Abstract
Motor neuron diseases (MNDs) are a group of neurodegenerative disorders with involvement of upper and/or lower motor neurons, such as amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA), progressive bulbar palsy, and primary lateral sclerosis. Recently, we have mapped a new locus for an atypical form of ALS/MND (atypical amyotrophic lateral sclerosis [ALS8]) at 20q13.3 in a large white Brazilian family. Here, we report the finding of a novel missense mutation in the vesicle-associated membrane protein/synaptobrevin-associated membrane protein B (VAPB) gene in patients from this family. Subsequently, the same mutation was identified in patients from six additional kindreds but with different clinical courses, such as ALS8, late-onset SMA, and typical severe ALS with rapid progression. Although it was not possible to link all these families, haplotype analysis suggests a founder effect. Members of the vesicle-associated proteins are intracellular membrane proteins that can associate with microtubules and that have been shown to have a function in membrane transport. These data suggest that clinically variable MNDs may be caused by a dysfunction in intracellular membrane trafficking.
Introduction
The motor neuron diseases (MNDs) are a heterogeneous group with involvement of upper and/or lower motor neurons. Spinal muscular atrophy (SMA [MIM 253300]) is one of the most common autosomal recessive disorders in childhood, with an incidence of 1 in 6,000–10,000 births. SMA is caused by degeneration of the anterior horn cells in the spinal cord associated with muscle paralysis and atrophy. On the basis of age at onset and clinical severity, SMA is subdivided into three forms: SMA-I (MIM 253300), SMA-II (MIM 253550), and SMA-III (MIM 253400). An autosomal dominant, familial, late-onset SMA (MIM 182980) has also been reported (Richieri-Costa et al. 1981). More recently, an X-linked recessive form of SMA with slow progression was mapped to Xq13.1-q21 (Takata et al. 2004).
Amyotrophic lateral sclerosis (ALS) is a heterogeneous, progressive, upper and lower motor neuron degenerative disease characterized by loss of motor neurons in the spinal cord, brainstem, and motor cortex. The combination of both upper and lower motor neuron loss and pyramidal tract degeneration creates a picture of progressive weakness with skeletal muscle wasting and a fatal disease course of <5 years. Approximately 10% of all cases are familial (FALS), and nine loci have been associated with this disease. To date, mutations in only three genes have been found to be associated with FALS. ALS1 (MIM 105400) was first identified as an adult autosomal dominant disease associated with mutations in the Cu/Zn superoxide dismutase gene (SOD1) on chromosome 21 (Rosen et al. 1993). More than 100 mutations in this gene have been found in familial and sporadic cases of ALS. SOD1 catalyzes the conversion of superoxide radicals to molecular oxygen and hydrogen peroxide.
The exact relationship of this activity to the disease process remains uncertain, since some of the disease-associated mutations retain normal levels of enzyme activity. ALS2 (MIM 606352) is a juvenile autosomal recessive disorder with slow progression and is associated with mutations in the Alsin gene on chromosome 2 (Hadano et al. 2001; Yang et al. 2001). Mutations in this gene have also been associated with spastic paraplegia (Eymard-Pierre et al. 2002; Gros-Louis et al. 2003), showing a clinical heterogeneity. The ALS4 form is a rare juvenile autosomal dominant disorder with slow progression, pyramidal signs, and severe muscle wasting, associated with mutation of the sentaxin gene (SETX [MIM 608465]) (Chen et al. 2004). SETX encodes a novel DNA/RNA helicase. It is interesting that some mutations in SETX lead to ataxia-oculomotor apraxia type 2 (AOA2), an autosomal recessive disorder (Moreira et al. 2004).
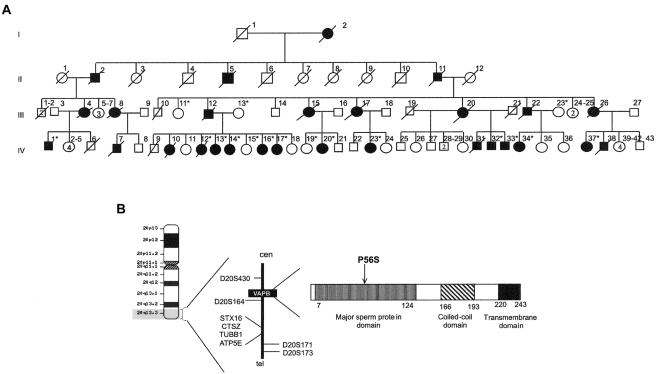
Recently, we mapped a new locus for ALS/MND at 20q13.3 (ALS8 [MIM 608627]) in a large white Brazilian family with 28 affected members distributed across four generations (fig. 1a) (Nishimura et al. 2004). ALS8 is an autosomal dominant slowly progressive disorder characterized by fasciculation, cramps, and postural tremor. Recombinant events in the marker D20S430 and the TUBB1 gene allowed us to locate the responsible gene within a region of 1.5 Mb. A missense mutation was found in the vesicle-associated membrane protein (VAMP)/synaptobrevin-associated membrane protein B gene (VAPB) in all patients of this family, as well as in six additional families with a different diagnosis. Although we were not able to link these families, the historical data pointed to a common Portuguese ancestor, indicating that they might belong to a large genealogy with >200 affected patients and at least 1,300 normal relatives.
Figure 1.
Mapping of the VAPB locus and mutation analyses. a, Pedigree from the first family reported with a diagnosis of ALS/MND (an asterisk [*] indicates DNA was available). b, VAPB locus at 20q13.3. Recombinant events reduced the region to 1.5 Mb, between marker D20S430 and the TUBB1 gene.
Patients, Material, and Methods
Families
We studied seven kindreds with patients who were affected by an MND but had different clinical courses: three had late-onset slowly progressive atypical ALS, three showed a late-onset SMA (type Finkel [MIM 182980]), and, in one kindred, some patients had typical ALS, whereas others had an atypical form of ALS. The first family was already reported in the mapping of the ALS8 locus (fig. 1b) (Nishimura et al. 2004). DNA from affected subjects and from normal controls was obtained after informed consent was given. The controls were selected on the basis of sex, age, and race.
Fine Mapping of ALS/MND Variant
To identify the gene involved in this form of late-onset MND, we first analyzed the genes located at 20q13.3, flanking the markers D20S430 and D20S173. We searched the public databases in MapView at the National Center for Biotechnology Information and compared them with the Ensembl Genome database.
Genomic PCR
We performed PCR amplification of the region containing both exons and intron-exon boundaries in the candidate-region genes. Genomic DNA was obtained from peripheral blood and was subsequently subjected to amplification using Taq DNA polymerase (Invitrogen) and comprising 35 cycles of 30 s at 95°C, 30 s at 58°C, and 30 s at 72°C followed by an extension of 6 min. The oligonucleotide primers for the VAPB gene were designed using Primer 3.0. The PCR products were analyzed by SSCP and by direct sequencing using the Amersham Mega Bace 1000 DNA Sequencers.
Multiple Alignments
The multiple amino acid sequence alignment was performed using the ClustalW program. We included the VAPB amino acid sequence from Homo sapiens, Mus musculus, and Rattus norvegicus; VAPA amino acid sequence from H. sapiens; and VAP-33 amino acid sequence from Aplysia californica, as well as VAP homologues and isoforms of VAPB in Drosophila melanogaster and Saccharomyces cerevisiae.
VAPB Models
Two VAPB structural models were generated for normal and mutant proteins, by use of the 3D-PSSM server and the UCLA-DOE fold recognition server (top solution score of 62), on the basis of the major sperm protein (MSP) structure of Ascaris suum (Protein Data Bank code 1MSP) (Bullock et al. 1996; Kelley et al. 2000; Mallick et al. 2002). The top five solutions were from the same family of immunoglobulin-like folds.
Cell Culture and Expression Constructs
HEK293 cells were maintained in Dulbecco's modified Eagle's medium (D-MEM) with glucose, l-glutamine, and 10% fetal calf serum and were transfected by standard calcium-phosphate precipitation. Primary hippocampal neurons were prepared from E18-19 rat em bryos, as described elsewhere (Bekkers and Stevens 1989) and were transfected by a modified form of calcium-phosphate precipitation (Kohrmann et al. 1999). Fluorescent images of living cells were acquired on an Olympus IX70 controlled by OpenLab software (Improvision) with the use of a Hammamtsu ORCA ER camera.
The full-length coding sequence of VAPB was generated by PCR on mouse brain cDNA (C57/BL6) and Pfu polymerase. Restriction sites for EcoRI and BamHI were incorporated into the PRC primers and were used to insert the cDNA into pEGFP-C1 (Clontech). The P56S mutation was introduced by PCR using Pfu and the primers 5′-TACTGCGTGCGGTCCAACAGTGGGG-3′ and 5′-CCCCACTGTTGGACCGCACGCAGTA-3′. For colocalization studies, cells were cotransfected with pEGFP-C1, pDsRed2-C1, pDsRed-ER, or pGolgi-ECFP (Clontech).
Results
Clinical Evaluation
The clinical evaluation of 24 patients from seven kindreds is summarized in table 1. In the studied cases, age at onset varied from 25 to 55 years, and both sexes were equally affected. Symptoms and signs, such as muscular cramps, fasciculation, and progressive limb and trunk weakness with decreased or absent deep-tendon reflexes, were present in all patients. However, a marked inter- and intrafamilial heterogeneity was seen. Three phenotypes could be distinguished: late-onset SMA, atypical ALS, and typical ALS. SMA was seen in eight patients, with an average age at onset of 46 years (range 36–55 years) and average age at ascertainment of 54 years (range 43–62 years). There was no bulbar or pyramidal involvement in this group. Atypical ALS was seen in 15 patients, with an average age at onset and age at ascertainment of 38 years (range 25–44 years) and 50 years (range 42–60 years), respectively. Bulbar signs (dysphagia) were seen in 11 (73%) subjects. After 2–60 years of progression, 5 (33%) of the 15 patients were wheelchair bound. In addition to SMA manifestations, these patients were classified as having ALS because of the presence of bulbar and pyramidal signs—but also as having atypical ALS because of essential tremor, present in all these patients, which is not seen in typical ALS. Finally, five patients from kindred 4 (fig. A1 [online only]) showed a progression characteristic of typical ALS, with a life span of <5 years after onset and a combination of pyramidal and peripheral motor neuron involvement. It is interesting that, in this same family, there are at least six patients with a slow progressive course with symptoms and signs comparable to atypical ALS.
Table 1.
Clinical Evaluation of Patients with VAPB Mutation in the ALS8 Locus[Note]
|
Finding in Patient from Kindred |
||||||||||||||||||||||||
| F1 |
F2 |
F3 | F4 |
F5 | F6 |
F7 | ||||||||||||||||||
| Characteristic | IV-1 | IV-12a | IV-13 | IV-14 | IV-16 | IV-17 | IV-20 | IV-32 | IV-33 | IV-34 | IV-37 | IV-1 | IV-23 | III-4 | IV-12b | V-13 | IV-14 | VI-26 | VI-91 | VI-93 | VI-97 | VI-96 | VI-119 | IV-14 |
| Age (years) | 42 | 50 | 49 | 47 | 44 | 42 | 55 | 56 | 49 | 45 | 50 | 60 | 57 | 58 | 56 | 49 | 58 | 51 | 62 | 48 | 61 | 55 | 43 | 52 |
| Age at onset (years) | 31 | 40 | 43 | 45 | 41 | 30 | 25 | 41 | 44 | 37 | 37 | 37 | 40 | 38 | 52 | 38 | 48 | 47 | 52 | 44 | 55 | 45 | 36 | 40 |
| Progression (years) | 11 | 10 | 6 | 2 | 3 | 12 | 30 | 15 | 5 | 8 | 13 | 23 | 17 | 20 | 4 | 11 | 10 | 4 | 10 | 4 | 6 | 10 | 7 | 12 |
| Fasciculation | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Cramps | + | + | + | + | + | + | − | + | + | + | + | + | + | + | ? | + | + | + | + | + | − | + | + | + |
| Tremor | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | + | − | − | − | − | − | − |
| DDS | + | − | − | + | − | + | + | − | − | + | + | + | + | − | − | + | − | − | − | − | − | + | − | − |
| DPS | + | − | + | + | − | + | + | + | − | + | + | + | + | − | + | + | + | + | + | − | + | + | + | + |
| Wheelchair (years) | − | 48 | 49 | − | − | − | 48 | 56 | − | − | − | 54 | − | − | − | − | − | − | − | − | − | 52 | − | − |
| Decreased/absent DTR | − | − | − | + | − | + | + | + | − | + | − | + | + | − | − | − | + | + | + | + | + | + | + | + |
| Bulbar signs | − | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | − | − | − | − | − | − | − | − |
| Pyramidal signs | + | − | − | − | + | − | − | − | − | − | − | − | + | − | + | + | − | − | − | − | − | |||
| Muscle atrophy | − | − | − | − | − | D | P/D | P/D | − | P/D | P/D | P/D | P | P | P | P | P/D | − | P | − | P | P | P | P |
| SMA | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | + | + | + |
| Atypical ALS | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | − | − | − | − | − | − | − |
| Typical ALS | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
Note.— F = family; DDS = decreased distal strength; DPS = decreased proximal strength; DTR = deep-tendon reflexes; P = proximal; D = distal; + = positive finding; − = negative finding.
Survival of 10 years.
Survival of 4 years.
Identification of a Missense Mutation in the VAPB Gene
We had previously found linkage at 20q13.3 within an interval of 2.7 Mb, using microsatellite markers D20S430-D20S164-D2094-D20S171-D20S173 in family 1 (fig. 1b). The maximum multipoint LOD score was 7.45, found close to marker D20S164. Mutation screening of candidate genes excluded the cathepsin Z (CTSZ), syntaxin 16 (STX 16), ATP synthase, epsilon subunit (ATP5E), and tubulin β1 (TUBB1) genes. It is notable that we found a polymorphism in the 3′ UTR of the TUBB1 gene, which was present in almost all patients but which was subsequently also found in Brazilian normal controls. Since RT-PCR analysis did not show any alteration in the expression of TUBB1, it was then used as an intrafamilial marker. In addition, this polymorphism reduced the interval to 1.5 Mb between the marker D20S430 and the TUBB1 gene. This region contains nine genes (a Z-DNA binding protein gene; a prostate androgen–induced RNA; an oncogene; and genes encoding a protein phosphatase, an aminopeptidase-like protein, a protein involved in imprinting, a Drosophila-like protein, and two proteins involved in intracellular trafficking) and seven hypothetical or predicted genes.
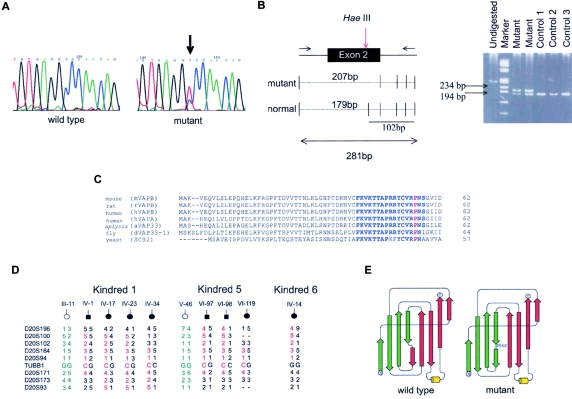
Mutation screening in these candidates genes led to the identification of a C→T substitution in exon 2 of the VAMP/synaptobrevin-associated membrane protein B (VAPB) gene (fig. 2a). This mutation at nucleotide 166 removes a HaeIII restriction site and results in a substitution of the conserved proline for a serine at codon 56 (Pro56Ser, or P56S) (fig. 2b) This proline is conserved in several species, such as H. sapiens, M. musculus, R. norvegicus, A. californica, D. melanogaster, and S. cerevisae (fig. 2c). This mutation was present in all affected members of family 1 but not in unaffected relatives or in 400 chromosomes of unrelated normal controls. Subsequently, we found the same mutation in 22 patients from six additional large Brazilian kindreds with inter- and intrafamilial clinical heterogeneity (fig. A1 [online only]). Although it was not possible to link all these families, the haplotype analysis with nine polymorphic markers flanking the VAPB gene suggests a common ancestor and, therefore, a founder effect (fig. 2d).
Figure 2.
Analyses of VAPB gene. a, Chromatogram showing the mutation in VAPB coding sequence and normal control. b, Segregation of the P56S mutation in heterozygous patients and normal controls (homozygous). The HaeIII enzyme cuts in several fragments, producing one major allele of 179 bp in normal subjects and one allele of 207 bp in heterozygous patients. c, Partial amino alignment of the VAP homologues genes. Gaps were introduced for optimal alignment. Lengths of each partial protein are indicated. The amino acid sequence FKVKTTAPRXYCVRPNS is highly conserved (blue), and the proline is indicated (red). d, Haplotype analysis of three kindred studied. All the patients have a common haplotype (red), which was not present in normal relatives (green). e, VAPB models. The top part of the β sandwich is made of green β strands, and the bottom part is made of red strands. Wild type induces a kink and splits half of the strand into the top part of the sandwich (green) and the other half of the strand into the bottom part (red). The P56S mutation disrupts the hydrogen bonds between strands on the same sheet of the β sandwich, favoring new hydrogen bonds that move the bottom half to the opposite side of the β sandwich.
Characterization of VAPB
The VAPB protein has three structural domains. The first 150 residues constitute an MSP domain conserved between all VAP family members (Weir et al. 1998; Nishimura et al. 1999). The central region of the protein contains an amphipathic helical structure and is predicted to form a coiled/coil protein-protein interaction motif and a hydrophobic carboxy terminus that acts as a membrane anchor (fig. 1b).
Using a three-dimensional model of VAPB based on the nematode 1MSP dimer structure, we examined the possible effect of this mutation on the protein structure. In wild-type VAPB, P56 lies within a particularly highly conserved motif, FKVKTTAPRRYCVRPNS, distal to the dimerization face (fig. 2c). The serine substitution would remove a kink between two short stretches of β strand, switching the hydrogen-bond pattern from one β sheet to another. This would make the strand more flexible and would greatly disrupt the hydrogen bonds between strands on the same sheet of the β sandwich, perhaps favoring new hydrogen bonds that move the bottom half of the strand to the opposite side of the β sandwich (fig. 2e).
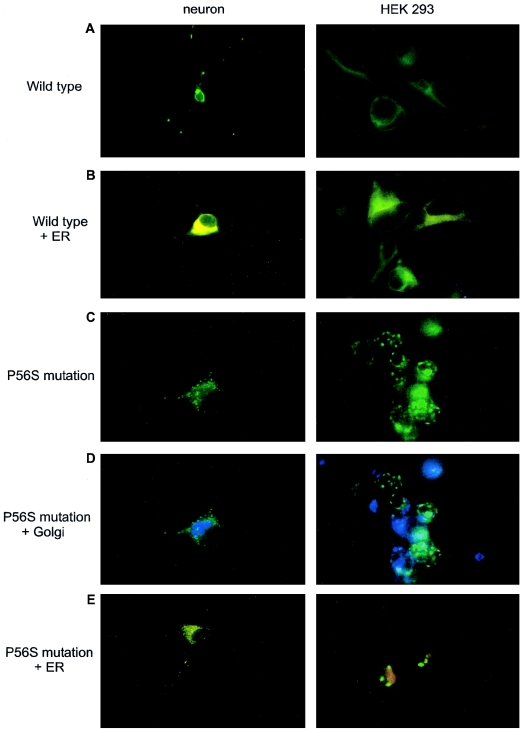
VAPB and the closely related VAPA proteins have been shown to associate with intracellular membranes, including the endoplasmic reticulum (ER) and the Golgi apparatus (Soussan et al. 1999; Skehel et al. 2000). To examine the effect of the P56S mutation in VAPB on the localization of the protein and on cellular membranes, wild-type and mutant VAPB was expressed as green fluorescent protein (GFP) fusion proteins in two different cell types, HEK 293 and primary cultured rat hippocampal neurons (fig. 3a). In comparison with ER-DsRed, wild-type VAPB-EGFP predominantly localized to the ER (Soussan et al. 1999) (fig. 3b). In contrast, the P56S mutation dramatically disrupts the subcellular distribution of the numerous protein-inducing intracellular aggregates (fig. 3c), which do not accumulate the luminal ER-DsRed or the Golgi apparatus marker (fig. 3d–3e). Thus, the P56S mutation in VAPB does not seem to be a normally functioning polymorphism.
Figure 3.
GFP expression in hippocampal neurons and HEK293 cell culture. a, Neurons and HEK cells expressing the VAPB wild type (green). b, Using an ER marker (red), we observed a colocalization of the VAPB and ER compartment (yellow), as reported elsewhere (only the colocalization is shown). c, P56S mutant disrupts the subcellular distribution, showing intracellular aggregates. d–e, P56S mutant does not colocalize either with Golgi apparatus (blue) or with the ER (red).
Discussion
Here, we describe the identification of a missense mutation in the VAPB gene in seven families with different diagnoses that range from typical ALS to mild SMA. The VAMP-associated protein of 33 kD (VAP-33) was first identified in A. californica (GenBank accession number Q16943) (Skehel et al. 1995). This protein is highly conserved in different species, with homologues in yeast (SCS2 [GenBank accession numberP40075]), fly (DVAP-33A [GenBank accession numberAY060395]), mouse (Ensembl Genome accession number ENSMUSP00000029026), rat (Ensembl Genome accession number ENSRNOP00000007554), and human (VAPA and VAPB [Ensembl Genome accession numbers ENSP00000217602 and ENSP00000265619]) (fig. 2c). The localization of these proteins appears to differ among the species. VAP proteins in mouse and SCS2 in yeast are localized on intracellular membranes, particularly the ER compartment (Kagiwada et al. 1998; Soussan et al. 1999; Skehel et al. 2000). Ultrastructural and biochemical analyses indicated that these VAP proteins can associate with microtubules and are often found at the junction between intracellular vesicles and cytoskeletal structures (Soussan et al. 1999; Skehel et al. 2000). In Drosophila, the DVAP-33A homologue seems to be localized in the neuromuscular junctions. Male mutants show severe locomotion defects and die early in larvae development (Pennetta et al. 2002). Moreover, the presynaptic microtubule assembly is severely compromised.
The VAPB has been shown to act during ER-Golgi transport and secretion (Soussan et al. 1999; Foster et al. 2000). It is possible that the mutation of the MSP domain disrupts this function, leading to the accumulation of transport intermediates in the form of cytosolic membranous aggregates. Although the expression of the mutant form of VAPB does not lead to gross changes in ER structure, we cannot exclude the possibility that more subtle perturbations in Golgi and ER membrane systems are occurring in cells expressing the mutant form of VAPB. Thus, the mutant VAPB protein may compromise intracellular membrane transport and secretion, which may subsequently lead to loss of extracellular trophic signals or to disruption of intracellular processes that ultimately result in the loss of motor neurons. It is also possible that VAPB is present on membrane structures distinct from the ER and Golgi and that the MSP mutation affects the accumulation of the protein at these sites. It is notable that, in older neuronal cultures, the closely related protein VAPA accumulates in additional areas distinct from ER and Golgi markers (data not shown).
It was recently shown that VAPA acts as a membrane receptor for lipid- and sterol-binding proteins (Weir et al. 1998; Wyles et al. 2002). This interaction is partly dependent on the MSP domain and might, therefore, be disrupted by the mutation causing a perturbation in the composition of intracellular membranes. Mutations in the yeast SCS2 affect phospholipid metabolism and disrupt the signaling pathway from the ER that regulates inositol biosynthesis and the unfolded protein stress response (Kagiwada et al. 1998; Loewen et al. 2003). This raises the interesting possibility that the MSP domain of VAPA and VAPB proteins serve both a structural and signaling function. It is notable that the MSP has a signaling function during fertilization in nematodes (Miller et al. 2001). VAPA and VAPB proteins can form multimeric protein complexes (Soussan et al. 1999; Foster et al. 2000; P.S., unpublished data). The P56S VAPB mutation lies outside the dimerization face of the MSP domain and most likely affects the protein's association with other polypeptides. The P56S mutation does not seem to result in a complete loss of function of the MSP domain, since the expression of a truncated form of the protein lacking the entire MSP domain does not induce the formation of similar intracellular aggregates (data not shown).
VAPB is ubiquitously expressed (Nishimura et al. 1999), yet the P56S mutation reported here predominantly affects motor neurons. This selective vulnerability also occurs with ALS-associated SOD-1, ALS2, and SETX mutations, and several hypotheses might explain this observation. Perhaps different types of cells do not require the same amount of VAPB for survival, or VAPB may have another function specific to as-yet unknown neurons. P56S VAPB could interfere with the stability of a VAPB-containing protein complex, and a haploinsufficiency or gain-of-function mechanism could result in neurotoxicity and, consequently, in motor neuron death.
MNDs are a heterogeneous group, and the subclassification is difficult, particularly when the lower motor neuron signs are the major evidence of the disorder. In three kindreds, there were patients with pyramidal tract or upper motor neuron signs comparable to ALS, whereas, in others, the diagnosis was more similar to late-onset SMA. It is not known why the same mutation is responsible for different phenotypes in these families. Intrafamilial clinical heterogeneity has been found for several human disorders, including the FALS forms in patients with ALS and SMA who belong to the same family (Corcia et al. 2002). A mutation in the Dysferlin gene was also found to cause distal Miyoshy myopathy in some patients and proximal limb-girdle muscular dystrophy type B in others from the same genealogy (Illarioshkin et al. 1996; Weiler et al. 1996; Zatz et al. 2000, 2003). These findings suggest that modifier genes or other factors probably play an important role in modulating the clinical course in affected patients carrying the same pathogenic mutation.
In summary, our data strongly suggest that the P56S mutation in a highly conservative domain of the VAPB gene is responsible for a variable form of MND in seven large Brazilian kindreds. These data bring new insight to understanding the mechanisms underlying motor neuron degeneration and may help provide novel therapeutic targets to rectify genetic errors.
Acknowledgments
We are extremely grateful for the collaboration of all family members. We also thank Dr. Rita e Cássia Pavanello, Dr. Ivo Pavanello, Dr. Mariz Vainzof, Dr. Lydia Yamamoto, Robson Sartorello, Dr. Célia Garcia, Marta Canovas, Constância Urbani, Camila Guindalini, Manuela Tonini, Antonia de Cerqueira, Patricia Arashiro, Dr. Andréa Bernardino, Dr. Esteban Dell’Angelica, Dr. George Jackson, Dr. Yama Akbari, Dr. Daniel Geschwind, Dr. Maria Rita Passos-Bueno, and the Centro de Estudos do Genoma Humano group, for their invaluable help. We would also like to thank Dr. Hugo Bellen and Dr. Giuseppa Pennetta, for helpful suggestions. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo–Centros de Pesquisa, Inovação e Difusão, Conselho Nacional de Desenvolvimento Científico e Tecnológico, The Wellcome Trust, and The Royal Society.
Appendix A: Supplemental Figure
Figure A1.
Pedigrees from families studied. a, Family 2: white family with diagnosis of atypical ALS. b, Family 3: African Brazilian family with diagnosis of atypical ALS. c, Family 4: white family with diagnosis of typical ALS and atypical ALS. d, Family 5: African Brazilian family with diagnosis of mild SMA. e, Family 6: white family with diagnosis of late-onset SMA type Finkel. f, Family 7: African Brazilian family with diagnosis of late-onset SMA type Finkel. It was not possible to link all these subjects to the same pedigree; however, all patients tested have the same P56S mutation in the VAPB gene. An asterisk (*) indicates DNA was available.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- 3D-PSSM server, http://www.sbg.bio.ic.ac.uk/servers/3dpssm/
- ClustalW, http://www.ebi.ac.uk/clustalw/
- Ensembl Genome database, http://www.ensembl.org/ (for mouse VAP [accession number ENSMUSP00000029026], rat VAP [accession number ENSRNOP00000007554], human VAPA [accession number ENSP00000217602], and human VAPB [accession number ENSP00000265619])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for D. melanogaster DVAP-33A [accession number AY060395], S. cerevisiae SCS2 [accession number P40075], and A. californica VAP-33 [accession number Q16943])
- MapView at the National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/mapview/
- Primer 3.0, http://www-genome.wi.mit.edu
- Protein Data Bank (PDB), http://pdb.org (for protein structure of A. suum [PDB code 1MSP])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- UCLA-DOE fold recognition server, http://fold.doe-mbi.ucla.edu/
References
- Bekkers JM, Stevens CF (1989) NMDA and non-NMDA receptors are co-localized at individual excitatory synapses in cultured rat hippocampus. Nature 341:230–233 10.1038/341230a0 [DOI] [PubMed] [Google Scholar]
- Bullock TL, Roberts TM, Stewart M (1996) 2.5 Å resolution crystal structure of the motile major sperm protein (MSP) of Ascaris suum. J Mol Biol 263:284–296 10.1006/jmbi.1996.0575 [DOI] [PubMed] [Google Scholar]
- Chen Y-Z, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, Nicholson GA, Auer-Grumbach M, Wagner K, De Jonghe P, Griffin JW, Fischbeck KH, Timmerman V, Cornblath DR, Chance PF (2004) DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am J Hum Genet 74:1128–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcia P, Khoris J, Couratier P, Mayeux-Portas V, Bieth E, De Toffol B, Autret A, et al (2002) SMN1 gene study in three families in which ALS and spinal muscular atrophy coexist. Neurology 59:1464–1466 [DOI] [PubMed] [Google Scholar]
- Eymard-Pierre E, Lesca G, Dollet S, Santorelli FM, di Capua M, Bertini E, Boespflug-Tanguy O (2002) Infantile-onset ascending hereditary spastic paralysis is associated with mutations in the alsin gene. Am J Hum Genet 71:518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster LJ, Weir ML, Lim DY, Liu Z, Trimble WS, Klip A (2000) A functional role for VAP-33 in insulin-stimulated GLUT4 traffic. Traffic 1:512–521 10.1034/j.1600-0854.2000.010609.x [DOI] [PubMed] [Google Scholar]
- Gros-Louis F, Meijer IA, Hand CK, Dube MP, MacGregor DL, Seni MH, Devon RS, Hayden MR, Andermann F, Andermann E, Rouleau GA (2003) An ALS2 gene mutation causes hereditary spastic paraplegia in a Pakistani kindred. Ann Neurol 53:144–145 10.1002/ana.10422 [DOI] [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 10.1038/ng1001-166 [DOI] [PubMed] [Google Scholar]
- Illarioshkin SN, Ivanova-Smolenskaya IA, Tanaka H, Vereshchagin NV, Markova ED, Poleshchuk VV, Lozhnikova SM, Sukhorukov VS, Limborska SA, Slominsky PA, Bulayeva KB, Tsuji S (1996) Clinical and molecular analysis of a large family with three distinct phenotypes of progressive muscular dystrophy. Brain 119:1895–1909 [DOI] [PubMed] [Google Scholar]
- Kagiwada S, Hosaka K, Murata M, Nikawa J, Takatsuki A (1998) The Saccharomyces cerevisiae SCS2 gene product, a homolog of a synaptobrevin-associated protein, is an integral membrane protein of the endoplasmic reticulum and is required for inositol metabolism. J Bacteriol 180:1700–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, MacCallum RM, Sternberg MJ (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol 299:499–520 [DOI] [PubMed] [Google Scholar]
- Kohrmann M, Haubensak W, Hemraj I, Kaether C, Lessmann VJ, Kiebler MA (1999) Fast, convenient, and effective method to transiently transfect primary hippocampal neurons. J Neurosci Res 58:831–835 [DOI] [PubMed] [Google Scholar]
- Loewen CJ, Roy A, Levine TP (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J 22:2025–2035 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick P, Weiss R, Eisenberg D (2002) The directional atomic solvation energy: an atom-based potential for the assignment of protein sequences to known folds. Proc Natl Acad Sci USA 99:16041–16046 10.1073/pnas.252626399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D (2001) A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291:2144–2147 10.1126/science.1057586 [DOI] [PubMed] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Nemeth AH, Le Ber I, Moniz JC, Tranchant C, et al (2004) Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet 36:225–227 10.1038/ng1303 [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Oliveira JR, Vainzof M, Zatz M (2004) A novel locus for late onset amyotrophic lateral sclerosis/motor neurone disease variant at 20q13. J Med Genet 41:315–320 10.1136/jmg.2003.013029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Hayashi M, Inada H, Tanaka T (1999) Molecular cloning and characterization of mammalian homologues of vesicle-associated membrane protein-associated (VAMP-associated) proteins. Biochem Biophys Res Commun 254:21–26 10.1006/bbrc.1998.9876 [DOI] [PubMed] [Google Scholar]
- Pennetta G, Hiesinger P, Fabian-Fine R, Meinertzhagen I, Bellen H (2002) Drosophila VAP-33A directs bouton formation at neuromuscular junctions in a dosage-dependent manner. Neuron 35:291–306 10.1016/S0896-6273(02)00769-9 [DOI] [PubMed] [Google Scholar]
- Richieri-Costa A, Rogatko A, Levisky R, Finkel N, Frota-Pessoa O (1981) Autosomal dominant late adult spinal muscular atrophy, type Finkel. Am J Med Genet 9:119–128 [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- Skehel PA, Fabian-Fine R, Kandel ER (2000) Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc Natl Acad Sci USA 97:1101–1106 10.1073/pnas.97.3.1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel PA, Martin KC, Kandel ER, Bartsch D (1995) A VAMP-binding protein from Aplysia required for neurotransmitter release. Science 269:1580–1583 [DOI] [PubMed] [Google Scholar]
- Soussan L, Burakov D, Daniels MP, Toister-Achituv M, Porat A, Yarden Y, Elazar Z (1999) ERG30, a VAP-33-related protein, functions in protein transport mediated by COPI vesicles. J Cell Biol 146:301–311 10.1083/jcb.146.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata RI, Speck Martins CE, Passosbueno MR, Abe KT, Nishimura AL, da Silva MD, Monteiro A Jr, Lima MI, Kok F, Zatz M (2004) A new locus for recessive distal spinal muscular atrophy at Xq13.1-q21. J Med Genet 41:224–229 10.1136/jmg.2003.013201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler T, Greenberg CR, Nylen E, Halliday W, Morgan K, Eggertson D, Wrogemann K (1996) Limb-girdle muscular dystrophy and Miyoshi myopathy in an aboriginal Canadian kindred map to LGMD2B and segregate with the same haplotype. Am J Hum Genet 59:872–878 [PMC free article] [PubMed] [Google Scholar]
- Weir ML, Klip A, Trimble WS (1998) Identification of a human homologue of the vesicle-associated membrane protein (VAMP)-associated protein of 33 kDa (VAP-33): a broadly expressed protein that binds to VAMP. Biochem J 333:247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyles JP, McMaster CR, Ridgway ND (2002) Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J Biol Chem 277:29908–29918 10.1074/jbc.M201191200 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 10.1038/ng1001-160 [DOI] [PubMed] [Google Scholar]
- Zatz M, Paula F, Starling A, Vainzof M (2003) The ten autosomal recessive limb-girdle muscular dystrophies. Neuromuscul Disord 13:532–544 10.1016/S0960-8966(03)00100-7 [DOI] [PubMed] [Google Scholar]
- Zatz M, Vainzof M, Passos-Bueno MR (2000) Limb-girdle muscular dystrophy: one gene with different phenotypes, one phenotype with different genes. Curr Opin Neurol 13:511–517 10.1097/00019052-200010000-00002 [DOI] [PubMed] [Google Scholar]