Abstract
3p deletion syndrome is a rare contiguous-gene disorder involving the loss of the telomeric portion of the short arm of chromosome 3 and characterized by developmental delay, growth retardation, and dysmorphic features. All reported cases have involved, at a minimum, the deletion of chromosome 3 telomeric to the band 3p25.3. Despite the presence of several genes in this region that are involved in neural development, a causative relationship between a particular transcript and the observed clinical manifestations has remained elusive. We have identified a child with characteristic physical features of 3p deletion syndrome and both verbal and nonverbal developmental delay who carries a de novo balanced translocation involving chromosomes 3 and 10. Fine mapping of this rearrangement demonstrates that the translocation breakpoint on chromosome 3 falls within the recently identified minimal candidate region for 3p deletion syndrome and disrupts the Contactin 4 (CNTN4) mRNA transcript at 3p26.2–3p26.3. This transcript (also known as BIG-2) is a member of the immunoglobulin super family of neuronal cell adhesion molecules involved in axon growth, guidance, and fasciculation in the central nervous system (CNS). Our results demonstrate the association of CNTN4 disruption with the 3p deletion syndrome phenotype and strongly suggest a causal relationship. These findings point to an important role for CNTN4 in normal and abnormal CNS development.
3p deletion syndrome is a rare contiguous-gene disorder involving the telomeric portion of the short arm of chromosome 3. Clinically, the syndrome is characterized by developmental delay, growth retardation, and dysmorphic features (Verjaal and De Nef 1978; Schwyzer et al. 1987; Narahara et al. 1990; Mowrey et al. 1993; Phipps et al. 1994; Drumheller et al. 1996; McClure et al. 1996; Angeloni et al. 1999; Benini et al. 1999; Cargile et al. 2002). Less commonly, renal cysts, gastrointestinal abnormalities, and congenital heart defects have been described. All reported cases involve, at a minimum, the loss of chromosomal material telomeric to 3p25.3. Terminal deletions involving the more centromeric regions of 3p have been thought to predispose to cardiac septal defects (Brand et al. 1987; Phipps et al. 1994; Drumheller et al. 1996; Green et al. 2000). However, a minimal candidate region conferring the common features of the syndrome has been identified recently in a patient with an interstitial deletion involving a 4.5-Mb interval between markers D3S3630 and D3S1304 (Cargile et al. 2002). Despite investigations of several genes in the 3p region involved in CNS development (Angeloni et al. 1999; Endris et al. 2002; Montag-Sallaz et al. 2003; Soderling et al. 2003), a causative relationship between any particular transcript and the range of observed clinical manifestations has remained elusive.
CT is a boy aged 7 years 11 mo with borderline intellectual functioning, distinctive facial (fig. 1) and other physical features, and behavioral problems who was seen in the Genetics Outpatient Clinic at Yale and evaluated under a research protocol approved by the Yale Human Investigation Committee with informed consent. The patient was the product of a nonconsanguineous union, delivered at 40 wk gestation from his 29 year-old primiparous mother by cesarean section performed because of failure to progress. There was no history of alcohol consumption or medication use during pregnancy and no known exposure to teratogens. He had a difficult facial presentation, with mild perinatal respiratory depression and an Apgar score of 3 at 1 min. Cord blood pH was 7.28. After brief positive pressure ventilation, his Apgar score was 9 at 5 min. Birth weight was 3,787 g and length was 19 inches. Cytogenetic analysis was performed shortly after birth, which revealed a balanced translocation (46, XY, t[3;10][p26;q26]) (fig. 2A). Both parents had normal karyotypes. No other family members were reported to have mental retardation or dysmorphic features.
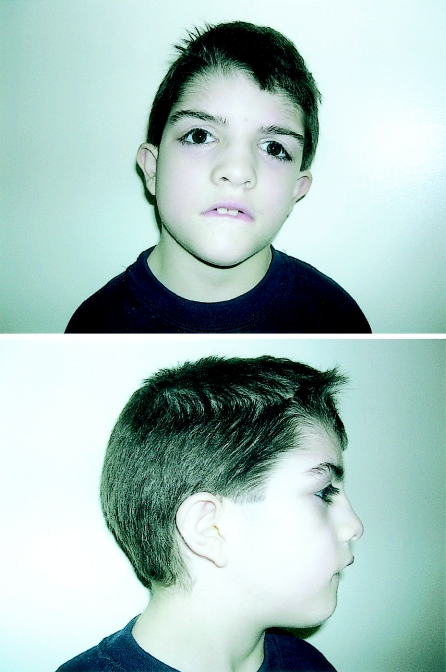
Figure 1.
Eight-year-old boy with developmental delay and features of 3p deletion syndrome. The child has had surgical correction of prominent ptosis.
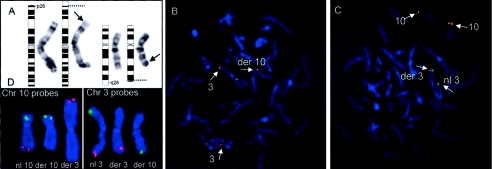
Figure 2.
Cytogenetic and molecular cytogenetic studies of a de novo t(3;10) translocation. A, G-banded karyotype of the patient’s lymphocytes showing a t(3p26;10q26) translocation. Arrows highlight the breakpoints on the respective chromosomes, and dotted lines show the corresponding position on the idiograms. 10/10 metaphase spreads examined showed the identical translocation. B, BAC probe 299N3 (red) spanning the chromosome 3p breakpoint. Hybridization signals are seen on both normal and derivative chromosomes 3. A third red hybridization signal is also seen on the derivative chromosome 10, as a result of the breakpoint transecting the region corresponding to the experimental probe. A control probe (green) maps to the q arm of chromosome 3. C, Probe 355F22 (red) spanning the chromosome 10q breakpoint. Hybridization signals are present on the normal and derivative chromosomes 10. A third red signal is seen on the telomeric region of chromosome 3. Control probes (green) map to the short arm of chromosome 3. D, Subtelomeric FISH results are shown. Chromosome 10 probes are in the left box—the normal (nl) chromosome 10 with both green and red probes, the derivative (der) 10 with only a green probe, and the derivative chromosome 3 with only the red probe are shown. The box on the right shows chromosome 3 probes, with the normal chromosome showing both red and green probes, the derivative 3 showing only the red probe, and the derivative 10 showing only the green probe.
The child’s early motor development was unremarkable: he rolled over at 4 mo and was sitting with support at 6.5 mo. At 7 mo, he was able to stand with assistance, and he walked at 14 mo. His development of language, in contrast, was markedly delayed, with the absence of distinct single words until 3.5 years. CT underwent a cognitive assessment at age 7 years 2 mo and was found to have a full scale IQ of 73 on the Wechsler Intelligence Scale for Children, Third Edition (WISC-III) (Wechsler 1992) (Mean = 100; SD=15), which places his cognitive skills within the borderline range of intellectual functioning. He obtained a verbal IQ of 81 and a performance (or nonverbal) IQ of 70. Significant weaknesses in his profile related to attention to visual detail (subtest score of 3; mean = 10, SD=3), visual-spatial reconstruction skills (subtest score of 3), and understanding of social mores and expectations (subtest score of 3). These deficits contrasted with his significant strength in the area of fund of knowledge (subtest score of 12). An assessment of adaptive functioning at age 7 years 11 mo using the Vineland Adaptive Behavior Scales, Expanded Edition (Sparrow et al. 1984), showed marked impairments in social functioning and daily living skills. In the Daily Living Skills Domain, his percentile rank score was <0.1, corresponding to an age equivalent score of 4 years. In the Socialization Domain, his percentile rank score was within the first percentile, corresponding to an age equivalent score of 4 years 1 mo. Taken together, these findings demonstrated developmental delay, with particular deficits in nonverbal learning.
CT’s past medical history was unremarkable. He was noted to be in excellent physical health and had required surgery only for correction of ptosis. He had been followed closely from a very young age by his pediatrician as a consequence of his dysmorphic features and known cytogenetic rearrangement and was not found to have any evidence of GI, cardiac, or renal abnormalities on physical exam or by history. A referral to a pediatric neurologist for behavioral issues at age 6 years revealed a normal neurological exam, except for the identification of mild hypotonia. Neither neuroimaging nor electroencephalogram was deemed to be clinically indicated.
Physical examination at age 7 years 11 mo revealed a height of 115.6 cm (5th–10th percentile), a weight of 21.4 kg (5th percentile), and a head circumference of 52 cm (50th–75th percentile). The patient showed high myopia and astigmatism, a low frontal hairline with right upsweep, forehead hypertrichosis, and bushy eyebrows without frank synophrys. He was also noted to have surgically improved ptosis and slight epicanthal folds, a broad nasal bridge with mild hypertelorism (measured at +1.5 SD at age 21 mo), downslanting palpebral fissures, downturned corners of the mouth, prominent but normal-sized ears with superior helices sagging downward, slight fifth-finger clinodactyly, branched transverse palmar crease of right hand, mild bilateral syndactyly of the second and third toes, hypertrichosis of his thighs and legs, and mild hypotonia. Deep tendon reflexes were normal.
The patient’s clinical presentation, including developmental delay, growth retardation, and ptosis in combination with the known cytogenetic abnormality, suggested a diagnosis of 3p deletion syndrome (table 1). Those physical signs not present in this case, such as trigonocephaly, microcephaly, and micrognathia, are not ubiquitous findings in other reported cases in the literature. The patient’s particular cognitive findings are mirrored in at least one other case, described by Angeloni et al. (1999), in which the patient was found to have a full scale IQ in the borderline range with what appeared to be significant weaknesses in adaptive functioning. In addition, a marked delay in early language acquisition, as is seen in the present case, frequently has been noted anecdotally. However, a more detailed comparison of CT’s cognitive findings with other reported cases is not possible, as a result of the informal descriptions of developmental delay that characterize the 3p deletion syndrome literature. What is apparent is that phenotypic variability in all aspects of the syndrome appears to be the rule rather than the exception. Moreover, genotype/phenotype relationships are not straightforward. For instance, as noted, there is a strong relationship between terminal deletions involving more proximal regions of chromosome 3 and cardiac findings (Brand et al. 1987; Phipps et al. 1994; Drumheller et al. 1996; Green et al. 2000). In contrast, the range and severity of dysmorphology and developmental delay may differ markedly in patients with similar, or even identical, deletions of the regions telomeric to 3p25.3 (Tazelaar et al. 1991; Knight et al. 1995; Benini et al. 1999).
Table 1.
Selected Features of 3p Deletion Syndrome
| Features of 3p Deletion Syndrome | Affected Frequencya(%) | Status in thePresented Caseb |
| Developmental delay | 86 | + |
| Postnatal growth retardation | 86 | + |
| Ptosis | 77 | + |
| Low birth weight | 73 | − |
| Malformed ears | 68 | + |
| Long philtrum | 68 | − |
| Broad nasal bridge | 64 | + |
| Hypotonia | 50 | + |
| Microcephaly | 50 | − |
| Micrognathia | 50 | − |
| Epicanthal folds | 41 | + |
| Post axial polydactyly | 40 | − |
| Hyperteliorism | 36 | + |
| Feeding problems | 29 | + |
| Clinodactyly | 27 | + |
| Trigonocephaly | 23 | − |
| Downturned corners of mouth | 24 | + |
| Synophoris | 18 | − |
| Low frontal hairline | 14 | + |
Frequency data is derived from the most recent and largest case review in the literature (Benini et al. 1999), consisting of 22 patients.
+ = present; − = absent.
Molecular studies were undertaken to fine map the (3;10) translocation using FISH, as described elsewhere (Lichter et al. 1990). Bacterial artificial chromosomes (BACs) corresponding to the observed cytogenetic bands involved in the apparently balanced translocation were identified using the UCSC genome browser and obtained from an RPCI-11 (Roswell Park Cancer Institute) library. These probes were hybridized to metaphase spreads consisting of the patient’s lymphocytes. Several probes mapping centromeric and telomeric to the translocation breakpoints on both chromosomes were identified (table 2). Subsequent to the identification of BACs mapping on both sides of the translocation breakpoint on each chromosome, BACs within the translocation intervals were used, eventually identifying probes spanning both the chromosome 3 and 10 breakpoints (figs. 2 and 3). Two BACs spanning the chromosome 3 breakpoint, RPCI-11 299N33 and RPCI-11 121K9, were found to map in their entirety in the first intron of the gene Contactin 4 (CNTN4 [MIM 607280]) (fig. 3), establishing that the translocation breakpoint disrupted the gene within the 5′ UTR of the mature CNTN4 mRNA. Importantly, CNTN4 maps both within the minimal candidate region for 3p deletion syndrome defined by Cargile et al. (2002) and within a 6.7-cM region found linked to nonspecific mental retardation in large kindred (Zmax=9.18 at marker D3S3050) (Higgins et al. 2000) (fig. 3A).
Table 2.
BAC Probes Investigated Using FISH
| BAC ID | Base Positiona | Chromosomal Band | Location Relativeto Breakpoint |
| RP11-1082A18 | chr3:10,298,689-10,516,466 | 3p25.3 | Centromeric |
| RP11-572M14 | chr3:10,011,785-10,180,797 | 3p25.3 | Centromeric |
| RP11-1020A11 | chr3:9,778,862-9,995,772 | 3p25.3 | Centromeric |
| RP11-19E8 | chr3:8,917,083-9,067,340 | 3p25.3 | Centromeric |
| RP11-128A5 | chr3:8,686,926-8,857,271 | 3p25.3 | Centromeric |
| RP11-34L16 | chr3:8,442,935-8,594,857 | 3p25.3 | Centromeric |
| RP11-470E10 | chr3:7,841,368-8,025,425 | 3p26.1 | Centromeric |
| RP11-507D6 | chr3:6,997,984-7,199,514 | 3p26.1 | Centromeric |
| RP11-318I14 | chr3:6,283,022-6,437,405 | 3p26.1 | Centromeric |
| RP11-161L3 | chr3:5,744,835-5,909,633 | 3p26.1 | Centromeric |
| RP11-129J10 | chr3:5,027,803-5,180,950 | 3p26.1 | Centromeric |
| RP11-453A3 | chr3:4,158,926-4,329,972 | 3p26.1 | Centromeric |
| RP11-245A6 | chr3:3,545,257-3,707,298 | 3p26.2 | Centromeric |
| RP11-198P17 | chr3:2,928,181-3,117,274 | 3p26.2 | Centromeric |
| RP11-94M9 | chr3:2,804,247-2,975,122 | 3p26.2 | Centromeric |
| RP11-785A7 | chr3:2,690,183-2,862,993 | 3p26.2–3p26.3 | Centromeric |
| RP11-82O3 | chr3:2,566,282-2,742,197 | 3p26.3 | Centromeric |
| RP11-63O1 | chr3:2,443,361-2,617,387 | 3p26.3 | Centromeric |
| RP11-211K13 | chr3:2,307,996-2,470,413 | 3p26.3 | Centromeric |
| RP11-762O12 | chr3:2,287,581-2,441,071 | 3p26.3 | Centromeric |
| RP11-299N3 | chr3:2,181,279-2,367,269 | 3p26.3 | Spanning |
| RP11-129K1 | chr3:2,132,133-2,290,752 | 3p26.3 | Spanning |
| RP11-176P14 | chr3:2,021,987-2,193,951 | 3p26.3 | Telomeric |
| RP11-416N8 | chr3:1,244,095-1,420,317 | 3p26.3 | Telomeric |
| RP11-392M7 | chr3:1,064,436-1,252,749 | 3p26.3 | Telomeric |
| RP11-114K9 | chr3:333,643-518,322 | 3p26.3 | Telomeric |
| RP11-306H5 | chr3:159,323-343,406 | 3p26.3 | Telomeric |
| RP11-338L11 | chr10:116,448,883-116,645,816 | 10q25.3 | Centromeric |
| RP11-169K19 | chr10:117,157,040-117,301,164 | 10q25.3 | Centromeric |
| RP11-96N16 | chr10:117,471,461-117,637,580 | 10q25.3 | Centromeric |
| RP11-295023 | chr10:117,733,868-117,949,898 | 10q25.3 | Centromeric |
| RP11-498B4 | chr10:118,011,104-118,209,364 | 10q25.3 | Centromeric |
| RP11-5G18 | chr10:118,364,598-118,528,414 | 10q25.3–10q26.11 | Centromeric |
| RP11-328K15 | chr10:119,130,080-119,172,006 | 10q26.11 | Centromeric |
| RP11-355F22 | chr10:119,170,007-119,301,205 | 10q26.11 | Spanning |
| RP11-136I7 | chr10:119,181,243-119,340,828 | 10q26.11 | Spanning |
| RP11-280F22 | chr10:119,299,202-119,400,731 | 10q26.11 | Telomeric |
| RP11-3H12 | chr10:119,316,263-119,394,906 | 10q26.11 | Telomeric |
| RP11-354M20 | chr10:119,398,732-119,592,400 | 10q26.11 | Telomeric |
Derived from the July 2003 freeze of the Human Genome Project.
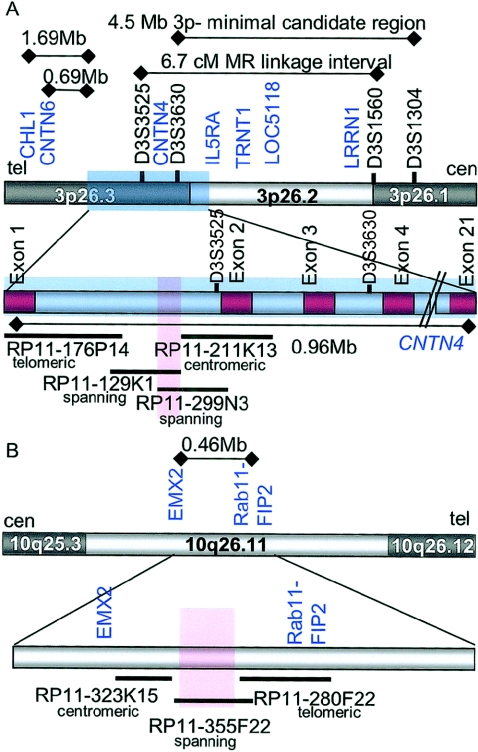
Figure 3.
Diagram of chromosomes 3 and 10 breakpoints. A, Telomeric region of chromosome 3 from 3p26.1 to 3p26.3, represented by the grey and white bar at the top of the diagram. Relevant STS markers are in black text, and known genes from the RefSeq database are in blue text. The region corresponding to CNTN4 is pictured as a blue shaded rectangle. Distances noted were derived from the July 2003 freeze of the Human Genome Project and include: (1) the 1.69 Mb-interval from the 3′-most exon of CHL1 to exon 1 of CNTN4; (2) the 0.69-Mb interval from the 3′-most exon of CNTN6 to exon 1 of CNTN4; (3) the 4.5-Mb interval defined by Cargile et al. (2002) as the smallest interstitial deletion identified in a 3p deletion syndrome case; and (4) the 6.7-cM interval identified by Higgins et al. (2002) in a large pedigree affected with nonspecific mental retardation (Zmax=9.18 at marker D3S3050). The bar below the genomic interval shows detail of the region, including exons of CNTN4 (in red). Selected BACs are noted as dark horizontal bars beneath. The breakpoint interval is shown as a red rectangle. The position of each BAC probe relative to the breakpoint on chromosome 3 is indicated below the RPCI-11 library identifier. The distance for the entire genomic interval corresponding to CNTN4 is indicated below the thin black line. B, Chromosome 10 in the region from 10q25.3-10q26.12, represented by the grey and white bar at the top of the diagram. Known genes from the RefSeq database are noted in blue. The 0.46-Mb distance between the 3′-most exon of EMX2 and the 5′-most exon of Rab11-FIP2 was determined by the July 2003 freeze of the Human Genome Project. The lower bar shows details of the interval, with the relative position of known genes in the region in blue text. The breakpoint region is highlighted with a red rectangle. No known genes or spliced ESTs were identified on the BAC spanning the chromosome 10 breakpoint. BACs from the region are identified as dark horizontal lines below the genomic region, with their position relative to the breakpoint on chromosome 10 noted below their RPCI identifiers.
BACs RPCI-11 355F22 (figs. 2 and 3) and RPCI-11 136I7 were found to span the chromosome 10 translocation breakpoint. This chromosome segment lies within a gene-poor region in the band 10q26.11(fig. 3B). No known or predicted transcripts or spliced ESTs were identified on these clones with the use of the UCSC genome browser. Five distinct ESTs without canonical splice sites were mapped to the BAC. Efforts to extend these fragments in silico with the use of the BLAST server at the National Center for Biotechnology Information were unsuccessful, and four of these five fragments showed no significant homology to other human ESTs or known genes. One region at the far telomeric extent of BAC 355F22 contained a block of 444 bp of DNA showing >95% homology at the nucleotide level to the tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide (YWHAQ [accession number BC050601]) on chromosome 2 as well as the YWHAQ pseudogene 2 on chromosome 22 (accession number NM_006826). No additional sequences with homology to YWHAQ were identified in the chromosome 10 interval via BLAST or BLAT searches. These analyses do not exclude the possibility of a rare transcript mapping to the 10q breakpoint region. However, the characteristic physical features in this case point to the chromosome 3 region as the proximate cause of the patient’s presentation.
Additional FISH studies were undertaken to evaluate the regions telomeric and centromeric to the 3p translocation (table 2). These resulted in no evidence of a deletion. Subtelomeric FISH with the use of commercially available probes was also performed to confirm the integrity of the terminal regions of the normal and derivative chromosomes 3 and 10 (fig. 2D). FISH mapping of BACs corresponding to the two closest transcripts on the telomeric aspect of the breakpoint, CHL1 (Close Homologue of L1, also known as CALL [MIM 607416]) and CNTN6 (MIM 607220), each showed the expected pattern of two hybridization signals, and direct sequencing of CNTN4 in the patient demonstrated no mutations in the coding regions of the remaining, intact transcript (data not shown).
The finding of disruption of CNTN4 is of particular interest as this gene encodes a neuronal adhesion molecule that is a member of the immunoglobulin (Ig) superfamily of genes. Similar to other members ofthe contactin family, which includes F3/F11/contactin (MIM 600316), TAG-1 (CNTN2 [MIM 190197]), BIG-1 (CNTN3 [MIM 601325]), NB-2 (CNTN5 [MIM 607219]), and NB-3 (CNTN6), full-length CNTN4 consists of six immunoglobulin domains and four fibronectin III (FNIII) domains and is anchored to the cell membrane via a glycosylphosphatidyl-inositol (GPI) domain. This family of proteins have been shown to be involved in axon growth, guidance, and fasciculation (Yoshihara et al. 1995; Ogawa et al. 1996; Saito et al. 1998; Kamei et al. 2000) and have been proposed to have a role in synaptic plasticity (Murai et al. 2002). A specific role for CNTN4 in neuronal development has not yet been clarified. However, knockouts of homologous neuronal adhesion molecules in mouse have resulted in viable mutants demonstrating morphological, neurological, and behavioral abnormalities (Berglund et al. 1999; Montag-Sallaz et al. 2002; Montag-Sallaz et al. 2003; Rolf et al. 2003).
At present, three isoforms of CNTN4 are described in the RefSeq database at NCBI. The identified breakpoint in this case disrupts only the longest variant which is highly expressed in human brain, particularly in cerebellum, thalamus, amygdala, and cerebral cortex (Kamei et al. 2000; Zeng et al. 2002). An intermediate-sized isoform, consisting of three Ig domains and three FNIII domains, has not been found to be expressed in the CNS (Kamei et al. 2000; Zeng et al. 2002; Hansford et al. 2003). A third, markedly truncated variant lacking all six Ig domains and two of the four FNIII domains has been shown to have low levels of expression in the brain, on the basis of northern blot and RT-PCR analysis (Zeng et al. 2002).
Given the inaccessibility of the relevant tissue, we were not able to test directly whether the translocation altered expression of the short brain-expressed variant, despite that transcript remaining structurally intact. Several characteristics of the full-length isoform, however, point to the clearly disrupted variant as the key determinant in the present case, irrespective of the status of the alternative transcript. First, the long variant is the only known isoform that codes for six Ig-like domains and four FNIII domains, reflecting the canonical structure of CNTN proteins. Studies of the contactin gene family have revealed a complex pattern of homophilic and heterophilic interactions that are required for axon growth and pathfinding. Such studies demonstrate that these essential functions are mediated by the combination and juxtaposition of multiple Ig and FNIII domains. Second, these neuronal adhesion molecules demonstrate highly regulated temporal and spatial expression patterns in the CNS. For this reason, the disruption of the regulatory region of the predominant brain-expressed isoform reasonably would be expected to have significant functional consequences. Third, the smaller brain-expressed variant, CNTN4a, demonstrates low levels of expression that are confined to the cerebral cortex, thus minimally overlapping with that of the long isoform (Zeng et al. 2002). These factors in combination with the lack of all Ig and several FNIII domains suggest that this truncated gene product would be a poor candidate to compensate for disruption of the full-length isoform. Finally, the clinical overlap in this case with findings from cases of frank deletion of the terminal region of chromosome 3p supports the hypothesis that the interruption of the long variant alone is sufficient to result in functional haploinsufficiency at this locus.
The region telomeric to the disrupted CNTN4 gene contains CNTN6 and CHL1, both neuronal cell adhesion molecules that also contain Ig and FNIII domains (fig. 3). Of note, a patient with nonspecific mental retardation and a balanced translocation disrupting CHL1 was described elsewhere (Frints et al. 2003). This patient was not found to have any other physical abnormalities associated with 3p deletion syndrome, and the translocation also involved the X chromosome, though no transcript was identified at that breakpoint. Analyses of CHL1 knockout mice, in this study and in others, have identified anomalies in axonal organization in various brain regions, as well as subtle behavioral abnormalities in homozygous null animals (Montag-Sallaz et al. 2002, 2003; Frints et al. 2003).
Given the proximity of these brain-expressed molecules with similar structures and putative functions, it is conceivable that the chromosomal rearrangement leading to disruption of CNTN4 could also be mediating functional consequences for neighboring transcripts and, thereby, leading to the observed phenotype. Although the possibility of position effect cannot be excluded on the basis of current data (State et al. 2003), the direct disruption of the CNTN4 transcript and the genomic distances separating CNTN6 and CHL1 from the breakpoint (∼0.7 Mb and ∼1.7Mb, respectively) suggest that the most likely effect of the rearrangement is on CNTN4 function. Moreover, two additional lines of evidence support a specific role for CNTN4 in conferring the observed phenotype. First, in the single reported case of a translocation disrupting CHL1, the patient did not show dysmorphic features consistent with 3p deletion syndrome (Frints et al. 2003), and, second, the 3p minimal candidate region (Cargile et al. 2002) includes the CNTN4 transcript, but not CNTN6 and CHL1 (fig. 3A), suggesting that position effect would have to be invoked as an explanation in both the interstitial deletion and translocation cases.
Our findings strongly suggest that disruption of a single copy of CNTN4 is sufficient to confer key aspects of the 3p deletion syndrome phenotype, including developmental delay. Thus, CNTN4 is one of a relatively small number of autosomal genes identified as contributing to cognitive deficits in a contiguous-gene syndrome. It is interesting to note that many features of a presumed multigene phenotype are manifested as the result of the disruption of only a single transcript. This finding provides a rationale for mutation screening of CNTN4 in selected patients with developmental delay and dysmorphic features who have normal karyotypes. Although, in the present case, there appears to be a strong relationship between alterations in CNTN4 expression and characteristic physical findings, nonetheless, it is noteworthy that a previous analysis of an extended pedigree identified evidence for a recessive locus conferring nonspecific mental retardation within a 7-cM region containing CNTN4 (Higgins et al. 2000). On the basis of the present findings, this gene should certainly be considered a candidate transcript in the linkage interval. Finally, it is particularly interesting that the patient’s developmental difficulties involve notable weaknesses in nonverbal aspects of learning as well as a delay in language acquisition. The identification of a likely etiological role for CNTN4 in these types of human cognitive deficits suggests that further studies of its expression and specific functions in the CNS are warranted.
Acknowledgments
The work was funded by a grant from the Shepherd Foundation (M.W.S.) as well as an National Institutes of Health career award (RR-16118 to M.W.S.). The authors wish to thank George and Lynn Bovenizer, whose support was instrumental in the conduct of this research. The authors would also like to thank Fred Volkmar for his continuing collaboration and Celine Saulnier, Eunice Huang, and Reid Warner of the Developmental Disabilities Clinic at the Yale Child Study Center for their highly skilled clinical assistance. Finally, we express our deepest gratitude to the patient and his family for their participation in this research.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Basic Local Alignment Search Tool (BLAST), http://www.ncbi.nlm.nih.gov/BLAST/ [DOI] [PubMed]
- BLAT Search Genome, http://genome.ucsc.edu/cgi-bin/hgBlat
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
- NCBI Reference Sequence (RefSeq), http://www.ncbi.nlm.nih.gov/RefSeq
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
- UCSC Genome Browser, http://genome.ucsc.edu
References
- Angeloni D, Lindor NM, Pack S, Latif F, Wei MH, Lerman MI (1999) CALL gene is haploinsufficient in a 3p− syndrome patient. Am J Med Genet 86:482–485 [DOI] [PubMed] [Google Scholar]
- Benini D, Vino L, Vecchini S, Fanos V (1999) 46, XY, del (3) (pter→p25) syndrome: further delineation of the clinical phenotype. Eur J Pediatr 158:955–957 10.1007/s004310051256 [DOI] [PubMed] [Google Scholar]
- Berglund EO, Murai KK, Fredette B, Sekerkova G, Marturano B, Weber L, Mugnaini E, Ranscht B (1999) Ataxia and abnormal cerebellar microorganization in mice with ablated contactin gene expression. Neuron 24:739–750 [DOI] [PubMed] [Google Scholar]
- Brand A, Reifen RM, Armon Y, Kerem E, Horenstein E, Gale R (1987) Double mitral valve, complete atrioventricular canal, and tricuspid atresia in chromosomal 3p− syndrome. Pediatr Cardiol 8:55–56 [DOI] [PubMed] [Google Scholar]
- Cargile CB, Goh DL, Goodman BK, Chen XN, Korenberg JR, Semenza GL, Thomas GH (2002) Molecular cytogenetic characterization of a subtle interstitial del(3)(p25.3p26.2) in a patient with deletion 3p syndrome. Am J Med Genet 109:133–138 10.1002/ajmg.10323 [DOI] [PubMed] [Google Scholar]
- Drumheller T, McGillivray BC, Behrner D, MacLeod P, McFadden DE, Roberson J, Venditti C, Chorney K, Chorney M, Smith DI (1996) Precise localisation of 3p25 breakpoints in four patients with the 3p− syndrome. J Med Genet 33:842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endris V, Wogatzky B, Leimer U, Bartsch D, Zatyka M, Latif F, Maher ER, Tariverdian G, Kirsch S, Karch D, Rappold GA (2002) The novel Rho-GTPase activating gene MEGAP/ srGAP3 has a putative role in severe mental retardation. Proc Natl Acad Sci USA 99:11754–11759 10.1073/pnas.162241099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frints SG, Marynen P, Hartmann D, Fryns JP, Steyaert J, Schachner M, Rolf B, Craessaerts K, Snellinx A, Hollanders K, D’Hooge R, De Deyn PP, Froyen G (2003) CALL interrupted in a patient with non-specific mental retardation: gene dosage-dependent alteration of murine brain development and behavior. Hum Mol Genet 12:1463–1474 10.1093/hmg/ddg165 [DOI] [PubMed] [Google Scholar]
- Green EK, Priestley MD, Waters J, Maliszewska C, Latif F, Maher ER (2000) Detailed mapping of a congenital heart disease gene in chromosome 3p25. J Med Genet 37:581–587 10.1136/jmg.37.8.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansford LM, Smith SA, Haber M, Norris MD, Cheung B, Marshall GM (2003) Cloning and characterization of the human neural cell adhesion molecule, CNTN4 (alias BIG-2). Cytogenet Genome Res 101:17–23 10.1159/000073412 [DOI] [PubMed] [Google Scholar]
- Higgins JJ, Rosen DR, Loveless JM, Clyman JC, Grau MJ (2000) A gene for nonsyndromic mental retardation maps to chromosome 3p25-pter. Neurology 55:335–340 [DOI] [PubMed] [Google Scholar]
- Kamei Y, Takeda Y, Teramoto K, Tsutsumi O, Taketani Y, Watanabe K (2000) Human NB-2 of the contactin subgroup molecules: chromosomal localization of the gene (CNTN5) and distinct expression pattern from other subgroup members. Genomics 69:113–119 10.1006/geno.2000.6310 [DOI] [PubMed] [Google Scholar]
- Knight LA, Yong MH, Tan M, Ng IS (1995) Del(3) (p25.3) without phenotypic effect. J Med Genet 32:994–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC (1990) High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247:64–69 [DOI] [PubMed] [Google Scholar]
- McClure RJ, Telford N, Newell SJ (1996) A mild phenotype associated with der(9)t(3;9) (p25;p23). J Med Genet 33:625–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag-Sallaz M, Baarke A, Montag D (2003) Aberrant neuronal connectivity in CHL1-deficient mice is associated with altered information processing-related immediate early gene expression. J Neurobiol 57:67–80 10.1002/neu.10254 [DOI] [PubMed] [Google Scholar]
- Montag-Sallaz M, Schachner M, Montag D (2002) Misguided axonal projections, neural cell adhesion molecule 180 mRNA upregulation, and altered behavior in mice deficient for the close homolog of L1. Mol Cell Biol 22:7967–7981 10.1128/MCB.22.22.7967-7981.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowrey PN, Chorney MJ, Venditti CP, Latif F, Modi WS, Lerman MI, Zbar B, Robins DB, Rogan PK, Ladda RL (1993) Clinical and molecular analyses of deletion 3p25-pter syndrome. Am J Med Genet 46:623–629 [DOI] [PubMed] [Google Scholar]
- Murai KK, Misner D, Ranscht B (2002) Contactin supports synaptic plasticity associated with hippocampal long-term depression but not potentiation. Curr Biol 12:181–190 10.1016/S0960-9822(02)00680-2 [DOI] [PubMed] [Google Scholar]
- Narahara K, Kikkawa K, Murakami M, Hiramoto K, Namba H, Tsuji K, Yokoyama Y, Kimoto H (1990) Loss of the 3p25.3 band is critical in the manifestation of del(3p) syndrome: karyotype-phenotype correlation in cases with deficiency of the distal portion of the short arm of chromosome 3. Am J Med Genet 35:269–273 [DOI] [PubMed] [Google Scholar]
- Ogawa J, Kaneko H, Masuda T, Nagata S, Hosoya H, Watanabe K (1996) Novel neural adhesion molecules in the Contactin/F3 subgroup of the immunoglobulin superfamily: isolation and characterization of cDNAs from rat brain. Neurosci Lett 218:173–176 10.1016/S0304-3940(96)13156-6 [DOI] [PubMed] [Google Scholar]
- Phipps ME, Latif F, Prowse A, Payne SJ, Dietz-Band J, Leversha M, Affara NA, Moore AT, Tolmie J, Schinzel A, Lerman MI, Ferguson-Smith MA, Maher ER (1994) Molecular genetic analysis of the 3p− syndrome. Hum Mol Genet 3:903–908 [DOI] [PubMed] [Google Scholar]
- Rolf B, Lang D, Hillenbrand R, Richter M, Schachner M, Bartsch U (2003) Altered expression of CHL1 by glial cells in response to optic nerve injury and intravitreal application of fibroblast growth factor-2. J Neurosci Res 71:835–843 10.1002/jnr.10533 [DOI] [PubMed] [Google Scholar]
- Saito H, Mimmack M, Kishimoto J, Keverne EB, Emson PC (1998) Expression of olfactory receptors, G-proteins and AxCAMs during the development and maturation of olfactory sensory neurons in the mouse. Brain Res Dev Brain Res 110:69–81 [DOI] [PubMed] [Google Scholar]
- Schwyzer U, Binkert F, Caflisch U, Baumgartner B, Schinzel A (1987) Terminal deletion of the short arm of chromosome 3, del(3pter-p25): a recognizable syndrome. Helv Paediatr Acta 42:309–315 [PubMed] [Google Scholar]
- Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, Scott JD (2003) Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Natl Acad Sci USA 100:1723–1728 10.1073/pnas.0438033100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow S, Balla D, Cicchetti D (1984) Vineland adaptive behavior scales, expanded ed. American Guidance Service, Circle Pines, MN [Google Scholar]
- State MW, Greally JM, Cuker A, Bowers PN, Henegariu O, Morgan TM, Gunel M, DiLuna M, King RA, Nelson C, Donovan A, Anderson GM, Leckman JF, Hawkins T, Pauls DL, Lifton RP, Ward DC (2003) Epigenetic abnormalities associated with a chromosome 18(q21-q22) inversion and a Gilles de la Tourette syndrome phenotype. Proc Natl Acad Sci USA 100:4684–4689 10.1073/pnas.0730775100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazelaar J, Roberson J, Van Dyke DL, Babu VR, Weiss L (1991) Mother and son with deletion of 3p25-pter. Am J Med Genet 39:130–132 [DOI] [PubMed] [Google Scholar]
- Verjaal M, De Nef MB (1978) A patient with a partial deletion of the short arm of chromosome 3. Am J Dis Child 132:43–45 [DOI] [PubMed] [Google Scholar]
- Wechsler D (1992) Wechsler intelligence scale for children, 3rd ed (WISC-III). Psychological Corporation, San Antonio, TX [Google Scholar]
- Yoshihara Y, Kawasaki M, Tamada A, Nagata S, Kagamiyama H, Mori K (1995) Overlapping and differential expression of BIG-2, BIG-1, TAG-1, and F3: four members of an axon-associated cell adhesion molecule subgroup of the immunoglobulin superfamily. J Neurobiol 28:51–69 [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang C, Xu J, Ye X, Wu Q, Dai J, Ji C, Gu S, Xie Y, Mao Y (2002) A novel splice variant of the cell adhesion molecule contactin 4 ( CNTN4) is mainly expressed in human brain. J Hum Genet 47:497–499 10.1007/s100380200073 [DOI] [PubMed] [Google Scholar]