Abstract
Weill-Marchesani syndrome (WMS) is characterized by the association of short stature; brachydactyly; joint stiffness; eye anomalies, including microspherophakia and ectopia of the lenses; and, occasionally, heart defects. We have recently mapped a gene for the autosomal recessive form of WMS to chromosome 19p13.3-p13.2, in a 12.4-cM interval. Here, we report null mutations in a member of the extracellular matrix protease family, the gene encoding ADAMTS10, a disintegrin and metalloprotease with thrombospondin motifs. A total of three distinct mutations were identified in two consanguineous families and in one sporadic WMS case, including one nonsense mutation (R237X) and two splice mutations (1190+1G→A and 810+1G→A). ADAMTS10 expression studies using reverse-transcriptase polymerase chain reaction, northern blot, and dot-blot analyses showed that ADAMTS10 is expressed in skin, fetal chondrocytes, and fetal and adult heart. Moreover, electron microscopy and immunological studies of the skin fibroblasts from the patients confirmed impairment of the extracellular matrix. We conclude, therefore, that ADAMTS10 plays a major role in growth and in skin, lens, and heart development in humans.
Introduction
Weill-Marchesani syndrome (WMS [MIM 277600]) is characterized by the association of short stature; brachydactyly; joint stiffness; eye anomalies, including microspherophakia, ectopia of the lenses, severe myopia, and glaucoma; and, occasionally, heart defects. Despite the disease's clinical homogeneity, autosomal recessive (AR) and autosomal dominant (AD) modes of inheritance have been reported, and we have recently identified an in-frame deletion of the fibrillin 1 gene in a family with AD WMS (Faivre et al. 2003a, 2003b). Using a homozygosity-mapping strategy in two consanguineous families from Lebanon and Saudi Arabia, we reported linkage of the AR WMS gene to chromosome 19p13.3-p13.2, in a 12.4-cM interval (Faivre et al. 2002). Several candidate genes involved in the extracellular matrix (ECM) structure were considered, including the gene encoding ADAMTS10 (GenBank accession number NM_030957), a disintegrin and metalloprotease with thrombospondin (TS) motifs. Here, we present three distinct mutations of the ADAMTS10 gene in these two families and in one sporadic WMS case, including one nonsense mutation and two splice mutations. Previous electron microscopy studies of skin fibroblasts from affected individuals suggested an impairment of the ECM, and our immunological data support the disorganization of the cytoskeleton in WMS. The data presented here provide the first evidence of the involvement of a member of the ADAMTS family in brachydactyly and microspherophakia.
Material and Methods
Patients
A total of six children belonging to three families were included in the present study. In two sibships, an AR mode of inheritance has been established, and the disease gene has been mapped to chromosome 19p13.3 (Faivre et al. 2002). The disease in the third subject is isolated and sporadic. All affected children fulfilled the criteria for WMS—that is, short stature, brachydactyly, limitation of joint movement, microspherophakia, dislocated lenses, severe myopia, and glaucoma. Additional features included pulmonary stenosis (in two patients) and aortic and pulmonary stenosis with dysplastic valves and hypertrophic obstructive cardiomyopathy (in two patients).
Molecular Studies
Blood samples were obtained with the written consent of the patients and unaffected relatives. Genomic DNA was extracted from leukocytes, according to standard procedures. A series of 48 intronic primers (available on request) were designed to amplify the 24 coding exons of the ADAMTS10 gene. Amplification products were purified and sequenced by use of the fluorescent dideoxy-terminator method on an ABI 3100 automatic sequencer.
Total RNAs were extracted from human primary cultured cells (fetal chondrocytes and skin fibroblasts) through use of the RNeasy Mini Kit (Qiagen). cDNA was synthesized by priming with random hexamers in the presence of MuLV reverse transcriptase through use of the manufacturer’s protocol (GeneAmp RNA PCR Core Kit [Roche]). Thirty-five to 40 PCR cycles were performed, at an annealing temperature of 56°C, to amplify a 470-bp fragment specific for the ADAMTS10 gene (forward: 5′-TGCAGCGTCAATGAGGACAT-3′; reverse: 5′-AGCACCACCCCTTGTCGAT-3′). Sequences of the sense and antisense primers used for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplification were as follows: 5′-AGACAGCCGCATCTTCTTGT and 3′-CCACAGTCTTCTGAGTGGCA (587 bp).
Premade northern blot containing 2 μg of poly(A)+RNA per lane from four human fetal tissues (brain, lung, liver, and kidney) and a human multiple-tissue expression array were used (ref 7756-1 and ref 7776-1 [Clontech Laboratories]). A 413-bp cDNA fragment of ADAMTS10 was obtained by amplification of fibroblast cDNA. The sense and antisense primer sequences were 5′-CCTAGCGGAAGGCTTCAACT and 3′-AGTCGCAGTTGAAAGGTGGT, respectively. This fragment was then purified, labeled by random priming (Amersham Pharmacia Biotech), and incubated overnight in a hybridization buffer at 42°C. The two blots were washed three times at 42°C with 2×SSC:0.1% SDS for 15 min and once under more stringent conditions, at 65°C with 0.1×SSC:0.1% SDS for 20 min. The blots were then exposed to Kodak X-Omat films with an intensifying screen at −80°C.
Immunological Studies
Subconfluent control and WMS fibroblasts grown on a plastic support were fixed with 4% paraformaldehyde in PBS for 30 min. Cells were then stained with Phalloidin-Alexa (Molecular Probes) and examined by confocal microscopy.
Results
More than 100 known genes were found to map to the WMS critical region on chromosome 19p13 through use of publicly available sequences (National Center for Biotechnology Information; the Human Genome Working Draft at the University of California–Santa Cruz Genome Bioinformatics Web site). Of these genes, five were regarded as possible candidate genes on the basis of their function—namely, COL5A3; intercellular adhesion molecule 1, 4, and 5 (ICAM1, -4, and -5); the SWI/SNF-related matrix-associated actin-dependent regulator of chromatin, subfamily A, member 4 (SMARCA4); the tubulin β 5 gene (TUBB5); and ADAMTS10.
No deleterious mutations were identified by direct sequencing of the COL5A3 gene. We then considered ADAMTS10 as a candidate gene, since it belongs to a family of proteases involved in the cleavage of procollagen amino propeptide and aggrecan (Tang 2001; Cal et al. 2002; Apte 2004). ADAMTS10 is composed of 24 coding exons and encodes a protein of 1,103 amino acids that is composed of one disintegrin-like domain, one metalloprotease domain, one cysteine-rich domain, and five TS type 1 repeats. By direct sequencing, three distinct mutations were identified, including one nonsense mutation (R237X) and two splice-site mutations (1190+1G→A and 810+1G→A) (fig. 1). The three mutations predicted premature termination of translation and were located in the predicted metalloprotease domain (table 1). The mutations and the flanking polymorphic markers were found to cosegregate with the disease (fig. 2) and were not identified in 210 control chromosomes.
Figure 1.
Structure of the ADAMTS10 gene. The three mutations are located in the metalloprotease domain.
Table 1.
ADAMTS10 Mutations Identified in Three Families with WMS
| Family | Countryof Origin | ParentsConsanguineous | Nucleotide Change | Amino Acid Change | MutatedExon(s) |
| 1 | Lebanon | Yes | 709C→T/709C→T | R237X/R237X | 6/6 |
| 2 | Saudi Arabia | Yes | 1190+1G→A/1190+1G→A | Frameshift causing a stop, 1 codon downstream | 10/10 |
| 3 | France | No | 709C→T/810+1G→A | R237X/frameshift causing a stop, 20 codons downstream | 6/6 |
Figure 2.
ADAMTS10 mutations and segregation of flanking markers in the two consanguineous families
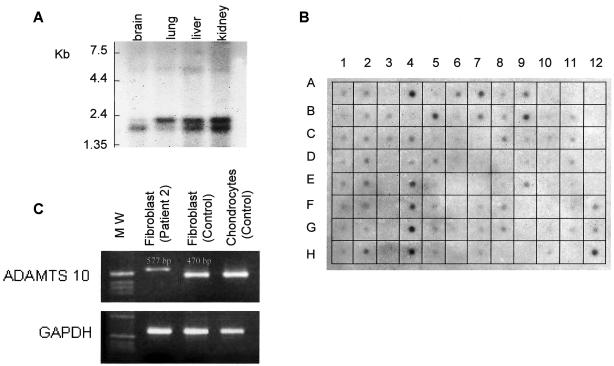
Dot-blot analyses detected a specific signal in a large number of tissues (except gonads, the majority of the digestive tract, and malignant cells) but particularly in adult heart, stomach, kidney, and skeletal muscle (fig. 3B). Moreover, northern and dot-blot analyses of fetal tissues showed a very weak signal in brain but a specific signal in lung, liver, kidney (fig. 3A and 3B), and heart (fig. 3B). RT-PCR analysis detected a specific signal in skin fibroblasts and fetal chondrocytes from controls (fig. 3C). By contrast, RT-PCR analysis of fibroblast RNA from affected individuals detected a weak signal in all cases and an abnormally long transcript in the two patients carrying splice-site mutations corresponding to intron inclusion (patient 2, fig. 3C)
Figure 3.
A, Northern analysis of the ADAMTS10 gene in various human fetal tissues. Note the presence of three distinct transcripts, of 5.5, 2.0, and 1.7 kb, probably corresponding to alternative splicing of the same RNA transcript. B, Human multiple-tissue expression array of ADAMTS10. 1A–1H, 2A–2H, and 3B, Brain. 4A, Heart. 4B, Aorta. 4C, Atrium, left. 4D, Atrium, right. 4G, Intraventricular septum. 4H, Apex of the heart. 5A–5H and 6A–6C, Digestive tract. 5B, Stomach. 7A, Kidney. 7B, Skeletal muscle. 8D–8G, Genitourinary tissues. 9A, Liver. 9B, Pancreas. 9E, Salivatory gland. 10A–10H, Leukemia and malignant cell lines. 11A, Fetal brain. 11B, Fetal heart. 11C, Fetal kidney. 11D, Fetal liver. 11E, Fetal spleen. 11F, Fetal thymus. 11G, Fetal lung. C, RT-PCR detection of ADAMTS10 RNA in skin fibroblasts from patient 2, control fibroblasts, and control fetal chondrocytes. The specific transcript is detected in fibroblasts and chondrocytes. Note the abnormal size and weak signal of ADAMTS10 transcript in patient 2, corresponding to unstable mRNA.
The study of actin distribution (by use of Phalloidin-Alexa) in cultured fibroblasts of patients with WMS showed large cytoplasmic extensions and a stronger fluorescent signal compared with controls (fig. 3). Moreover, staining of WMS fibroblasts revealed the presence of thick and straight bundles of actin filaments aligned in parallel and penetrating deeply within long cytoplasmic extensions. By contrast, actin filaments of controls were less numerous, shorter, and located mainly in the central part of the cells (fig. 4).
Figure 4.
Actin organization in the cultured fibroblasts of patients and controls (Phalloidin-Alexa staining). Note the reduced intensity of actin staining in control cells and the large actin bundles in a long cytoplasmic extension of patient fibroblasts.
Discussion
We report here three distinct nonsense and frameshift mutations of the ADAMTS10 gene in three families with the AR form of WMS. The mutations were located in the catalytic domain of the protein and predicted the generation of a markedly truncated protein.
ADAMTS is a family of proteins believed to be anchored to the ECM through interactions with aggregan or other matrix components, via one or more TS type 1 motifs. This family of metalloproteases has structural and functional properties related to (but distinct from) the ADAMS family by the presence of several TS-like domains in the C-terminal region and the lack of transmembrane domain. They are therefore secreted and associated with the ECM, which is proteolytically processed. At least 19 enzymes (1–20) have been characterized to date (Tang 2001; Cal et al. 2002; Apte 2004). They all share the same genomic organization, consisting of a signal sequence, a propeptide domain, a metalloproteinase domain containing a zinc-binding active-site motif (similar to that of the ADAMS proteases), a disintegrin-like domain, a cysteine-rich region, and a variable number of TS-1 repeats. Overall, the mammalian ADAMTS genes show 20%–40% similarity to each other, with different recognizable subfamilies. ADAMTS10 differs from the others by the presence of five TS domains and a unique C-terminal PLAC (protease and lacunin) domain and is closely related to ADAMTS6.
The ADAMTS protein family has previously been shown to be involved in various human biological processes (normal or pathological), including connective tissue organization, coagulation, inflammation, arthritis, angiogenesis, and cell migration. ADAMTS2, -3 (procollagen N-propeptidase), and -14 process the amino peptide of the procollagens I, II, and III, and ADAMTS2 mutations are known to cause autosomal recessive Ehlers-Danlos syndrome type VIIC (Colige et al. 1999). ADAMTS4, -5, and -11 are aggreganases involved in articular cartilage aggregan degradation in arthritic disease (Caterson et al. 2000; Sandy and Verscharen 2001). ADAMTS4 also has the ability to process brevican in the brain. Decreased ADAMTS13 (i.e., von Willebrand factor–cleaving protease) activity is responsible for thrombotic thrombocytopenic purpura (Kokame et al. 2002). Finally, ADAMTS1, -8 and, presumably, -15 have angio-inhibitory activities (Cal et al. 2002). The other members of this family (ADAMTS6, -7, -9, -12, and -16 through -19) and ADAMTS10 have been characterized only at the structural level, and their putative role and substrate remain unknown.
Since WMS is a connective tissue disorder characterized by microspherophakia, lens dislocation, progressive joint stiffness, and short stature with brachydactyly, it is important to note that the specific transcript was detected not only in skin fibroblasts but also in chondrocytes, which accounts for the short stature and brachydactyly in WMS. ADAMTS10 was also expressed in fetal lung, liver, and kidney, as previously shown for other ADAMTS gene products (Cal et al. 2002); however, at variance with the other gene products, ADAMTS10 was expressed in fetal and adult heart, a feature that is likely to be related to the cardiac defects observed in WMS. The members of the ADAMTS family have nonredundant functions in organogenesis, and the specific pattern of expression in heart, chondrocytes, and skin is probably a distinct characteristic of ADAMTS10.
Previous studies (using light microscopic examination and electron micrographs) of skin fibroblasts of a child with WMS have shown normal diameter and striation of collagen fibrils but abnormal extrafibrillary space and unusually large bundles of microfilaments within the cells (Faivre et al. 2002). These findings were highly suggestive of an impaired ECM. In fact, our immunological studies show the presence of abnormally large bundles of actin filaments, which are a reflection of cytoskeleton anomalies in WMS. The relationships between these cytoskeletal defects and ADAMST10 deficiencies remain unknown, but the abnormal actin distribution and organization in WMS fibroblasts could be the result of impaired connections between ECM components and cytoskeleton. Moreover, the observation of fibrillin-1 mutation in AD WMS (which is clinically undistinguishable from AR WMS) suggests a close interaction between ADAMTS10 and fibrillin 1. It is even tempting to hypothesize that fibrillin 1 could act a substrate for ADAMTS10 in the ECM. The extensive identification of ADAMTS10 ECM substrates would be particularly valuable for further understanding these complex interactions.
In conclusion, our study shows that null mutations in ADAMTS10 are responsible for the AR form of WMS with major impairment of the ECM. This study emphasizes the specific role of ADAMTS10 not only in skin development but also in growth and in lens and heart development. Additional studies will be important to further define the specific function of ADAMTS10 in ECM remodeling during cell migration and invasion.
Acknowledgments
We thank C. Esculpavit for technical assistance. Part of this work was supported by European Skeletal Dysplasia Network grant number CT2001-02188.
Electronic-Database Information
The URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for ADAMTS10 [accession number NM_030957])
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for WMS) [PubMed]
- University of California–Santa Cruz Genome Bioinformatics, http://genome.ucsc.edu/ (for the human genome working draft)
References
- Apte S (2004) A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motifs: the ADAMTS family. Int J Biochem Cell Biol 36:981–985 10.1016/j.biocel.2004.01.014 [DOI] [PubMed] [Google Scholar]
- Cal S, Obaya AJ, Llamazares M, Garabaya C, Quesada V, Lopez-Otin C (2002) Cloning, expression analysis and structural characterization of seven novel human ADAMTS, a family of metalloproteinases with disintegrin and thrombospondin-1 domains. Gene 283:49–62 10.1016/S0378-1119(01)00861-7 [DOI] [PubMed] [Google Scholar]
- Caterson B, Flannery CR, Hughes CE, Little CB (2000) Mechanisms involved in cartilage proteoglycan catabolism. Matrix Biol 19:333–344 10.1016/S0945-053X(00)00078-0 [DOI] [PubMed] [Google Scholar]
- Colige A, Sieron AL, Li S-W, Schwarze U, Petty E, Wertelecki W, Wilcox W, Krakow D, Cohn DH, Reardon W, Byers P, Lapière CM, Prockop D, Nusgens BV (1999) Human Ehlers-Danlos syndrome type VII C and bovine dermatoisparasis are caused by mutations in the procollagen I N-proteinase gene. Am J Hum Genet 65:308–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre L, Dollfus H, Lyonnet S, Mégarbané A, Samples J, Gorlin RJ, Alswaid A, Feingold J, Le Merrer M, Munnich A, Cormier-Daire V (2003a) Clinical homogeneity and genetic heterogeneity in Weill-Marchesani syndrome. Am J Med Genet 123:204–207 10.1002/ajmg.a.20289 [DOI] [PubMed] [Google Scholar]
- Faivre L, Gorlin RJ, Wirtz MK, Godfrey M, Dagoneau N, Samples JR, Le Merrer M, Collod-Beroud G, Boileau C, Munnich A, Cormier-Daire V (2003b) In frame fibrillin-1 gene deletion in autosomal dominant Weill-Marchesani syndrome. J Med Genet 40:34–36 10.1136/jmg.40.1.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre L, Megarbane, A, Alswaid A, Zylberberg L, Aldohayan N, Campos-Xavier AB, Bacq D, Legeai-Mallet L, Bonaventure J, Munnich A, Cormier-Daire V (2002) Homozygosity mapping of a Weill-Marchesani syndrome locus to chromosome 19p13.3-p13.2. Hum Genet 110:366–370 10.1007/s00439-002-0689-3 [DOI] [PubMed] [Google Scholar]
- Kokame K, Matsumoto M, Soejima K, Yagi H, Ishizashi H, Funato M, Tamai H, Konno M, Kamide K, Kawano Y, Miyata T, Fujimura Y (2002) Mutations and common polymorphisms in ADAMTS 13 gene responsible for von Willebrand factor-cleaving protease activity. Proc Natl Acad Sci USA 99:11902–11907 10.1073/pnas.172277399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandy JD, Verscharen C (2001) Analysis of agregan in human knee cartilage and synovial indicates that aggreganase (ADAMTS) activity is responsible for the catabolic turnover and loss of whole aggregan whereas other protease activity is required for C-terminal processing in vivo. Biochem J 358:615–626 10.1042/0264-6021:3580615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL (2001) ADAMTS: a novel family of extracellular matrix proteases. Int J Biochem Cell Biol 33:33–44 10.1016/S1357-2725(00)00061-3 [DOI] [PubMed] [Google Scholar]