Abstract
Beckwith-Wiedemann syndrome (BWS), which causes prenatal overgrowth, midline abdominal wall defects, macroglossia, and embryonal tumors, is a model for understanding the relationship between genomic imprinting, human development, and cancer. The causes are heterogeneous, involving multiple genes on 11p15 and including infrequent mutation of p57KIP2 or loss of imprinting of either of two imprinted gene domains on 11p15: LIT1, which is near p57KIP2, or H19/IGF2. Unlike Prader-Willi and Angelman syndromes, no chromosomal deletions have yet been identified. Here we report a microdeletion including the entire LIT1 gene, providing genetic confirmation of the importance of this gene region in BWS. When inherited maternally, the deletion causes BWS with silencing of p57KIP2, indicating deletion of an element important for the regulation of p57KIP2 expression. When inherited paternally, there is no phenotype, suggesting that the LIT1 RNA itself is not necessary for normal development in humans.
Introduction
Beckwith-Wiedemann syndrome (BWS [MIM 130650]) rarely involves mutation of the imprinted cyclin-dependent kinase inhibitor gene p57KIP2 (MIM 600856), which is normally expressed from the maternal allele (Matsuoka et al. 1996; Lee et al. 1997; O’Keefe et al. 1997). However, 40% of patients with BWS show loss of imprinting (LOI) of LIT1 (MIM 604115), an antisense RNA within the KVLQT1 gene (MIM 192500) and 220 kb telomeric to p57KIP2 on chromosome 11p15 (Lee et al. 1999; Smilinich et al. 1999). An additional 15% of patients show LOI of IGF2 (MIM 147470), located in a second imprinted subdomain 280 kb telomeric to KVLQT1 (Weksberg et al. 1993; Steenman et al. 1994). LOI of LIT1 involves aberrant hypomethylation and activation of the normally silent maternal allele. The target of LOI of LIT1 presumably is p57KIP2, although it is not clear whether LIT1 serves as a transcriptional repressor of p57KIP2 or as an insulator separating p57KIP2 from its enhancer. Deletion of the differentially methylated region (DMR) leads to activation of the paternal allele of p57KIP2, as well as those of Tssc3, Tssc5, Kcnq1, Tssc4, and Ascl2 (Fitzpatrick et al. 2002). Similarly, targeted deletion of the LIT1 CpG island on a human paternal chromosome in chicken DT40 cells leads to activation of the paternal allele of Kvlqt1 and p57KIP2 (Horike et al. 2000). Presumably, the DMR contains elements important for silencing on the paternal allele. An engineered translocation in the mouse, disrupting the imprinted cluster with a targeted translocation between p57KIP2 and Kvlqt1, results in loss of expression of the maternal allele of p57KIP2, Tssc3, and Tssc5 (Cleary et al. 2001), suggesting that there are also elements within or proximal to LIT1 important for gene activation on the maternal allele. It should be noted, however, that there is no mouse model involving LIT1 leading to BWS.
We sought to identify patients with BWS who have microdeletions on 11p15, since similar studies had clarified the role of imprinting regulatory elements in the Prader Willi/Angelman syndrome gene region of 15q11-15q13 (Buiting et al. 1995). To screen for LIT1 deletions, we used real-time quantitative PCR of genomic DNA for copy number analysis, normalizing to a reference locus, and combined these efforts with FISH of metaphase and interphase cells.
Subjects and Methods
Subjects
Patients with BWS were identified as part of a BWS registry first established in 1994. Participating families provided detailed clinical information and written informed consent approved by the respective institutional review boards. BWS in the registry is defined by a clinical diagnosis by a experienced physician but requires at least two of the five most common features: (1) macroglossia, (2) birth weight and length >90th percentile, (3) hypoglycemia in the 1st mo of life, (4) ear creases or ear pits, and (5) midline abdominal wall defects.
Real-Time Karyotyping
Genomic DNA was prepared from peripheral-blood lymphocytes by standard proteinase K digestion and phenol extraction (Cui et al. 1998). Genomic copy numbers of sequences within KVLQT1 and flanking genes were measured using quantitative real-time PCR amplification with Taqman probes. Reactions were performed using 1× Taqman master mix (ABI), 300 nM each primer, 200 nM probe, and 10 ng of peripheral white blood cell genomic DNA in a total volume of 25 μl. All reactions were performed in triplicate. For each sequence amplified, triplicate reactions of a no template control and a standard curve were amplified at the same time. The standard curve was prepared from normal control genomic DNA at 100× (100 ng per reaction), 10× (10 ng per reaction), and 1× (1 ng per reaction). Reactions were run on an ABI 7700 instrument under standard conditions (50°C for 2 min; 95°C for 10 min; 95°C for 15 s, 60°C for 1 min for 40 cycles). All triplicate cycle threshold (Ct) values were checked to ensure that they were within 1 Ct of each other. The log input amount of the standard curve was plotted versus the output Ct values; only amplifications that produced a slope of −3.3 to −3.8 were accepted as quantitative. The log input amount of each sample was calculated according to the formula (Ct-b)/m, where b is the Y-intercept, and m is the slope. The log input amount was converted to input amount according to the formula 10^(log input amount), and triplicate input amounts were averaged for each sample. The average input amount of each sequence amplified was normalized to the average input amount of peripheral myelin protein 22 (PMP22); primer and probe sequences for PMP22 were reported by Wilke et al. (2000). PMP22 is located on chromosome 17 and is involved in Charcot-Marie-Tooth syndrome, a sensory and motor neuropathy disease unrelated to BWS. All primers are listed in the appendix (online only).
FISH
To confirm the deletion localization in lymphoblast cells derived from patients II-2 and III-4, two-color FISH was performed using several BAC and PAC clones, including KVLQT1 genomic DNA as a probe. The probes were labeled with digoxigenin-11-UTP (Roche) and biotin-16-dUTP (Roche), respectively, by nick translation. Probes were purified by ethanol precipitation, dissolved in 5 μl of formamide, mixed, and denatured. Hybridization solution (BSA [Roche]:10× SSC:50% dextran sulfate [Sigma], 1:2:2) and labeled probes were mixed 1:1, dropped onto denatured chromosomes, covered with parafilm, and incubated at 37°C for 15 h in a humidified chamber. After hybridization, the slides were washed sequentially at 37°C in 50% formamide/2× SSC, 2× SSC, 1× SSC for 15 min each and once in 4× SSC for 5 min. The slides were immersed in 70 μl of 3 μg/ml fluorescein isothiocyanate (FITC)–avidin (Vector Laboratories), and 1.6 μg/ml anti-digoxigenin-rhodamine (Roche), 4× SSC, and 1% BSA for 45 min at 37°C. The slides were then washed for 5 min each in 4× SSC, 4× SSC containing 0.05% Triton X-100, and then 4× SSC. The slides were incubated with 70 μl of 5 μg/ml biotinylated anti-avidin (Vector Laboratories), 4× SSC, and 1% BSA at 37°C for 60 min. After washing, another layer of FITC-avidin was added for amplification. The slides were washed and mounted in antifade solution (1% diazabicyclooctane [Sigma] in glycerol with 10% PBS) containing 0.2 μg/ml 4′,6′-diamidino-2-phenylindole (Sigma) and 100 μg/ml p-phenylenediamine (Sigma). Signals were observed with a Nikon fluorescence microscope. One hundred nuclei and metaphases were analyzed.
Real-Time Quantitative Analysis of Gene Expression
RNA isolated from primary lymphocytes was converted to cDNA through use of the Superscript II kit (Invitrogen) according to the manufacturer’s protocol. A standard curve was constructed from pooled normal control cDNA. Reactions were performed using 1× Taqman master mix (Applied Biosystems), 270 nM each primer, and 60 nM probe in a total volume of 25 μl. GUS reactions were performed using 24 ng of original input RNA, and p57KIP2 reactions were performed using 192 ng of original input RNA. All reactions were performed in triplicate, including, for each sequence amplified, a no-template control. The RT-negative reactions were used as template for one p57KIP2 reaction, to ensure that no amplification occurred. Reactions were run on an ABI 7700 instrument according to standard conditions (50°C for 2 min; 95°C for 10 min; 95°C for 15 s, 60°C for 1 min for 40 cycles). All triplicate Ct values were checked to ensure that they were within 1 Ct of each other. For p57KIP2, the average slope of the standard curve was 3.73, and for GUS, the average slope of the standard curve was 3.68; both of these are within the desired quantitative range. Data analysis converting triplicate Ct values into average input amount ratios was performed as described above in the “Real-Time Karyotyping” subsection. The SD was, on average, 23% of the ratio value. A human endogenous control plate (Applied Biosystems) was used, according to the manufacturer’s instructions, to choose a housekeeping gene for expression normalization in primary lymphocytes.
Results
We used real-time quantitative PCR of genomic DNA to screen for microdeletions in BWS. We examined 34 patient samples by this approach. Of these, two patients had familial BWS, each with at least one fully affected sib. We detected a microdeletion in one of the familial cases, as described in detail below. The other familial case showed evidence of a complex rearrangement (not discussed here).
At the time of ascertainment, patient III-4 (fig. 1A) was a 6-year-old boy with hemihypertrophy with asymmetric enlargement of the right leg and arm, hypospadias, cleft palate, midline abdominal wall defect (diastasis recti and umbilical hernia), characteristic ear pits and creases, undescended testes, and macroglossia requiring surgery. Two siblings also had BWS: a sister (patient III-1, fig. 1A) with prenatal overgrowth, ear creases and pits, and macroglossia requiring surgery; and a brother (patient III-2, fig. 1A), stillborn at 28 wk, with prenatal overgrowth and macroglossia. The father was 37 years old and the mother 36 years old at the proband’s birth, and both parents showed no signs of BWS in childhood or later.
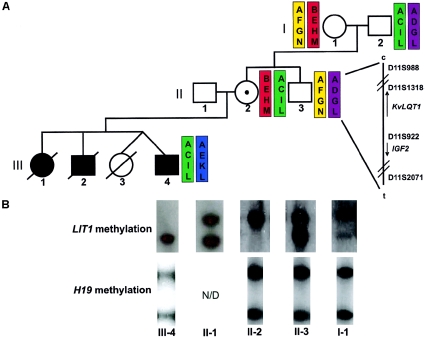
Figure 1.
Haplotype and methylation analysis of LIT1 microdeletion. A, Haplotype mapping using 11p15 microsatellite markers D11S988, D11S1318, D11S922, D11S2071, which were amplified using FAM-labeled primers and detected using an ABI 3100 capillary electrophoresis instrument. Haplotypes were constructed using peak size data (see fig. A [online only]). The haplotypes of individuals I-1 and II-3 are inferred. B, Methylation at LIT1 and H19, assayed by methyl-sensitive Southern blotting using genomic DNA from the indicated individuals. For LIT1, the upper band (6.0 kb) is methylated and represents the maternal allele, and the lower band (4.2 kb) is unmethylated and represents the paternal allele. For H19, the upper band (1.8 kb) is methylated and represents the paternal allele, and the lower band (1.0 kb) is unmethylated and represents the maternal allele. LIT1 shows gain of methylation in II-2 and loss of methylation in III-4. H19 is unaffected.
The deletion was mapped by real-time karyotyping with probes spanning KVLQT1 (fig. 2A). The deleted region of ∼250 kb removes all of LIT1 and is within KVLQT1; the centromeric breakpoint is located between exons 1b and 1c, and the telomeric breakpoint is located between exons 11 and 14 (KVLQT1 exon numbering from GenBank sequence AJ006345). This deletion was detected in individuals II-2 and III-4; II-2 is the mother of III-4 (fig. 1A), indicating that it had been transmitted from the mother to the proband and likely to the other two deceased sibs with BWS as well.
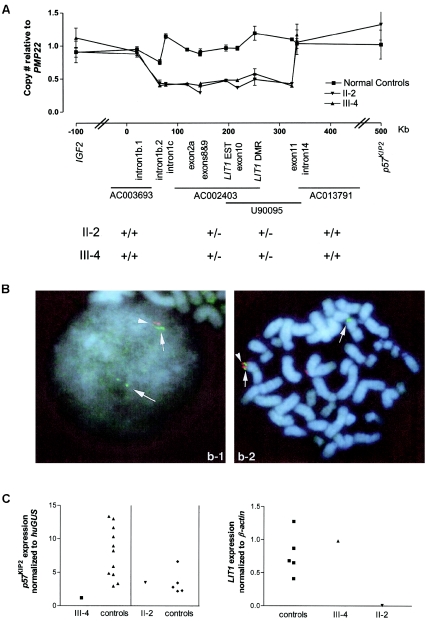
Figure 2.
Deletion of LIT1 and altered expression of p57KIP2. A, Genomic copy number of sequences in KVLQT1 in individuals II-2 and III-4 and in four normal control individuals. Genomic DNA was amplified in the presence of a Taqman probe and normalized to PMP22 on chromosome 17. The X-axis indicates KVLQT1 genomic sequence (GenBank accession number AJ006345), and the Y-axis indicates the relative copy number of each probed sequence. See the appendix (online only) for primer and probe sequences. B, Two-color FISH using PAC probes in lymphoblastoid nuclei from patient III-4. Probe AC003693 (blue) generated two chromosome signals in interphase (left) and metaphase (right) nuclei, whereas probe U90095 (red) generated one chromosome signal in interphase and metaphase nuclei. A pair of spots was scored as one signal, since it represents the two sister chromatids after S phase. Interphase nuclei and chromosomes were counterstained with DAPI. One hundred interphase and metaphase nuclei were analyzed, 90 and 85 of which, respectively, showed similar patterns to those described above. LIT1 is deleted in individuals II-2 and III-4. C, Real-time mRNA expression analysis of p57KIP2 and LIT1 in deletion carriers. Values represented are the average input amount of p57KIP2, normalized to the average input amount of the β-glucuronidase (GUS) gene in triplicate samples, compared with age-matched normal lymphocyte controls; and the average input amount of LIT1, normalized to the average input amount of the β-actin gene in triplicate samples, compared with normal lymphoblastoid controls. The housekeeping gene chosen for normalization in cases was the gene with the least variance among eight control samples. p57KIP2 expression is decreased in the proband (III-4), and LIT1 expression is absent in the mother (II-2).
The presence and localization of the deletion was independently confirmed cytogenetically through use of two-color interphase and metaphase FISH with BAC and PAC probes spanning KVLQT1 (fig. 2B). PAC probes AC002403 and U90095, spanning KVLQT1 from approximately exon 2a to exon 11, generated one chromosomal signal in a majority of interphase and metaphase nuclei in diploid lymphoblast cells derived from patients II-2 and III-4, whereas flanking probes AC003693 (PAC) and AC013791 (BAC) generated two chromosomal signals (fig. 2B).
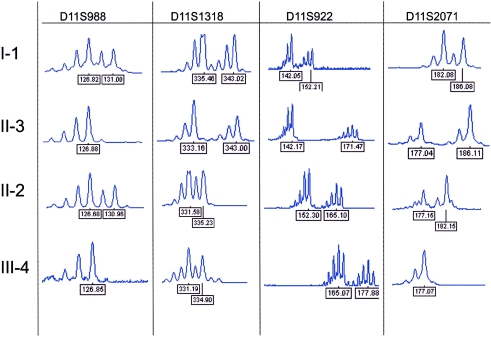
To determine the identity of the parental allele carrying the deletion, we performed haplotype analysis, using microsatellite markers across 11p15 (fig. A [online only]). Through use of these data, haplotype analysis of the available individuals placed the deletion on the “ACIL” haplotype (fig. 1A). Individual II-2 inherited the chromosome carrying this haplotype from her father, individual I-2, and passed this haplotype to her son, individual III-4, indicating that the deletion was present on a paternal chromosome in individual II-2 and on a maternal chromosome in individual III-4. Methylation analysis of the LIT1 DMR by methyl-sensitive Southern blotting (fig. 1B) was used to confirm the parental origin and indicated the absence of an unmethylated paternal band in individual II-2 and the absence of a methylated maternal band in individual III-4. Methylation analysis of the H19 DMR by methyl-sensitive Southern blotting (fig. 1B) was used to confirm that the deletion had no effect on the distal, independently regulated H19/IGF2 cluster and indicated the normal presence of both parental bands in all tested individuals.
Figure A.
Microsatellite analysis. The indicated family members were typed for microsatellites across 11p15 using FAM-labeled primers. PCR products were separated and detected using an ABI3100 capillary electrophoresis instrument, and analysis was performed using Genescan software (ABI). The peaks represent the different size alleles in the individual; the number below indicates the peak size in base pairs.
The patient’s mother inherited the deletion from her father, and thus we could compare the effect on gene expression of a maternally inherited deletion with that of a paternally inherited deletion. If the deletion removed a p57KIP2 enhancer, then the patient would be expected to show reduced expression with no change in expression in the mother. In contrast, if LIT1 acts as a silencer—that is, as a repressor of p57KIP2 over a distance—then the paternally inherited allele would be expected to be activated. To determine the relationship between the microdeletion and expression of p57KIP2, we performed real-time quantitative RT-PCR of p57KIP2 (fig. 2C). We could not measure allele-specific expression of p57KIP2 directly, since there were no polymorphisms in the exons, introns, or promoter of p57KIP2. TSSC3 and TSSC5 are not consistently imprinted in humans postnatally, so the effect on imprinting of more-distant genes could not be assessed.
The expression level of p57KIP2 was measured in peripheral blood lymphocyte RNA from individuals III-4 and II-2 and compared with normal age-matched controls. These data indicated that p57KIP2 expression is reduced in the proband (relative expression level 1.2, versus a mean ± SD of 7.9±3.8 in age-matched controls [fig. 2C]). In contrast, expression of p57KIP2 was normal in the patient’s mother, compared with that in age-matched controls (fig. 2C). Therefore, the microdeletion decreased p57KIP2 expression when inherited maternally but had no effect when inherited paternally, suggesting a mechanism of enhancer deletion. In addition, LIT1 itself showed normal expression in the proband but no detectable expression in the proband’s mother (fig. 2C), consistent with loss of her paternally inherited copy, which is normally the only active allele. Since the mother had no growth or developmental abnormalities, the absence of LIT1 RNA had no discernible phenotypic effect on her.
Discussion
In summary, we have described a microdeletion on 11p15 in two related individuals, one in whom the deletion is maternally inherited and the other in whom the deletion is paternally inherited. Although the mechanism of regulation of the imprinted cluster of the genes IGF2 and H19 has been well studied and related to the BWS phenotype, the regulation and disease-causing mechanism of the imprinted cluster of the genes p57KIP2, KVLQT1, LIT1, TSSC3, and TSSC5 is less well understood. In humans, it is clear that inactivating mutations in p57KIP2 cause BWS, and LOI and aberrant methylation of LIT1 has been associated with a third of all BWS cases. A number of studies in mice have indirectly shown that LIT1 plays a role in regulating imprinted expression of the nearby genes p57KIP2, TSSC3, and TSSC5. In mice, disruption of the imprinted cluster with a targeted translocation between p57KIP2 and KVLQT1 results in loss of expression and loss of imprinting of the genes p57KIP2, TSSC3, and TSSC5 (Cleary et al. 2001), indicating that KVLQT1 harbors important regulatory elements. However, knockout of the DMR of LIT1 in mice results in derepression of nearby imprinted genes and a microsomic phenotype when inherited paternally, although there is no affect on growth or imprinted gene expression when inherited maternally (Fitzpatrick et al. 2002).
Here, we provide the first human genetic evidence that the LIT1 domain contains regulatory elements that play a mechanistic role in the BWS phenotype and regulates the imprinting and expression of nearby imprinted genes. This microdeletion provides genetic confirmation of the importance of regulation of p57KIP2 at a distance in the pathogenesis of BWS. The model that best fits these data is that, in the case of the maternal deletion, p57KIP2 expression is lost because of loss of an enhancer within or telomeric to LIT1, leading to BWS, which supports the prediction of an engineered translocation in the mouse (Cleary et al. 2001). Consistent with this idea, Diaz-Meyer et al. (2003) found incomplete silencing of p57KIP2 in several patients with BWS who had LOI of LIT1, compared with patients without BWS, although patients with BWS who didn't have LOI were not examined in that study.
In addition, absence of an LIT1 transcript, as found in the proband’s mother, had no phenotypic consequence, indicating that the LIT1 RNA has no significant physiological function when inherited paternally. The deletion in this family is also of practical importance that should be brought to the attention of clinical geneticists performing molecular diagnosis of BWS. Several centers worldwide are performing methylation analysis for BWS, the results of which will be abnormal for these individuals. However, unlike LOI of LIT1, in which the recurrence risk and transmissibility are unknown, definitive counseling can be provided to patients with the deletion by using methylation analysis and either digital karyotyping or FISH with the appropriate probes. A mother carrying the deletion would have a 50% risk of transmission to subsequent offspring, and similar counseling could be provided to the probands as well.
Acknowledgments
We thank the families who participated in the study and Marcia Cruz-Correa for preparing the Spanish language consent. We thank Shirley Tilghman for many helpful conversations and discussion of unpublished data in the mouse. This work was supported by National Institutes of Health grant CA54358 (to A.P.F.) and by the Birth Defects March of Dimes Foundation.
Appendix: Primer and Probe Sequences
IGF2
IGF2-19F-GTTTAGCTTCTCTCCTCCTGATGG
IGF2-89R-TCTCTGTGCCAGGGAGGCT
IGF2-47T-VIC-CCTGGTCCCCCTTGCTGCTCTCC-TAMRA
KVLQT1 intron 1b.1
KvL-20to25KF-GCCAGGCGGGTCTAGATG
KvL-20to25KR-GGACAGGGCAGGTCATGAG
KvL-20to25KT-6-FAM-CAGCCAGGAACCAACC-TAMRA
KVLQT1 intron 1b.2
KvL-65to70KF-CTGCCGTGTTCAGAGTTCCA
KvL-65to70KR-CCAGTAGGCCCCAGCAT
KvL-65to70KT-6-FAM-CAGGCACCAGTCCACC-TAMRA
KVLQT1 intron 1c
KvL-int1c-426F-TCTCGGGCTATGACCCTGG
KvL-int1c-508R-TCCGACATCATTTCAACATCTCA
KvL-int1c466T-6-FAM-AGGGTTCCTGCATTCTCCTGTGTTCCC-TAMRA
KVLQT1 exon 2a
KvL-ex2a-3F-CCCTTTGCACTGCCTGCTA
KvL-ex2a-73R-GCAAATGGCATGCAGACCT
KvL-ex2a-23T-6-FAM-TTGTTTTCTGTCTTCATCCTGAGAGGGCTC-TAMRA
KVLQT1 exon 8
KvLQT1ex8-143544F-CCGGAGCCACACTCTGCT
KvLQT1ex8-143606R-GGGCTACTCACCACCACAGAC
KvLQT1ex8-143563T-6-FAM-TCACCCAGCCCCAAACCCAAGA-TAMRA
KVLQT1 exon 9
KvLQT1ex9-55F-AAGATGCTCACAGTCCCCCA
KvLQT1ex9-117R-GAAGTGGTCCAGCCGCC
KvLQT1ex9-76T-6-FAM-ATCACGTGCGACCCCCCAGAAG-TAMRA
LIT1 EST
AA602136-227F-TTACCCACTAACCAACCTCTCTTCA
AA602136-311R-TCATGGAGGTAGAGAGTAGAAGGATAGA
AA602136-253T-6-FAM-CCTCCAGACATACCTTTCACAGCCTTTGG-TAMRA
KVLQT1 exon 10
KvLQT1ex10-28F-TGAGCATGCCCCATTTCAT
KvLQT1ex10-95R-CAGAGTCTCCCCTTCCAGGTC
KvLQT1ex10-50T-6-FAM-AACCAACAGCTTCGCCGAGGACCT-TAMRA
LIT1 DMR
LIT1SEQ673F-CTCTGGGTATGTTCTCGAAAGTTG
LIT1SEQ743R-TCCCCCTTCCTCCTGTGTTT
LIT1SEQ699T-VIC-ACAACCCCAACCCAGGGTTGACCT-TAMRA
KVLQT1 exon 11
KvLQT1ex11flank-167F-TCCTCTCTCCACTGCAGGCT
KvLQT1ex11flank-230R-GCATGCGCGAATGACCTTA
KvLQT1ex11flank-188T-6-FAM-CGGGAACACCATCGGGCCAC-TAMRA
KVLQT1 intron 14
KvLQT1int14-128F-TCGCGTGCTGATCTGTTTG
KvLQT1int14-194R-CGTGGGTACCTGGAGGCC
KvLQT1int14-148T-VIC-CCCCTCCCCCACCACTTCTAGGTCTTTC-TAMRA
p57KIP2
KIP-2207F-AGGCGTGGACCGCTCTG
KIP-2276R-TGTGGGAGATGGAGAGTGCC
KIP-2227T-6-FAM-CGCACTAGCTCGGTTATTGGTTATGCCAA-TAMRA
PMP22
PMP22ex4F-CCCTTCTCAGCGGTGTCATC
PMP22ex4R-ACAGACCGTCTGGGCGC
PMP22ex4T-VIC-TCATTCGCGTTTCCGCAAGATCACA-TAMRA
(Wilke et al. 2000)
p57KIP2 for Real-Time Quantitative PCR
p57-1044F-GCGGCGATCAAGAAGCTG
p57-1124R-CGACGACTTCTCAGGCGC
p57-1069T-VIC-CTCTGATCTCCGATTTCTTCGCCAAGC-TAMRA
β-glucuronidase (GUS): purchased from Applied Biosystems.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for KVLQT1 genomic sequence [accession number AJ006345])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BWS, p57KIP2, LIT1,KVLQT1, and IGF2)
References
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B (1995) Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting centre on human chromosome 15. Nat Genet 9:395–400 10.1038/ng0495-395 [DOI] [PubMed] [Google Scholar]
- Cleary MA, van Raamsdonk CD, Levorse J, Zheng B, Bradley A, Tilghman SM (2001) Disruption of an imprinted gene cluster by a targeted chromosomal translocation in mice. Nat Genet 29:78–82 10.1038/ng715 [DOI] [PubMed] [Google Scholar]
- Cui H, Horon IL, Ohlsson R, Hamilton SR, Feinberg AP (1998) Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat Med 4:1276–1280 10.1038/3260 [DOI] [PubMed] [Google Scholar]
- Diaz-Meyer N, Day CD, Khatod K, Maher ER, Cooper W, Reik W, Junien C, Graham G, Algar E, Der Kaloustian VM, Higgins MD (2003) Silencing of CDKN1C (p57KIP2) is associated with hypomethylation at KvDMR1 in Beckwith-Wiedemann syndrome. J Med Genet 40:797–801 10.1136/jmg.40.11.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ (2002) Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32:426–431 10.1038/ng988 [DOI] [PubMed] [Google Scholar]
- Horike S, Mitsuya K, Meguro M, Kotobuki N, Kashiwagi A, Notsu T, Schulz TC, Shirayoshi Y, Oshimura M (2000) Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith-Wiedemann syndrome. Hum Mol Genet 9:2075–2083 10.1093/hmg/9.14.2075 [DOI] [PubMed] [Google Scholar]
- Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S, Oshimura M, Feinberg AP (1999) Loss of imprinting of a paternally expressed transcript, with antisense orientation to KvLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA 96:5203–5208 10.1073/pnas.96.9.5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MP, DeBaun M. Randhawa G, Reichard BA, Elledge SJ, Feinberg AP (1997) Low frequency of p57KIP2 mutation in Beckwith-Wiedemann syndrome. Am J Hum Genet 61:304–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Thompson JS, Edwards MC, Bartletta JM, Grundy P, Kalikin LM, Harper JW, Elledge SJ, Feinberg AP (1996) Imprinting of the gene encoding a human cyclin-dependent kinase inhibitor, p57KIP2, on chromosome 11p15. Proc Natl Acad Sci USA 93:3026–3030 10.1073/pnas.93.7.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe D, Dao D, Zhao L, Sanderson R. Warburton D, Weiss L, Anyane-Yeboa K, Tycko B (1997) Coding mutations in p57KIP2 are present in some cases of Beckwith-Wiedemann syndrome but are rare or absent in Wilms’ tumors. Am J Hum Genet 61:295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, Nicholls RD, Weksberg R, Driscoll DJ, Maher ER, Shows TB, Higgins MJ (1999) A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci USA 96:8064–8069 10.1073/pnas.96.14.8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenman MJ, Rainier S. Dobry CJ, Grundy P, Horon IL, Feinberg AP (1994) Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumor. Nat Genet 7:433–439 10.1038/ng0794-433 [DOI] [PubMed] [Google Scholar]
- Weksberg R, Shen DR, Fei YL, Song QL, Squire J (1993) Disruption of insulin-like growth factor 2 imprinting in Beckwith-Wiedemann syndrome. Nat Genet 5:143–150 10.1038/ng1093-143 [DOI] [PubMed] [Google Scholar]
- Wilke K, Duman B, Horst J (2000) Diagnosis of haploidy and triploidy based on measurement of gene copy number by real-time PCR. Hum Mutat 16:431–436 [DOI] [PubMed] [Google Scholar]