To the Editor:
In the January 2004 issue of AJHG, Groman et al. published a collaborative study on the disease penetrance of a common abbreviated tract of five thymidines (T5) in intron 8 of the CFTR gene (MIM 602421). By analyzing a large number of affected individuals presenting with male infertility or nonclassical cystic fibrosis (CF [MIM 219700]), they found that the pathogenic or benign effect of T5 correlated with variations in a TG repeat sequence just upstream of the polypyrymidines. In particular, longer TG repeats (12 or 13) are associated with disease phenotypes, and 91% of affected individuals, as opposed to 22% of unaffected ones, have repetitions of 12 or 13 TGs. Although the modifier effect of the TG repeats on the T5 variant has been proposed elsewhere (Cuppens et al. 1998), the principal novelty of the Groman et al. study lies in the high number of patients analyzed from different regions. As the T5 allele is found in ∼10% of the general population, the results of this study strongly indicate that determination of the TG repeats may have a significant diagnostic value.
The pathologies studied by Groman et al. are in line with the growing number of human diseases resulting from pre-mRNA splicing alterations and, specifically, those diseases caused by mutations in cis-acting elements (the TG and T repeats) that disrupt the use of alternative splice sites, as recently reviewed by Faustino and Cooper (2003). In this specific case, the pathogenetic role of these repeats is owing to their effects on CFTR exon 9 at the level of pre-mRNA splicing, with longer UG repeats (and short U tracts) increasing exon 9 skipping and leading to the production of a nonfunctional protein (Delaney et al. 1993; Strong et al. 1993).
An obvious question, not taken into consideration in the study by Groman et al., is what factor(s) or mechanism(s) determine the TG repeat number to be a reliable predictor of penetrance for the T5 allele. Indeed, some mechanisms have been proposed already, and, >2 years ago, we reported that TDP-43 binds specifically to the UG repeat sequence and, in this way, promotes skipping of CFTR exon 9 (Buratti et al. 2001). In the same study, overexpression of TDP-43 increased CFTR exon 9 skipping in a minigene system, and ablation of its expression by antisense oligonucleotides resulted in increased inclusion of the same exon. Moreover, TDP-43’s binding to the UG repeat sequence has been shown to correlate well with repeat length, and this has provided a clear rationale as to why longer UG tracts determine a higher rate of exon 9 skipping (Buratti and Baralle 2001). In this respect, it may be worthwhile also to point out that an independent and recent study confirmed that the mouse homologue of human TDP-43 (mTDP-43) also inhibits human CFTR exon 9 splicing in a minigene system (Wang et al. 2004).
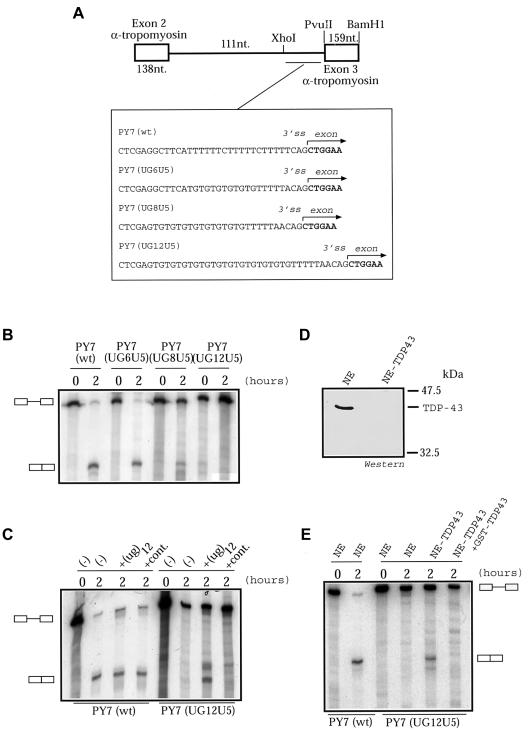
To demonstrate that the UG–TDP-43 interaction plays a direct role in splicing, we set up an in vitro splicing assay, a useful system to define the details of the molecular mechanisms involved and the role played by individual splicing factors. To this end, we introduced different TG repeats followed by 5T in the 3′ splice site in a heterologous context composed by two tropomyosin exons separated by a 111-nt intron sequence (see boxed sequences in fig. 1A). In vitro analysis of the splicing pattern of the transcribed RNA from these plasmids shows that UG elements repress splicing. In fact, increasing the number of UG repeats (UG6, UG8, and UG12) results in progressive splicing inhibition (fig. 1B). This direct role of UG repeats in repressing splicing also can be observed following the addition of an unlabeled (UG)12 RNA competitor to the splicing mix: as shown in figure 1C, this action can restore splicing activity of the UG12U5 RNA. As a control, this figure also shows that addition of cold (UG)12 RNA oligonucleotide does not have any effect on the splicing of the PY7(wt) construct (which does not contain any UG repeats) and that the addition of equal amounts of a control 30-mer RNA oligonucleotide has no effect on both the PY7(wt) and the PY7(UG12U5) RNAs.
Figure 1.
Interaction of UG repeats with TDP-43, causing splicing inhibition. A, Diagram of the in vitro splicing assay. Boxes indicate exons, and the horizontal line indicates intronic sequences. In the UG6U5 construct, an A residue was removed in an attempt to keep the distance to the branch point. This was not possible for UG8 and UG12, and the original CFTR 3′ splice site was maintained. All constructs were transcribed by use of SP6 RNA polymerase and assayed with HeLa cells nuclear extracts (C4, Biotech) at time 0 hours and after 2 hours, in accordance with standard protocols. Unprocessed and spliced RNAs (left) were resolved on 6% denaturing gels. B, Spliced RNAs, present in the PY7(wt) and PY7(UG6U5) transcripts, reduced in the PY7(UG8U5) construct, and absent in PY7(UG12U5). C, Binding competition analysis. Unlabeled (UG)12 RNA was added in a 10-fold molar excess to the splicing mix, successfully rescuing splicing in the PY7(UG12U5) construct but having no effect on the PY7(wt) RNA. D, Western blot of normal (NE) and TDP-43 depleted (NE-TDP43) nuclear extracts. TDP-43 is completely absent after depletion. E, Depletion of TDP-43 from the nuclear extract induces the appearance of the spliced form (lane NE-TDP43) in the UG12-containing construct. The addition of purified GST-TDP-43 (300 ng) completely restores the splicing inhibition (lane NE-TDP43+ GST-TDP43).
Having determined that (UG)12 sequences can promote splicing inhibition in vitro, we then checked whether depletion of TDP-43 from the splicing mix (fig. 1D) results in activation of splicing. Figure 1E shows that, indeed, depletion of TDP-43 from the nuclear extract results in activation of splicing in the UG12U5 RNA, an activation that can be abolished in the depleted extracts by the addition of recombinant TDP-43 (GST-TDP43). In conclusion, all these results are consistent with the model in which the TG repeats in the CFTR intron 8 bind to TDP-43, and this protein, in turn, inhibits splicing of exon 9.
Taken together, our results provide not only a mechanistic explanation for the association data of Groman et al. but also may provide a basis to explain the variable phenotypic penetrance of the TG repeats. In fact, individual and tissue-specific variability in the concentration of this inhibitory splicing factor (Buratti et al. 2001) may explain the not-yet-complete penetrance of the TG tract in 22% of the unaffected individuals with the T5 variant and a high TG number (Groman et al. 2004) and may even represent one of the possible factors that determine whether an individual will develop multisystemic (nonclassical CF) or monosymptomatic (CBAVD) disease.
Acknowledgments
This study was supported by grants from the Italian Telethon (GGP02453), Fondo per gli Investimenti della Ricerca di Base (RBNE01W9PM), and the Italian Cystic Fibrosis Research Foundation.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM) http://www.ncbi.nlm.nih.gov/Omim
References
- Buratti E, Baralle FE (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276:36337–36343 10.1074/jbc.M104236200 [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE (2001) Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J 20:1774–1784 10.1093/emboj/20.7.1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppens H, Lin W, Jaspers M, Costes B, Teng H, Vankeerberghen A, Jorissen M, Droogmans G, Reynaert I, Goossens M, Nilius B, Cassiman JJ (1998) Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes: the polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J Clin Invest 101:487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney SJ, Rich DP, Thomson SA, Hargrave MR, Lovelock PK, Welsh MJ, Wainwright BJ (1993) Cystic fibrosis transmembrane conductance regulator splice variants are not conserved and fail to produce chloride channels. Nat Genet 4:426–431 [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA (2003) Pre-mRNA splicing and human disease. Genes Dev 17:419–437 10.1101/gad.1048803 [DOI] [PubMed] [Google Scholar]
- Groman JD, Hefferon TW, Casals T, Bassas L, Estivill X, Des Georges M, Guittard C, et al (2004) Variation in a repeat sequence determines whether a common variant of the cystic fibrosis transmembrane conductance regulator gene is pathogenic or benign. Am J Hum Genet 74:176–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong TV, Wilkinson DJ, Mansoura MK, Devor DC, Henze K, Yang Y, Wilson JM, et al (1993) Expression of an abundant alternatively spliced form of the cystic fibrosis transmembrane conductance regulator (CFTR) gene is not associated with a cAMP-activated chloride conductance. Hum Mol Genet 2:225–230 [DOI] [PubMed] [Google Scholar]
- Wang HY, Wang IF, Bose J, Shen CK (2004) Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics 83:130–139 10.1016/S0888-7543(03)00214-3 [DOI] [PubMed] [Google Scholar]