, defective somitogenesis occurs resulting in a consistent and distinctive pattern of AVS affecting the entire spine. From our study cohort of cases of AVS, we have identified individuals and families with abnormal segmentation of the entire spine but no mutations in DLL3, and, in some of these, linkage to the DLL3 locus at 19q13.1 has been excluded. Within this group, the radiological phenotype differs mildly from that of DLL3 mutation–positive SCD and is variable, suggesting further heterogeneity. Using a genomewide scanning strategy in one consanguineous family with two affected children, we demonstrated linkage to 15q21.3-15q26.1 and furthermore identified a 4-bp duplication mutation in the human MESP2 gene that codes for a basic helix-loop-helix transcription factor. No MESP2 mutations were found in a further 7 patients with related radiological phenotypes in whom abnormal segmentation affected all vertebrae, nor in a further 12 patients with diverse phenotypes.
The spondylocostal dysostoses (SCD [MIM 277300]) are a heterogeneous group of disorders with severe axial skeletal malformation characterized radiologically by multiple vertebral segmentation defects (MVSD). In addition, the ribs are frequently malaligned, with points of fusion and sometimes a reduction in number. The diverse SCD phenotypes have been reviewed elsewhere (Turnpenny and Kusumi 2003). Sporadic cases occur more commonly than familial ones and are more likely to be associated with multiple congenital abnormalities, whereas monogenic forms more commonly demonstrate autosomal recessive (AR) rather than autosomal dominant inheritance. We previously demonstrated linkage in some families with AR SCD to chromosome 19q13.1 (Turnpenny et al. 1999), a region syntenic to the Dll3 homologue on mouse chromosome 7. Mutations in murine Dll3 result in the “Pudgy” phenotype (Kusumi et al. 1998), and we subsequently identified mutations in the human Delta-like 3 (DLL3 [MIM 602768]) gene (Bulman et al. 2000). We and others have characterized 17 familial mutations in DLL3, including nonsense, missense, frameshift, and splice-site mutations, all of which are likely to lead to functional null alleles either by premature truncation of the protein or by the substitution of essential amino acid residues (Bulman et al. 2000; Sparrow et al. 2002; Bonafé et al. 2003; Turnpenny et al. 2003).
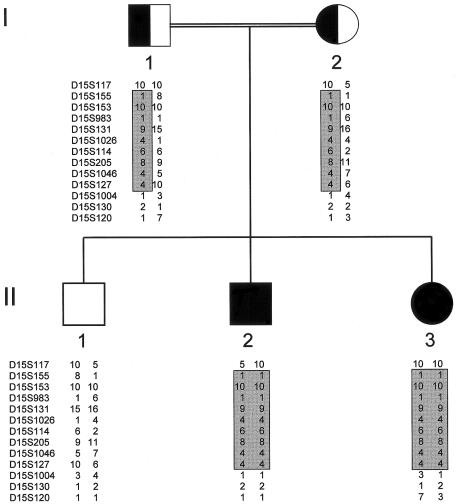
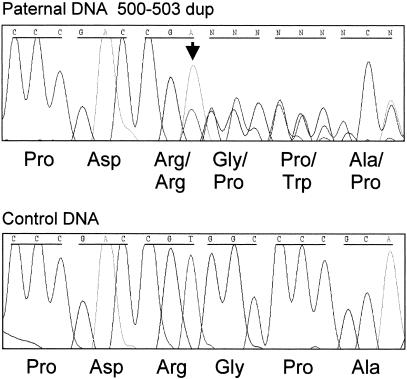
Here we have studied a consanguineous family of Lebanese Arab origin with two offspring affected with SCD (fig. 1). Affected individuals presented with truncal shortening and short necks but no other abnormalities. On radiological examination, thoracic vertebrae bear a resemblance to those seen in SCD due to mutated DLL3, but the lumbar vertebrae appear more angular and irregular. The vertebral morphology, with regional differences throughout the spine, is highlighted by MR imaging (fig. 2). By use of genomic DNA extracted from blood samples from all five available family members, flanking microsatellite markers excluded linkage to the 19q13.1/DLL3 locus. Subsequent genomewide homozygosity mapping, performed using the Perkin Elmer Biosystems Linkage Mapping Set (version 1) with affected genomic DNA from individual II.2, revealed 84 homozygous markers scattered throughout the genome, of which 6 were concentrated in a block on 15q (D15S153 to D15S120) and 5 were concentrated in a block on 20q (D20S117 to D20S186). Subsequent mapping, performed using the second affected individual II.3 and the unaffected sibling II.1, excluded linkage to the 20q region but demonstrated linkage to the 15q markers—namely, D15S153, D15S131, D15S205, and D15S127. Subsequent fine mapping, performed using additional markers, demonstrated a 36.6-Mb region (according to The Human Genome Working Draft) on 15q21.3-15q26.1 between markers D15S117 and D15S1004 with a maximum two-point LOD score of 1.588 at θ=0 for markers D15S131, D15S205, D15S1046, and D15S127 (table 1). The region between markers D15S117 and D15S1004 contains >50 genes and is syntenic to mouse chromosome 7, which contains the Mesp2 gene (MESP2 [MIM 605195]). The Mesp2-knockout mouse manifests altered rostro-caudal polarity, resulting in axial skeletal defects (Saga et al. 1997). The mouse gene was located using the Blat algorithm at The Human Genome Working Draft with MesP2 cDNA accession no. NM_008589, and the human gene was located, again using the Blat and Comparative Genome algorithms of The Human Genome Working Draft. Analysis of the predicted human gene, MESP2, reveals a two-exon gene spanning ∼2 kb of 15q26.1. Direct sequencing of MESP2 gene in the two affected individuals, by use of the primers M1 1F (5′-GAC ACC TCT CTG CAA CC TG-3′), M1 1R (5′-CCT GGA GTA GAT AAG CTG GG-3′), M1 2F (5′-CCA GCC ATA CCA TGG CAA C-3′), and M2 2R (5′-CCA AGC TAC AGG ACT GAT TC-3′), demonstrated a homozygous 4-bp (ACCG) duplication mutation in exon 1, termed “500–503dup” (fig. 3). The parents, I.1 and I.2, were shown to be heterozygous, and the unaffected sibling, II.1, was shown to be homozygous normal. Thus, the duplication segregates with the disease in this family. In addition, fluorescent PCR excluded this mutation from 68 ethnically matched control chromosomes. In analysis of the genomic structure of the MESP2 gene, we note that there is a discrepancy between the Sanger Centre and NCBI human genomic assembly databases. In the latter, there is an additional short intron located after base 502 of the MESP2 coding region. This does not appear in the Ensembl gene prediction, and, due to a lack of consensus splice sites within this proposed intronic sequence, we do not believe this intron exists. Clarification of this issue would necessitate transcript isolation and sequence analysis. This would be a difficult and ethically challenging task, since MESP2 is only expressed for a brief period during embryogenesis, in a highly restricted domain in the presomitic mesoderm. However, the presence or absence of such an intron does not effect our conclusions about the effects of the 4-bp insertion on MESP2 protein production. The insertion is located overlapping the NCBI proposed 5′ splice site and would interrupt splicing, leading to a frameshift at the same point in the MESP2 protein.
Figure 1.
Pedigree of the family affected with SCD, MESP2 type, demonstrating disease-linked haplotype. Affected members II.2 and II.3 are indicated by fully blackened symbols.
Figure 2.
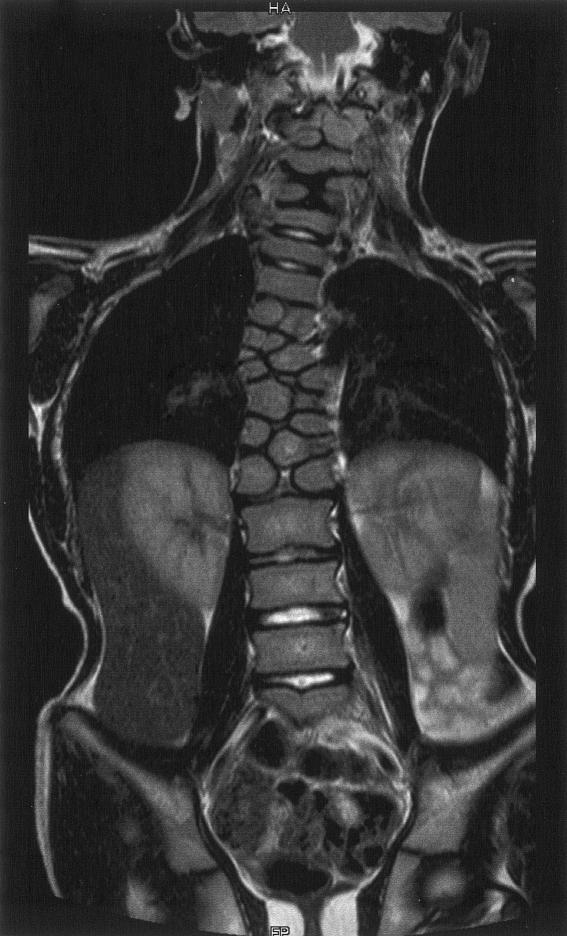
Anteroposterior MRI of the spine of individual II.2. The morphology of the thoracic spine and the upper cervical spine is severely disrupted, with multiple hemivertebrae, whereas the remaining regions show less severe disruption.
Table 1.
Two-Point LOD Scores between ARSCD Type 2 and Chromosome 15q Markers[Note]
|
Z at θ = |
|||||||
| Marker | .00 | .05 | .10 | .20 | .30 | .40 | .50 |
| D15S117 | −∞ | −.282 | −.085 | .020 | .273 | .015 | .000 |
| D15S155 | 1.125 | .991 | .857 | .594 | .353 | .151 | .000 |
| D15S153 | .034 | .026 | .019 | .095 | .004 | .001 | .000 |
| D15S983 | 1.125 | .991 | .857 | .594 | .353 | .151 | .000 |
| D15S131 | 1.588 | 1.420 | 1.248 | .895 | .550 | .239 | .000 |
| D15S1026 | 1.125 | .991 | .857 | .594 | .353 | .151 | .000 |
| D15S114 | 1.125 | .991 | .857 | .594 | .353 | .151 | .000 |
| D15S205 | 1.588 | 1.420 | 1.248 | .895 | .550 | .239 | .000 |
| D15S1046 | 1.588 | 1.420 | 1.248 | .895 | .550 | .239 | .000 |
| D15S127 | 1.588 | 1.420 | 1.248 | .895 | .550 | .239 | .000 |
| D15S1004 | −∞ | −1.137 | −.653 | −.266 | −.103 | −.025 | .000 |
| D15S130 | −∞ | −.282 | −.085 | .020 | .273 | .015 | .000 |
| D15S120 | −∞ | −1.929 | −1.148 | −.488 | −.200 | −.059 | .000 |
Note.— All values are calculated under the assumption of 100% penetrance, a disease allele frequency of 0.0001, and a disease haplotype frequency of 0.1.
Figure 3.
Detection of the mutation 500–503dup by direct nucleotide sequencing. The sequence of a heterozygous individual (as indicated by the arrow) (top) is compared with the normal homozygous sequence (bottom).
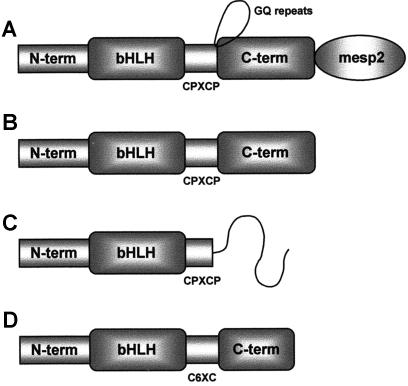
The MESP2 gene is predicted to produce a transcript of 1,191 bp encoding a protein of 397 amino acids with a predicted molecular weight of 41,744 Da and isoelectric point (pI) of 7.06. The human MesP2 protein has 58.1% identity with mouse MesP2, and 47.4% identity with human MesP1. Human MesP2 amino terminus contains a basic helix-loop-helix (bHLH) region encompassing 51 amino acids divided into an 11-residue basic domain, a 13-residue helix I domain, an 11-residue loop domain, and a 16-residue helix II domain. The loop region is slightly longer than that found in homologues such as paraxis. The length of the loop region is conserved between mouse and human MesP1, MesP2, and Thylacine 1 and 2, as well as chick mesogenin. In addition, both MesP1 and MesP2 contain a unique CPXCP motif immediately carboxy-terminal to the bHLH domain. The amino- and carboxy-terminal domains are separated in human MesP2 by a GQ repeat region also found in human MesP1 (2 repeats) but expanded in human MesP2 (13 repeats). Mouse MesP1 and MesP2 do not contain GQ repeats, but they do contain two QX repeats in the same region: mouse MesP1 QSQS; mouse MesP2 QAQM. Hydrophobicity plots indicate that MesP1 and MesP2 share a carboxy-terminal region that is predicted to adopt a similar fold, although MesP2 sequences do contain a unique region at the carboxy-terminus.
Sequence analysis of 20 ethnically matched and 10 nonmatched individuals revealed the presence of a variable length polymorphism in the GQ region of human MesP2, beginning at nucleotide 535. This region contains a series of 12-bp repeat units. The smallest GQ region detected contains two type A units (GGG CAG GGG CAA, encoding the amino acids GQGQ), followed by two type B units (GGA CAG GGG CAA, encoding GQGQ) and one type C unit (GGG CAG GGG CGC, encoding GQGR). The polymorphism comprises a variation in the number of type A units, with 2, 3, or 4 being present in the sequenced individuals. The fluorescent PCR used to confirm the familial mutation in the 68 control chromosomes revealed 23 alleles of 2 repeats, 44 alleles of 3 repeats, and 1 allele of 4 repeats, yielding a polymorphism information content (PIC) value of 0.3707, thus demonstrating its usefulness in linkage analysis. The 20 nonethnically matched chromosomes consisted of 7 alleles of 2 repeats, 12 alleles of 3 repeats, and 1 allele of 4 repeats. Therefore, allelic frequencies are not statistically significantly different between these two populations (P=.41).
Here we have shown that a frameshift mutation in the human homologue of the mesoderm posterior 2 gene, MESP2, results in AR SCD in man, and we suggest that this association be designated “SCD, MESP2-type” (or “SCD type 2,” to be distinguished from SCD type 1 by mutated DLL3). MesP2 is a member of the bHLH family of transcriptional regulatory proteins essential to a vast array of developmental processes (reviewed in Massari and Murre 2000). Murine Mesp1 and Mesp2, located on chromosome 7, are separated by ∼23 kb. They are positioned head to head and are transcribed from the interlocus region (Saga et al. 1996). At least two enhancers are involved in the expression of these genes in mouse (Haraguchi et al. 2001), one in early mesoderm expression and the other in presomitic mesoderm (PSM) expression. In addition, a suppressor responsible for the rostrally restricted expression in the PSM has been identified (Haraguchi et al. 2001). These enhancers are essential to the specific and coordinated expression of the MesP proteins. The expression of MesP1 and Mesp2 is first detected in the mouse embryo at the onset of gastrulation (∼6.5 d post coitum [dpc]) and is restricted to early nascent mesoderm (Saga et al. 1996; Kitajima et al. 2000). At this stage, the expression domain of Mesp1 is broader than that of Mesp2, and lineage analysis of Mesp2 expressing cells shows that they contribute to cranial, cardiac, and extra-embryonic mesoderm (Saga et al. 1999). Expression is then downregulated, as Mesp transcripts are not detected later in development (Saga et al. 1999). A second site of Mesp expression is detected at 8.0 dpc immediately prior to somitogenesis. A pair of MesP-expressing bands appear on each side of the embryonic midline, at the anterior part of the PSM where the somites are anticipated to form (Saga et al. 1997, 1999). During somitogenesis MesP1 and MesP2 continue to be expressed in single bilateral bands in the anterior PSM. MesP2 expression within the PSM continues until about the time when somite formation ceases (∼13.5 dpc) (Saga et al. 1997), after which Mesp expression is rapidly downregulated, as transcripts are not detected in newly formed somites.
Alignment of MesP2 homologues from human, mouse, Xenopus, and chick demonstrates a highly divergent carboxy-terminus between species (fig. 4), and it is unknown whether functional domains are similarly arranged in all Mesp2 orthologues, though this is well characterized for the Xenopus Mesp2 orthologue, Thylacine (Sparrow et al. 1998). If the carboxy-terminus in human MesP2 is required for transcriptional activation, then the mutant form of the protein described here, lacking the carboxy-terminus, would lose this function. Mouse pups lacking MesP2 die within 20 min of birth, presenting with short tapered trunks and abnormal segmentation affecting all but a few caudal (tail) vertebrae (Saga et al. 1997). This resembles the phenotype of Dll3-null mice, in which the rib and vertebral architecture is disturbed along the entire axis.
Figure 4.
Comparison of MesP2 (A) and MesP1 (B), both containing a bHLH domain. MesP1 and MesP2 contain a unique CPXCP motif immediately C-terminal to the bHLH domain. C, Predicted product of the human MesP2 frameshift mutant. Xenopus and zebrafish sequences (D) are shorter at the C-terminus and have the residues C6XC in place of the CPXCP motif.
Unlike the mouse null MesP2 allele, the human mutant protein retains its bHLH region and, although truncated, might still dimerize, bind DNA, and thus act in a dominant negative manner. However, the heterozygous parents in this family demonstrate no axial skeletal defects, from which we deduce that this MESP2 mutation is recessive. Similarly, heterozygous mice are normal and fertile (Saga et al. 1997). It is possible that a single functional carboxy-terminus is sufficient to activate transcription. Alternatively, MESP1 may compensate for the absence of MESP2 function, just as MesP1 can rescue the axial skeletal defects in MesP2-deficient mice in a dosage-dependent manner (Saga 1998). It is also possible that the mutant MESP2 transcript, containing a premature stop codon at 1099–1101 bp, is degraded by nonsense-mediated RNA decay, with no truncated protein being produced (see Culbertson 1999).
In murine somitogenesis, MesP2 has a key role in establishing rostrocaudal polarity by participating in distinct Notch-signaling pathways (Takahashi et al. 2003). Mesp2 expression is induced by Dll1-mediated (DLL1 [MIM 606582]) Notch signaling (presenilin1-independent (PSEN1 [MIM 104311]) and Dll3-mediated Notch signaling (presenilin1-dependent), whereas inhibition of Mesp2 expression is achieved through presenilin1-independent Dll3-Notch signaling. Since Mesp2 can inhibit Dll1 expression, this complex signaling network results in stripes of Dll1, Dll3, and Mesp2 gene expression in the anterior PSM. The extent to which this represents somitogenesis in man is unknown, except that the phenotypes of the Mesp2 and Dll3 mutant mice closely resemble SCD (Saga et al. 1997; Kusumi et al. 1998; Dunwoodie et al. 2002). Here we provide the first evidence that MESP2 is critical for normal somitogenesis in man. Affected individuals in the family studied have short trunks and abnormal segmentation of all vertebrae, though the disruption to normal morphology appears most marked in the thoracic spine. The reasons for regional variations in disruption pattern throughout the spine are not known. Mutated MESP2 appears to be a rare cause of SCD, since we have identified only one affected family from a panel of 20 cases that were DLL3 mutation–negative, including 8 cases in whom abnormal vertebral segmentation affects the entire spine, 4 cases of spondylothoracic dysplasia, and a further 8 cases with diverse phenotypes.
Acknowledgments
We thank Action Medical Research for funding this project (SP3751) on the genetic basis of congenital spinal deformities (P.D.T., S.E., N.V.W.), and the Royal Devon and Exeter National Health Service Healthcare Trust for ongoing support. S.L.D. is funded by National Health and Medical Research Council project grant #142006 and is a Pharmacia Foundation Australia Fellow. D.B.S. is a Westfield-Belconnen Fellow, and M.W. is a Freedman Foundation Fellow.
Electronic-Database Information
URLs for data presented herein are as follows:
- Human Genome Working Draft, The, http://genome.ucsc.edu/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Bonafé L, Giunta C, Gassner M, Steinmann B, Superti-Furga A (2003) A cluster of autosomal recessive spondylocostal dysostosis caused by three newly identified DLL3 mutations segregating in a small village. Clin Genet 64:28–35 10.1034/j.1399-0004.2003.00085.x [DOI] [PubMed] [Google Scholar]
- Bulman MP, Kusumi K, Frayling TM, McKeown C, Garrett C, Lander ES, Krumlauf R, Hattersley AT, Ellard S, Turnpenny PD (2000) Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat Genet 24:438–441 10.1038/74307 [DOI] [PubMed] [Google Scholar]
- Culbertson MR (1999) RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends in Genetics 15:74–80 10.1016/S0168-9525(98)01658-8 [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS (2002) Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129:1795–1806 [DOI] [PubMed] [Google Scholar]
- Haraguchi S, Kitajima S, Takagi A, Takeda H, Inoue T, Saga Y (2001) Transcriptional regulation of Mesp1 and Mesp2 genes: differential usage of enhancers during development. Mech Dev 108:59–69 10.1016/S0925-4773(01)00478-6 [DOI] [PubMed] [Google Scholar]
- Kitajima S, Takagi A, Inoue T, Saga Y (2000) MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development 127:3215–3226 [DOI] [PubMed] [Google Scholar]
- Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi DC, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES (1998) The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat Genet 19:274–278 10.1038/961 [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20:429–440 10.1128/MCB.20.2.429-440.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y (1998) Genetic rescue of segmentation defect in MesP2-deficient mice by MesP1 gene replacement. Mech Dev 75:53–66 10.1016/S0925-4773(98)00077-X [DOI] [PubMed] [Google Scholar]
- Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM (1996) MesP1: a novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development 122:2769–2778 [DOI] [PubMed] [Google Scholar]
- Saga Y, Hata N, Koseki H, Taketo MM (1997) Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev 11:1827–1839 [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T (1999) MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 126:3437–3447 [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Clements M, Withington SL, Scott AN, Novotny J, Sillence D, Kusumi K, Beddington RS, Dunwoodie SL (2002) Diverse requirements for Notch signalling in mammals. Int J Dev Biol 46:365–374 [PubMed] [Google Scholar]
- Sparrow DB, Jen WC, Kotecha S, Towers N, Kintner C, Mohun TJ (1998) Thylacine 1 is expressed segmentally within the paraxial mesoderm of the Xenopus embryo and interacts with the Notch pathway. Development 125:2041–2051 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Inoue T, Gossler A, Saga Y (2003) Feedback loops comprising Dll1, Dll3 and Mesp2, and differential involvement of Psen1 are essential for rostrocaudal patterning of somites. Development 130:4259–4268 10.1242/dev.00629 [DOI] [PubMed] [Google Scholar]
- Turnpenny PD, Bulman MP, Frayling TM, Abu-Nasra TK, Garrett C, Hattersley AT, Ellard S (1999) A gene for autosomal recessive spondylocostal dysostosis maps to 19q13.1-q13.3. Am J Hum Genet 65:175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnpenny PD, Kusumi K (2003) DLL3 and spondylocostal dysostosis. In: Epstein CJ, Erickson RP, Wynshaw-Boris A (eds) Inborn errors of development. Oxford University Press, New York, pp 470–481 [Google Scholar]
- Turnpenny PD, Whittock N, Duncan J, Dunwoodie S, Kusumi K, Ellard S (2003) Novel mutations in DLL3, a somitogenesis gene encoding a ligand for the Notch signalling pathway, cause a consistent pattern of abnormal vertebral segmentation in spondylocostal dysostosis. J Med Genet 40:333–339 10.1136/jmg.40.5.333 [DOI] [PMC free article] [PubMed] [Google Scholar]