Abstract
Myotonic dystrophy (DM) is caused by either an untranslated CTG expansion in the 3′ untranslated region of the DMPK gene on chromosome 19 (dystrophia myotonica type 1 [DM1]), or an untranslated CCTG tetranucleotide repeat expansion in intron 1 of the ZNF9 gene on chromosome 3 (dystrophia myotonica type 2 [DM2]). RNA-binding proteins adhere to transcripts of the repeat expansions that accumulate in the nucleus, and a trans-dominant dysregulation of pre-mRNA alternative splicing has been demonstrated for several genes. In muscle from patients with DM1, altered insulin-receptor splicing to the nonmuscle isoform corresponds to the insulin insensitivity and diabetes that are part of the DM phenotype; because of insulin-receptor species differences, this effect is not seen in mouse models of the disease. We now demonstrate that comparable splicing abnormalities occur in DM2 muscle prior to the development of muscle histopathology, thus demonstrating an early pathogenic effect of RNA expansions.
Myotonic dystrophy type 1 (DM1 [MIM 160900]) and myotonic dystrophy type 2 (DM2 [MIM 602668]) are complex multisystemic disorders with a constellation of unusual and seemingly unrelated clinical features, including myotonia (inability of muscle to relax), muscular dystrophy with characteristic histological features, distinctive iridescent cataracts, cardiac conduction defects, and various endocrine abnormalities, including insulin insensitivity and testicular failure (Harper 2001; Day et al. 2003). DM1 is caused by an expanded CTG trinucleotide repeat in the 3′ UTR of the dystrophia myotonica-protein kinase (DMPK) gene (Brook et al. 1992); DM2 is caused by a CCTG tetranucleotide expansion within intron 1 of the zinc finger protein 9 (ZNF9) gene (Liquori et al. 2001). In DM1, RNA containing the CUG expansion accumulates in ribonuclear foci and has been associated with dysregulation of RNA-binding proteins that have been proposed to result in a novel pathogenic mechanism via a trans-dominant effect on pre-mRNA alternative splicing (Philips et al. 1998; Savkur et al. 2001; Charlet et al. 2002; Mankodi et al. 2002; Kanadia et al. 2003). This mechanism underlies misregulated splicing of the insulin receptor (IR) and the muscle-specific chloride-channel pre-mRNAs in DM1, leading to insulin insensitivity and myotonia (Savkur et al. 2001; Charlet et al. 2002; Mankodi et al. 2002). Our discovery—that a transcribed but untranslated CCTG expansion causes DM2 and that, similar to DM1, CCUG repeat–containing RNA accumulates in ribonuclear inclusions of affected DM2 tissues—strongly suggests that pathogenic effects of RNA cause the clinical features common to both diseases (Liquori et al. 2001; Ranum and Day 2002). To further investigate myotonic dystrophy (DM) pathogenesis, we have now investigated alternative splicing in DM2.
Alternative splicing of the IR transcripts generates two isoforms: isoform A (IR-A), which lacks exon 11, and isoform B (IR-B), which includes exon 11 (Seino and Bell 1989). Normally, IR-B predominates in insulin-responsive tissues such as skeletal muscle, liver, and adipose tissue. IR-B has greater insulin responsiveness than does IR-A (Moller et al. 1989). A consistent finding in individuals with DM1 is expression of the nonmuscle IR-A isoform in skeletal-muscle tissue and primary-muscle cultures (Furling et al. 2001; Savkur et al. 2001). To investigate the role of misregulated alternative splicing in DM2 pathogenesis, we investigated IR splicing in muscle from subjects with DM2.
Subjects with DM2 were enrolled from the MN1 family in which the DM2 locus was initially mapped (Ranum et al. 1998). All patients had the cardinal features of DM2: muscle weakness (often restricted to neck flexors, deep-finger flexors, or hip extensors), electrical myotonia, muscular dystrophy, and cataracts. All muscle biopsies were performed on the vastus lateralis, except for a single diagnostic biceps brachii biopsy done of a subject who, 8 years later, also had a research biopsy of the vastus lateralis. Glucose-tolerance testing was performed following a 12-h fast, with measurement of the glucose and insulin levels before and at .5-h intervals after ingestion of 200 g of glucose. Measurements of glucose intolerance included fasting glucose, fasting insulin (Laakso 1993), glucose measurement 2 h after glucose ingestion, and the homeostasis model assessment (HOMA) ratio (Matthews et al. 1985). Results for subjects with DM2 were compared with results in normal subjects; subjects with DM1 underwent biopsies but were not recruited for the glucose-tolerance test. All studies were done under approval of the University of Minnesota institutional review board for human research.
DNA from blood cells was used to verify the presence of a DM2 expansion, as reported elsewhere (Day et al. 2003). Muscle specimens for both biochemical and histological analyses were quick-frozen in methyl butane that was suspended in liquid nitrogen. Routine histology on frozen sections of muscles was graded for each of four features (fibrosis, necrosis, central nuclei proliferation, and presence of pyknotic nuclear clump fibers), grading each feature as absent (0), moderately present (1), or diffusely present (2); the sum of scores for all four features is reported in table 1 as the biopsy score (Bx Score) (0–8, 8 being maximally abnormal). To verify that expanded CCUG transcripts accumulate as ribonuclear inclusions, muscle biopsies were also evaluated, by RNA-FISH, as described elsewhere (Liquori et al. 2001). RNA was extracted from muscle, and the percentage of IR mRNAs containing exon 11 was determined by RT-PCR (fig. 1), as described elsewhere (Savkur et al. 2001). IR RT-PCR results are expressed in tables 1 and 2 as percentage of IR-B, which was calculated from the RT-PCR results: [CPMIR-B/(CPMIR-A+CPMIR-B)]×100. Mean and SEs are given for the different subject groups, and P values are given, as determined by Student's t test comparisons between DM2 and DM1 and between DM2 and normal muscle. Reference ranges shown in the table define the various glucose and insulin measurements for normal subjects, subjects with impaired glucose tolerance, and subjects with non–insulin-dependent diabetes mellitus.
Table 1.
Comparison of Glucose Tolerance, Muscle Histology, and IR Isoform Expression for Subjects with DM2, Subjects with DM1, and Normal Subjects[Note]
| Subject | Sex | Age at Biopsy(years) | Fasting Glucosea(mg/dl) | Fasting Insulinb(mU/liter) | 2-h Glucosec(mg/dl) | HOMA Ratiod | Bx Score | IR-B(%) |
| DM2 1 | M | 52 | 107 | 9 | 135 | 2.4 | 5 | 12 |
| DM2 2 | M | 35 | 94 | 22 | 187 | 5.1 | 4 | 18 |
| DM2 3 | M | 33 | 95 | 13 | 109 | 3.1 | 3 | 20 |
| DM2 4 | M | 66 | 89 | 16 | 165 | 3.5 | 6 | 23 |
| DM2 5 | M | 43 | 86 | 4 | 23 | |||
| DM2 6 | M | 64 | 84 | 20 | 132 | 4.2 | 6 | 26 |
| DM2 7a | F | 26 | 0 | 26 | ||||
| DM2 7b | F | 34 | 96 | 23 | 90 | 5.5 | 6 | 4 |
| DM2 8 | M | 35 | 93 | 8 | 114 | 1.9 | 3 | |
| DM2 9 | M | 55 | 98 | 9 | 93 | 2.2 | 6 | |
| DM2 10 | M | 42 | 89 | 11 | 121 | 2.4 | 7 | |
| DM2 11 | M | 52 | 176 | 13 | 279 | 5.7 | 7 | |
| DM2 12 | F | 65 | 229 | 18 | 379 | 10.3 | 4 | |
| DM2 13 | F | 62 | 102 | 67 | 153 | 17.0 | 4 | |
| Mean±SD | 47±14 | 111±43 | 19±16 | 163±85 | 5.3±4.4 | 5±2 | 19±8 | |
| DM1 A | M | 46 | 75 | 6 | 10 | |||
| DM1 B | F | 59 | 86 | 1 | 35 | |||
| DM1 C | M | 30 | 102 | 0 | 41 | |||
| Mean±SD | 45±15 | 88±14 | 2±3 | 29±16 | ||||
| Normal A | F | 48 | 83 | 3 | 123 | .6 | 0 | 60 |
| Normal B | F | 70 | 87 | 3 | 118 | .6 | 0 | |
| Normal C | F | 37 | 88 | 6 | 112 | 1.3 | 0 | |
| Mean±SD | 52±17 | 86±3 | 4±2 | 118±6 | .8±.4 | 0±0 |
Note.— For DM2 versus DM1: age at biopsy P=.84; fasting-glucose level P=.24; Bx Score P=.43; IR-B% P=.42. For DM2 versus for normal subjects: age at biopsy P=.71; fasting-glucose level P=.06; fasting-insulin level P=.008; 2-h glucose level P=.09; HOMA Ratio P=.005; Bx Score P=.000.
Expected fasting glucose levels: normal (Nl) <110 mg/dl; impaired glucose tolerance (IGT) 110–126 mg/dl; non–insulin-dependent diabetes mellitus (NIDDM) >126 mg/dl.
Expected fasting insulin levels: Nl <20 mU/liter; IGT ⩾20 mU/liter.
Expected 2-h glucose levels: Nl <140 mg/dl; IGT 140–200 mg/dl; NIDDM >200 mg/dl.
Expected HOMA ratio: Nl 0–3.2; IGT 3.3–6.8; NIDDM >6.8.
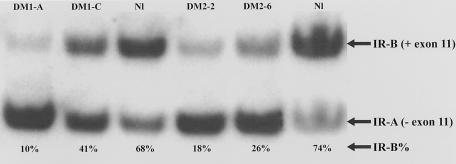
Figure 1.
Measurement of IR isoforms: RT-PCR analyses of IR isoforms in RNA from DM1, DM2, and normal muscle biopsies. The upper band represents the insulin-sensitive IR-B isoform (containing exon 11), and the lower band shows the insulin-insensitive IR-A isoform (without exon 11). In DM1 and DM2 muscle, the IR-A isoform predominates, as evidenced by the low percentage of IR-B relative to that in unaffected control.
Table 2.
Comparison of IR Isoform Expression in Subjects with DM1, Subjects with DM2, and Normal Subjects
| Subjects | Percentage of Insulin-Sensitive IR-B (Mean±SD) |
| Unaffected (n=9) | 66±7 |
| With DM1 (n=4) | 29±12 |
| With DM2 (n=8) | 19±8 |
Of the 13 subjects with DM2 who were tested for glucose intolerance, 7 showed some degree of insulin insensitivity (table 1). The most sensitive indicator of insulin insensitivity, the HOMA ratio, was abnormal in seven subjects. Two subjects had abnormal fasting glucose levels, and five had abnormal glucose levels 2 h after glucose administration (table 1). As indicated in table 1, fasting insulin levels and the HOMA ratio significantly (P<.01) differed between subjects with DM2 and normal subjects, with a trend in subjects with DM2 toward abnormal fasting glucose and 2-h glucose determinations during the glucose-tolerance test.
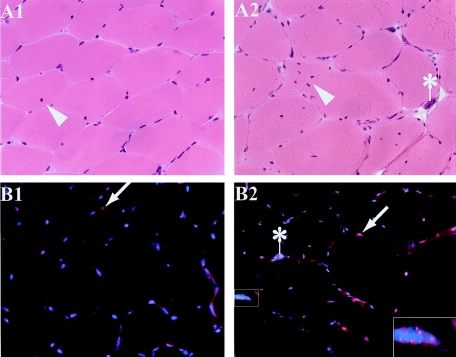
Biopsy results of 13 subjects were analyzed. One subject (DM2 7) (table 1) had two biopsies 8 years apart; results of her first biopsy (DM2 7a), obtained from the biceps brachii at the time of initial diagnosis at age 26 years, were histologically normal, but the second (DM2 7b), from the vastus lateralis at age 34 years, had histological changes typical of DM2. All other biopsy results from patients with DM2 showed the typical pathology of DM, including atrophic angulated fibers, severely atrophic fibers with pyknotic nuclei, and proliferation of centrally located nuclei. When examined by RNA-FISH, all muscle specimens, including the histologically normal 1993 biopsy from individual 7 (specimen DM2 7a), had ribonuclear inclusions detectable by FISH (fig. 2). The detection of ribonuclear inclusions in sample 7a before development of pathological changes indicates that the ribonuclear inclusions occur before histopathology rather than being an indirect consequence of the dystrophic changes.
Figure 2.
Histological and FISH features in DM2: histological sections of skeletal-muscle biopsies on the same patient, performed 8 years apart. The earlier biopsy from biceps brachii (A1 and B1) shows normal histology, with only one fiber in the entire section having a single central nucleus (arrowhead). The later biopsy from vastus lateralis (A2 and B2) shows characteristic histological features of DM, with angulated fibers, severely atrophic fibers with pyknotic nuclei (“nuclear bag fibers”) (indicated by an asterisk [*]), and proliferation of centrally located nuclei (arrowhead). RNA-FISH shows ribonuclear inclusions (red foci, small arrows) within blue DAPI-stained nuclei in both samples (B1 and B2), including within all nuclei of a severely atrophic fiber (inset).
The percentage of IR-A and IR-B isoforms of the IR in skeletal muscle from subjects with DM1, subjects with DM2, and control individuals was determined by RT-PCR by use of 32P-labeled forward PCR oligo, as described elsewhere (Savkur et al. 2001). The percentage of the IR-B splice form was reduced in all DM2 specimens, compared with specimens from normal control individuals (fig. 1; table 2). The percentage of IR-B in normal control individuals (table 2) differed significantly from that in DM2 muscle (normal range 60%–80%, mean 66±7%; DM2 range 8%–40%, mean 19±8%; P<10-8). Muscle biopsy 7a, which had normal histology (Bx score 0) (table 1) but did contain ribonuclear inclusions, had a clearly abnormal percentage of IR-B (26%) (table 1). The sample from this same individual 8 years later showed abnormal histology (Bx score 6) (table 1) and only 4% IR-B (table 1). These results demonstrate that IR splicing abnormalities and ribonuclear inclusions both precede development of dystrophic changes.
Our results demonstrate that IR splicing is altered in DM2; exon 11 inclusion is significantly lower in muscle from subjects with DM2 than in normal subjects. Similar results have been reported elsewhere in developing muscle and in adult skeletal muscle from subjects with DM1 (Savkur et al. 2001). This splice change results in reduced expression of insulin-sensitive receptors in skeletal muscle and corresponds to clinical observation of insulin insensitivity and diabetes in DM1 and DM2. The degree of IR-splicing change in DM2 (mean 25% IR-B) is comparable to the change found in DM1 (mean 38% IR-B), although other variables—such as age, muscle histopathology, or obesity—may also affect the ratio. The role of altered IR splicing has not been evaluated in typical patients with type 2 diabetes mellitus who do not have muscular dystrophy, in whom insulin insensitivity can result from other changes in receptor function. We have shown elsewhere that muscle specimens from subjects with other forms of muscular dystrophy and other neuromuscular disorders do not have abnormal IR splicing (Savkur et al. 2001). The data reported here of IR splicing in DM2, taken together with other evidence of altered gene splicing in DM1 and DM2 (Philips et al. 1998; Savkur et al. 2001; Charlet et al. 2002; Mankodi et al. 2002; Kanadia et al. 2003), support the model that altered RNA processing underlies DM pathogenesis, though additional functionally significant changes in IR could also occur.
Evaluation of the histologically normal DM2 muscle biopsy shows that pathogenic splicing alterations occur prior to development of dystrophic changes, verifying that aberrant RNA processing is an early feature of DM rather than being secondary to muscle-fiber degeneration or regeneration. Ribonuclear inclusions were present in the histologically normal specimen, as they were in all histologically abnormal specimens. The role of these inclusions in disease pathogenesis remains unclear, but inclusions and splicing changes are clearly present prior to the development of muscle degeneration. Finding that splicing changes precede histological abnormalities supports the view that splicing abnormalities are a primary event of the disease rather than being secondary to expansion of the satellite cell population during regeneration. Overt regeneration was not observed in these specimens that showed splicing changes, consistent with studies on DM1 tissues that showed the splicing switch in tissues in which markers of skeletal-muscle regeneration were not elevated (Savkur et al. 2001). Our finding that IR splicing changes in DM2 are an early molecular change in the disease is consistent with the observation that insulin resistance is an early manifestation of DM1 (Moxley et al. 1978, 1980).
Although abnormal chloride-channel splicing has been demonstrated in muscle both from patients with DM and from transgenic mice that carry a pathogenic CTG expansion, IR splicing can be investigated only in humans, because mouse skeletal muscle does not regulate IR splicing in the same way that human muscle does (Kanadia et al. 2003). Consequently, our identification of abnormal alternative IR splicing in skeletal muscle from subjects with DM2 and DM1 verifies the importance of this mechanism in development of clinical insulin insensitivity in a way that cannot be studied in the mouse models. The fact that DM2 and DM1 result in similar IR-splicing abnormalities provides additional evidence that these genetically distinct disorders share a common molecular mechanism. Since the genes at the DM1 and DM2 loci have no evident functional or mechanistic similarities, and because both disorders are caused by untranslated microsatellite expansions, the most likely explanation is that the multisystemic features of both disorders result from the deleterious effects of RNA transcripts containing CUG or CCUG repeat expansions.
Acknowledgments
Support from National Institutes of Health (NIH) grant R01AR45653, Muscular Dystrophy Association USA, and the Hunter Research Fund (to T.C.) and support from University of Minnesota General Clinical Research Center grant MO1-RR00400, the Muscular Dystrophy Association USA, and NIH grant R01NS35870 (to L.P.W.R. and J.W.D.) is gratefully acknowledged. We are also very grateful to members of the MN-1 family and other DM-affected families, without whose involvement this work would have been impossible.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM) http://www.ncbi.nlm.nih.gov/Omim/ (for DM1 and DM2) [PubMed]
References
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, Sohn R, Zemelman B, Snell RG, Rundle SA, Crow S, Davies J, Shelbourne P, Buxton J, Jones C, Juvonen V, Johnson K, Harper PS, Shaw DJ, Housman DE (1992) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 69:385 [DOI] [PubMed] [Google Scholar]
- Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA (2002) Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell 10:45–53 10.1016/S1097-2765(02)00572-5 [DOI] [PubMed] [Google Scholar]
- Day JW, Ricker K, Jacobsen J, Rasmussen L, Dick K, Kress W, Schneider C, Koch M, Beilman G, Harrison A, Dalton J, Ranum L (2003) Myotonic dystrophy type 2: molecular, diagnostic and clinical spectrum. Neurology 60:657–664 [DOI] [PubMed] [Google Scholar]
- Furling D, Coiffier L, Mouly V, Barbet JP, St Guily JL, Taneja K, Gourdon G, Junien C, Butler-Browne GS (2001) Defective satellite cells in congenital myotonic dystrophy. Hum Mol Genet 10:2079–2087 10.1093/hmg/10.19.2079 [DOI] [PubMed] [Google Scholar]
- Harper PS (2001) Myotonic dystrophy. Vol 37. W. B. Saunders, London [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS (2003) A muscleblind knockout model for myotonic dystrophy. Science 302:1978–1980 10.1126/science.1088583 [DOI] [PubMed] [Google Scholar]
- Laakso M (1993) How good a marker is insulin level for insulin resistance? Am J Epidemiol 137:959–965 [DOI] [PubMed] [Google Scholar]
- Liquori C, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor S, Day JW, Ranum LPW (2001) Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 293:864–867 10.1126/science.1062125 [DOI] [PubMed] [Google Scholar]
- Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA (2002) Expanded CUG Repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell 10:35–44 10.1016/S1097-2765(02)00563-4 [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC (1985) Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28:412–419 [DOI] [PubMed] [Google Scholar]
- Moller DE, Yokota A, Caro JF, Flier JS (1989) Tissue-specific expression of two alternatively spliced insulin receptor mRNAs in man. Mol Endocrinol 3:1263–1269 2779582 [DOI] [PubMed] [Google Scholar]
- Moxley RT, Griggs RC, Goldblatt D (1980) Muscle insulin resistance in myotonic dystrophy: effect of supraphysiologic insulinization. Neurology 30:1077–1083 6999379 [DOI] [PubMed] [Google Scholar]
- Moxley RT III, Griggs RC, Goldblatt D, VanGelder V, Herr BE, Thiel R (1978) Decreased insulin sensitivity of forearm muscle in myotonic dystrophy. J Clin Invest 62:857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280:737–741 10.1126/science.280.5364.737 [DOI] [PubMed] [Google Scholar]
- Ranum LP, Day JW (2002) Myotonic dystrophy: clinical and molecular parallels between myotonic dystrophy type 1 and type 2. Curr Neurol Neurosci Rep 2:465–470 [DOI] [PubMed] [Google Scholar]
- Ranum LPW, Rasmussen P, Benzow K, Koob M, Day JW (1998) Genetic mapping of a second myotonic dystrophy locus. Nat Genet 19:196–198 10.1038/570 [DOI] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA (2001) Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet 29:40–47 10.1038/ng704 [DOI] [PubMed] [Google Scholar]
- Seino S, Bell GI (1989) Alternative splicing of human insulin receptor messenger RNA. Biochem Biophys Res Commun 159:312–316 [DOI] [PubMed] [Google Scholar]