Abstract
Familial exudative vitreoretinopathy (FEVR) is a hereditary eye disorder that affects both the retina and vitreous body. Autosomal recessive FEVR was diagnosed in multiple individuals from three consanguineous families of European descent. A candidate-locus–directed genome scan shows linkage to the region on chromosome 11q flanked by markers D11S905 and D11S1314. The maximum LOD score of 3.6 at θ=0 is obtained with marker D11S987. Haplotype analysis confirms that the critical region is the 22-cM (311-Mb) interval flanked by markers D11S905 and D11S1314. This region contains LRP5 but not FZD4; mutations in both of these genes cause autosomal dominant FEVR. Sequencing of LRP5 shows, in all three families, homozygous mutations R570Q, R752G, and E1367K. This suggests that mutations in this gene can cause autosomal recessive as well as autosomal dominant FEVR.
Familial exudative vitreoretinopathy (FEVR [MIM 133780]) is a rare childhood hereditary eye disorder that affects both the retina and the vitreous body (Criswick and Schepens 1969). It is clinically similar to advanced retinopathy of prematurity, but the affected individuals do not have a history of premature birth and oxygen therapy. The condition is characterized by the premature arrest of the vascularization of the peripheral retina, leading to incomplete vascularization of the peripheral retina. This then results in the development of hyperpermeable blood vessels, neovascularization, and, hence, retinal folds, retinal exudates, and tractional retinal detachment in 20% of affected individuals. The disorder is bilateral and has a high degree of penetrance and a variable expressivity (Shastry and Hiraoka 2000). Minimally affected individuals do not show any visual symptoms. FEVR tends to be a slowly progressive disorder, with retinal detachment often not occurring until after the 1st decade of life.
FEVR is inherited in autosomal dominant (adFEVR [MIM 133780]), X-linked (MIM 305390), and autosomal recessive (arFEVR [MIM 601813]) fashions, although the autosomal dominant form is the most common. X-linked FEVR has been mapped to Xp11.4 and has been associated with mutations in the Norrie disease gene (ND), although genetic heterogeneity has been suggested by the failure to identify mutations in some patients (Chen et al. 1993; Fullwood et al. 1993; Shastry et al. 1995, 1997a, 1997b). adFEVR was initially mapped to two loci: EVR1 on chromosome 11q13-q23 (Li et al. 1992; Muller et al. 1994; Price et al. 1996; Shastry et al. 2000; Kondo et al. 2001) and EVR3 on chromosome 11p13-p12 (Downey et al. 2001).
Whereas the EVR3 locus has not been associated with mutations in any specific gene, the more common EVR1 locus has been associated with mutations in two linked genes: the Wnt-receptor frizzled-4 gene (FZD4) and the low-density lipoprotein receptor–related protein 5 gene (LRP5) (Robitaille et al. 2002; Toomes et al. 2004). Both FZD4 and LRP5 are involved in the Wnt signaling pathway, combining to bind Wnt proteins and to form a functional ligand-receptor complex (Pinson et al. 2000). It is interesting that norrin, the product of ND, is also a specific high-affinity ligand for FZD4 in concert with an LRP coreceptor, activating the Wnt signaling pathway (Xu et al. 2004). Mutations associated with Norrie disease decrease Wnt signaling to 20%–80% of control levels.
LRP5 is a member of the low-density lipoprotein receptor (LDLR) family, with four EGF precursor spacer domains—each comprising five YWTD repeats and one EGF repeat that together form a β-propeller structure—followed by three LDLR ligand-binding domain motifs, a single transmembrane domain, and a cytoplasmic tail (Hey et al. 1998). In addition to the interactions of LRP5 with FZD4 and Wnt, the intracellular domain of LRP5 binds Axin. This binding is facilitated by Wnt and is a part of Wnt signaling (Mao et al. 2001). LRP5 mutants lacking the extracellular domains bind Axin constitutively, inducing lymphoid enhancer-binding factor 1 (LEF1) activation by destabilizing Axin and stabilizing β-catenin. In animal models, LRP5 has been shown to be essential for bone accrual, eye development, and metabolism of cholesterol and glucose (Kato et al. 2002; Fujino et al. 2003). In humans, LRP5 is expressed by osteoblasts and affects bone mass (Gong et al. 2001), with specific LRP5 alleles associated with osteoporosis (Mizuguchi et al. 2004). Conversely, mutations in the amino-terminal part of LRP5 have been associated with a variety of conditions with increased bone density that are diagnosed as endosteal hyperostosis, Van Buchem disease, and osteopetrosis (Van Wesenbeeck et al. 2003). In addition to adFEVR, mutations in LRP5 have been associated with osteoporosis-pseudoglioma syndrome (OPPG [MIM 259770]), characterized by abnormal retinal vascularization often resulting in vitreal changes and pseudoglioma (Gong et al. 2001). Thus, mutations in LRP5 can result in either dominant or recessive disease affecting bone density, retinal vascularization, or both. It is notable that a number of individuals diagnosed with adFEVR and shown by Toomes et al. (2004) to have LRP5 mutations also had low bone density or a history of fractures.
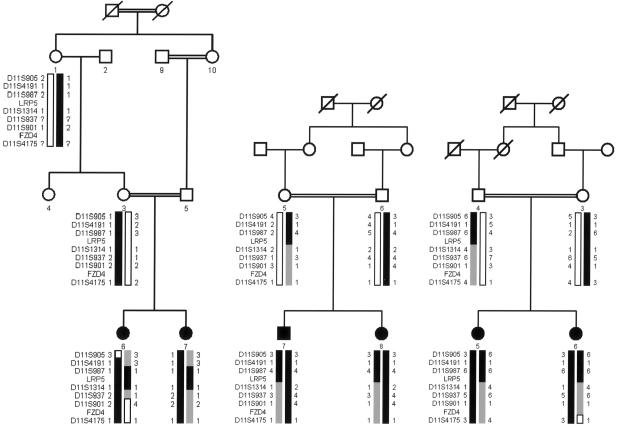
DNA samples were collected from three consanguineous families of European descent (families 68001, 68002, and 68003) (fig. 1) with 14 individuals, of whom 6 are affected. Informed consent was obtained from each individual studied. This study has been approved by the ethics review board of William Beaumont Hospital (Royal Oak, MI) and the institutional review board of the National Eye Institute and is consistent with the tenets of the Declaration of Helsinki. Patients were examined using indirect ophthalmoscopy and fluorescein angiography. Visual acuity was assessed in all patients.
Figure 1.
Pedigree of families 68001, 68002, and 68003. The blackened boxes correspond to affected haplotypes with alleles that cosegregate with the disease and that are homozygous in affected individuals.
Clinical descriptions of these families have been published elsewhere by Shastry and Trese (1997) (family 68001) and by de Crecchio et al. (1998) (families 68002 and 68003). In brief, the two affected sisters in family 68001 also have an affected paternal uncle (a product, along with five unaffected siblings, of the consanguineous mating of individuals 9 and 10, as shown in fig. 1). A fourth affected individual in this family is an affected nephew of individual 9, who is himself the product of a consanguineous mating (fig. 1) (see Shastry and Trese [1997]). Although the affected nephew—the son of an unaffected brother of individual 9—is not the product of a known consanguineous mating, the family belongs to an isolated Amish community, and it seems likely that his parents are distantly related. Both sisters were diagnosed at 3 years of age, at which time they manifested typical signs of FEVR, including large retinal folds, peripheral traction and exudates, and an avascular demarcation line near the equator and vitreous detachments. Unaffected relatives, including the parents, were examined and showed no funduscopic abnormalities (fig. 2) (see Shastry and Trese [1997]). Neither of the affected sisters, both 10 years old at the time of the study, has a history of bone fractures.
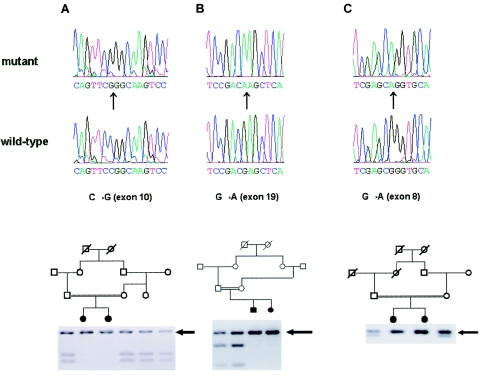
Figure 2.
Mutation screening of LRP5 in families; sequencing traces and restriction digests of amplified exons. A, Family 68001, showing a 2302C→G sequence change in exon 10 that eliminates an MspI site, so that affected siblings show a single 386-bp fragment and heterozygotes also show digested 212-bp and 174-bp fragments. The maternal aunt and grandmother are carriers, having both alleles. B, Family 68002, showing a 4147G→A sequence change in exon 19 that eliminates a SacI site, so that affected individuals show a single 269-bp fragment and heterozygotes also show digested 182-bp and 87-bp fragments. C, Family 68003, showing a 1757G→A sequence change in exon 8 that eliminates an AciI recognition site, so that affected individuals show a single 252-bp fragment and heterozygotes also show a 179-bp fragment (the remaining 73-bp fragment was not seen on the gel). Mutant and control (wild-type) sequence traces are shown above the restriction-enzyme-digestion products of exons 10, 19, and 8.
The affected brother and sister in family 68002 (family 2 in de Crecchio et al. 1998) were diagnosed with retinal detachments at ages 6 years and 8 years, respectively, and also showed other typical signs of FEVR. They showed no systemic symptoms, including fractures and traumatic injuries, by ages 12 years and 10 years, respectively. No additional family members are known to be affected, including seven maternal and seven paternal aunts and uncles. The affected sisters in family 68003 (family 1 in de Crecchio et al. 1998) were diagnosed at ages 5 years and 7 years and have shown typical signs of FEVR that required multiple procedures, including photocoagulation, cryopexy, and vitrectomy. They show no signs of systemic disease, including fractures, at ages 29 years and 31 years. Consanguinity in this family was disclosed after publication of the article by de Crecchio et al. (1998). No additional family members are known to be affected, including five paternal and four maternal aunts and uncles. In families 68002 and 68003, unaffected relatives, including both pairs of parents, underwent full ophthalmological examinations that included fluorescein angiography and showed no abnormalities, including no vascular tortuosity. Both the presence of consanguinity and the normal results of examination of the unaffected parents in each family strongly support autosomal recessive inheritance in these families. In addition, although we are unable to address bone density specifically, the absence of fractures in the six affected individuals in these three families, with an age range of 10–31 years, would be highly atypical for patients with OPPG.
For linkage analysis, DNA was extracted directly from blood by standard phenolchloroform protocols (Smith et al. 1989). A candidate-oriented genomewide scan was begun with 382 fluorescently labeled microsatellite markers (ABI Linkage Mapping Set MD-10) with the use of only a subset of samples, to conserve the limited amounts of DNA available from some individuals. Multiplexed PCR was performed as described elsewhere (Jiao et al. 2000). Two-point linkage analyses were performed using the FASTLINK implementation of the MLINK program of the LINKAGE program package (Lathrop and Lalouel 1984; Cottingham et al. 1993). For screening, equal allele frequencies were used for all markers. For fine mapping on chromosome 11, the allele frequencies were calculated using 15 control individuals (30 chromosomes) of European ancestry.
Evidence of significant linkage in the genomewidescan was first observed with markers D11S987 and D11S4191, which gave maximum LOD scores of 3.3 at θ=0 and 2.43 at θ=0, respectively. For fine mapping using markers taken from the Genethon database, additional individuals were added to the families, estimated allele frequencies were used, and the maximum LOD score for D11S987 increased to 3.6 at θ=0, whereas, for D11S4191, the LOD score decreased to −1.1 at θ=0, with a LOD score of 1.1 at θ=0.1 (table 1). By use of LOD scores ⩽−2 to exclude linkage in these families, arFEVR mapped to a 21.7-cM (311-Mb) interval flanked by D11S905 and D11S1314. Markers D11S901 and D11S4175, which flank FZD4, both show obligate recombinations.
Table 1.
LOD Scores of Markers on Chromosome 11q
|
Position |
LOD Score at θ = |
||||||||
| Markers | cM | Mb | 0 | .01 | .05 | .1 | .2 | .3 | .4 |
| D11S905 | 51.95 | 409.38 | −∞ | −2.2 | −.4 | .1 | .3 | .2 | 0 |
| D11S4191 | 60.09 | 597.75 | −1.1 | .6 | 1 | 1.1 | .8 | .5 | .2 |
| D11S987 | 67.48 | … | 3.6 | 3.5 | 3.1 | 2.7 | 1.8 | 1 | .4 |
| D11S1314 | 73.64 | 720.49 | −4.7 | −2 | −.8 | −.3 | 0 | 0 | 0 |
| D11S937 | 79.98 | 775.8 | −7.8 | −2.9 | −1 | −.4 | 0 | 0 | 0 |
| D11S901 | 85.48 | 815.71 | −∞ | −.4 | .6 | .8 | .8 | .5 | .2 |
| D11S4175 | 91.47 | 899.4 | −∞ | −.8 | .3 | .5 | .5 | .3 | .1 |
Examination of the haplotypes cosegregating with arFEVR in the three families confirms and extends the linkage results. In family 68001, a recombination event between marker D11S905 and the arFEVR locus in individual 6 provides the centromeric boundary to the linked region. In addition, lack of homozygosity for markers D11S905 and D11S4191 in both affected individuals suggests that the arFEVR locus might lie distal to D11S4191 (fig. 1). In family 68002, lack of homozygosity in marker D11S1314 suggests that the arFEVR locus lies proximal to that marker. Similarly, in family 68003, lack of homozygosity in markers D11S905 and D11S1314 suggests that arFEVR lies in the same interval suggested by the two-point linkage analysis: the linked region between D11S905 and D11S1314 (fig. 1). This critical interval contains LRP5, in which mutations have recently been shown to cause adFEVR. FZD4, the other gene on chromosome 11q implicated in adFEVR and also part of the Wnt signaling pathway, lies between markers D11S901 and D11S4175 and is excluded from the linked region by an obligate recombination event in individual 7 of family 68001 and by lack of homozygosity in all families.
For mutation screening of LRP5, coding exons and adjacent intronic sequences were amplified from genomic DNA of two affected patients and one unaffected individual. The reference genomic sequence is available from NCBI (accession number NM_002335). PCR amplification of 23 exons by use of primers listed in table 1 of Toomes et al. (2004) was performed in 20-μl reactions involving 2.5 U of Taq Gold (ABI), 10× 2 μl of Buffer Gold Taq, 1.9 mM of MgCl2, 0.25 mM of dNTP, 0.25 μM of primers, and 40 ng of genomic DNA. PCR cycling consisted of an initial 10-min denaturation step at 95°C for 5 min; 32 cycles at 94°C for 40 s, 55°C for 30 s, and 72°C for 30 s; and a final elongation step at 72°C for 5 min. PCR products were purified using either a Qiagen PCR purification kit or an Edge Biosystem QuickStepTm2 PCR purification kit. The PCR template was sequenced using ABI Dye Terminator, version 1, in a final reaction of 10 μl. Products of the sequencing reactions were purified using the Edge Biosystem Performa TM DTR gel-filtration system and were run on a 3100 ABI genetic analyzer. Sequences were aligned using the Seqman program of the DNASTAR program package.
In family 68001, exon 10 of the LRP5 gene shows a c2302C→G transversion resulting in an R752G mutation in the third YWTD domain of the protein. The 2302C→G sequence change eliminates an MspI site, so that amplification using the sequencing primers and digestion with MspI yields 212-bp and 174-bp fragments in control alleles but yields a 386-bp fragment in mutant alleles (fig. 2A). This nonconservative change occurs in an arginine residue evolutionarily conserved, from human to Drosophila, in LRP4, LRP5, and LRP6, when sequences are aligned using the Clustal W algorithm as implemented in the MegAlign program (DNASTAR). In family 68002, exon 19 shows a c4147G→A transition resulting in a E1367K change in the third LDLR ligand-binding domain of the protein. The 4147G→A sequence change eliminates a SacI site, so that amplification with the exon 19 sequencing primers, followed by digestion with SacI, yields 182-bp and 87-bp fragments from control alleles but yields a single 269-bp fragment from the mutant allele (fig. 2B). In the protein, this change from a negatively to a positively charged amino acid occurs at a glutamate residue evolutionarily conserved, from human to Drosophila, in LRP5 and LRP6 but not in LRP4. Finally, affected individuals in family 68003 show a c1757G→A transition in exon 8, resulting in a R570Q substitution in the second YWTD domain of the protein. This is the same residue which, when mutated to tryptophan (R570W) results in the osteoporosis-pseudoglioma syndrome. Although bone-density measurements are not available for these affected siblings, they have had no fractures by ages 29 years and 31 years, which would be distinctly unusual for OPPG, and their ophthalmic course is consistent with FEVR rather than OPPG, which often presents with blindness in the neonatal period (Beighton et al. 1985; De Paepe et al. 1993). The 1757G→A sequence change eliminates an AciI recognition site, so that PCR amplification with the distal exon 8 sequencing primers followed by digestion with AciI yields 73-bp and 179-bp fragments in control individuals but yields a single 252-bp fragment from the mutant allele (fig. 2C). In the protein, this substitution of a polar for a charged amino acid changes an arginine residue evolutionarily conserved, from human to Drosophila, in LRP5 and LRP6 proteins but not in LRP4 proteins from human and mouse. None of these changes were seen in 100 control individuals (200 chromosomes) of European ethnic extraction, when these individuals were tested using PCR amplification followed by digestion with the appropriate restriction endonuclease.
The combination of linkage and mutation analysis provides strong evidence that mutations in LRP5 can cause arFEVR. Thus, mutations in LRP5 can result in a variety of clinical presentations and can affect bone density and retinal vascularization. Mutations that truncate the protein or cause a frameshift early in the protein—in either the EGF or LDLR repeat domains—have been described in individuals with OPPG (Gong et al. 2001). Truncation or frameshift mutations more distal in the protein have been described in patients with adFEVR (although one family had associated osteoporosis) (Toomes et al. 2004). This was true in one mutation that occurred before the transmembrane domain, although, in this mutation, 52 novel amino acids of uncertain effect were encoded. Missense mutations in LRP5 can also cause a variety of clinical presentations. A set of moderate to conservative mutations (e.g., G171V, G171R, A214T, A214V, A242T, and T253I) in the first EGF spacer domain have been shown to cause disorders of increased bone mass (Boyden et al. 2002; Little et al. 2002; Van Wesenbeeck et al. 2003). Other missense mutations in the EGF repeat domains or LDLR ligand-binding domains have been found in individuals with OPPG (Gong et al. 2001) or adFEVR (Toomes et al. 2004). Here, we describe three additional missense mutations in the second and third EGF repeat and in the third LDLR ligand-binding domain that are associated with arFEVR. Although one would suspect that these mutations cause milder decreases in function of the LRP5 protein than the functional decrease associated with adFEVR, the proof of this must await functional studies of the mutant proteins. There also remains the possibility that the effects of mutations in LRP5 might vary with the genetic background, especially with regard to other genes in the Wnt signaling pathway. Finally, it seems likely that just as adFEVR can be caused by mutations in more than one member of the Wnt signaling pathway, arFEVR might show similar locus heterogeneity, which this study does not address.
Acknowledgments
We thank the members of the three families, for their participation in this study.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- Genethon, http://www.genethon.fr/php/index_us.php
- NCBI, http://www.ncbi.nlm.nih.gov/ (for LRP5 reference genome sequence [accession number NM_002335])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FEVR and OPPG)
References
- Beighton P, Winship I, Behari D (1985) The ocular form of osteogenesis imperfecta: a new autosomal recessive syndrome. Clin Genet 28:69–75 [DOI] [PubMed] [Google Scholar]
- Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP (2002) High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med 346:1513–1521 10.1056/NEJMoa013444 [DOI] [PubMed] [Google Scholar]
- Chen ZY, Battinelli EM, Fielder A, Bundey S, Sims K, Breakefield XO, Craig IW (1993) A mutation in the Norrie disease gene (NDP) associated with X-linked familial exudative vitreoretinopathy. Nat Genet 5:180–183 10.1038/ng1093-180 [DOI] [PubMed] [Google Scholar]
- Cottingham RW, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Criswick VG, Schepens CL (1969) Familial exudative vitreoretinopathy. Am J Ophthalmol 68:578–594 [DOI] [PubMed] [Google Scholar]
- de Crecchio G, Simonelli F, Nunziata G, Mazzeo S, Greco GM, Rinaldi E, Ventruto V, Ciccodicola A, Miano MG, Testa F, Curci A, D’Urso M, Rinaldi MM, Cavaliere ML, Castelluccio P (1998) Autosomal recessive familial exudative vitreoretinopathy: evidence for genetic heterogeneity. Clin Genet 54:315–320 10.1034/j.1399-0004.1998.5440409.x [DOI] [PubMed] [Google Scholar]
- De Paepe A, Leroy JG, Nuytinck L, Meire F, Capoen J (1993) Osteoporosis-pseudoglioma syndrome. Am J Med Genet 45:30–37 [DOI] [PubMed] [Google Scholar]
- Downey LM, Keen TJ, Roberts E, Mansfield DC, Bamashmus M, Inglehearn CF (2001) A new locus for autosomal dominant familial exudative vitreoretinopathy maps to chromosome 11p12-13. Am J Hum Genet 68:778–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T, Asaba H, Kang MJ, Ikeda Y, Sone H, Takada S, Kim DH, Ioka RX, Ono M, Tomoyori H, Okubo M, Murase T, Kamataki A, Yamamoto J, Magoori K, Takahashi S, Miyamoto Y, Oishi H, Nose M, Okazaki M, Usui S, Imaizumi K, Yanagisawa M, Sakai J, Yamamoto TT (2003) Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci USA 100:229–234 10.1073/pnas.0133792100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood P, Jones J, Bundey S, Dudgeon J, Fielder AR, Kilpatrick MW (1993) X linked exudative vitreoretinopathy: clinical features and genetic linkage analysis. Br J Ophthalmol 77:168–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, et al (2001) LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell 107:513–523 10.1016/S0092-8674(01)00571-2 [DOI] [PubMed] [Google Scholar]
- Hey PJ, Twells RC, Phillips MS, Yusuke N, Brown SD, Kawaguchi Y, Cox R, Guochun X, Dugan V, Hammond H, Metzker ML, Todd JA, Hess JF (1998) Cloning of a novel member of the low-density lipoprotein receptor family. Gene 216:103–111 10.1016/S0378-1119(98)00311-4 [DOI] [PubMed] [Google Scholar]
- Jiao X, Munier FL, Iwata F, Hayakawa M, Kanai A, Lee J, Schorderet DF, Chen MS, Kaiser-Kupfer M, Hejtmancik JF (2000) Genetic linkage of Bietti crystalline corneoretinal dystrophy to chromosome 4q35. Am J Hum Genet 67:1309–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, Hartmann C, Li L, Hwang TH, Brayton CF, Lang RA, Karsenty G, Chan L (2002) Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol 157:303–314 10.1083/jcb.200201089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Ohno K, Tahira T, Hayashi H, Oshima K, Hayashi K (2001) Delineation of the critical interval for the familial exudative vitreoretinopathy gene by linkage and haplotype analysis. Hum Genet 108:368–375 10.1007/s004390100503 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM (1984) Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 36:460–465 [PMC free article] [PubMed] [Google Scholar]
- Li Y, Müller B, Fuhrmann C, van Nouhuys CE, Laqua H, Humphries P, Schwinger E, Gal A (1992) The autosomal dominant familial exudative vitreoretinopathy locus maps on 11q and is closely linked to D11S533. Am J Hum Genet 51:749–754 [PMC free article] [PubMed] [Google Scholar]
- Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, et al (2002) A mutation in the LDL receptor–related protein 5 gene results in the autosomal dominant high–bone-mass trait. Am J Hum Genet 70:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B, Pan W, Farr GH III, Flynn C, Yuan H, Takada S, Kimelman D, Li L, Wu D (2001) Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell 7:801–809 10.1016/S1097-2765(01)00224-6 [DOI] [PubMed] [Google Scholar]
- Mizuguchi T, Furuta I, Watanabe Y, Tsukamoto K, Tomita H, Tsujihata M, Ohta T, Kishino T, Matsumoto N, Minakami H, Niikawa N, Yoshiura K (2004) LRP5, low-densitylipoprotein-receptor-related protein 5, is a determinant for bone mineral density. J Hum Genet 49:80–86 10.1007/s10038-003-0111-6 [DOI] [PubMed] [Google Scholar]
- Müller B, Orth U, van Nouhuys CE, Duvigneau C, Fuhrmann C, Schwinger E, Laqua H, Gal A (1994) Mapping of the autosomal dominant exudative vitreoretinopathy locus (EVR1) by multipoint linkage analysis in four families. Genomics 20:317–319 10.1006/geno.1994.1176 [DOI] [PubMed] [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC (2000) An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407:535–538 10.1038/35035124 [DOI] [PubMed] [Google Scholar]
- Price SM, Periam N, Humphries A, Woodruff G, Trembath RC (1996) Familial exudative vitreoretinopathy linked to D11S533 in a large Asian family with consanguinity. Ophthalmic Genet 17:53–57 [DOI] [PubMed] [Google Scholar]
- Robitaille J, MacDonald ML, Kaykas A, Sheldahl LC, Zeisler J, Dube MP, Zhang LH, Singaraja RR, Guernsey DL, Zheng B, Siebert LF, Hoskin-Mott A, Trese MT, Pimstone SN, Shastry BS, Moon RT, Hayden MR, Goldberg YP, Samuels ME (2002) Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat Genet 32:326–330 10.1038/ng957 [DOI] [PubMed] [Google Scholar]
- Shastry BS, Hejtmancik JF, Hiraoka M, Ibaraki N, Okubo Y, Okubo A, Han DP, Trese MT (2000) Linkage and candidate gene analysis of autosomal-dominant familial exudative vitreoretinopathy. Clin Genet 58:329–332 10.1034/j.1399-0004.2000.580412.x [DOI] [PubMed] [Google Scholar]
- Shastry BS, Hejtmancik JF, Plager DA, Hartzer MK, Trese MT (1995) Linkage and candidate gene analysis of X-linked familial exudative vitreoretinopathy. Genomics 27:341–344 10.1006/geno.1995.1052 [DOI] [PubMed] [Google Scholar]
- Shastry BS, Hejtmancik JF, Trese MT (1997a) Identification of novel missense mutations in the Norrie disease gene associated with one X-linked and four sporadic cases of familial exudative vitreoretinopathy. Hum Mutat 9:396–401 [DOI] [PubMed] [Google Scholar]
- Shastry BS, Hiraoka M (2000) Molecular genetics of familial exudative vitreoretinopathy and Norrie disease. Curr Genomics 1:259–269 [Google Scholar]
- Shastry BS, Liu X, Hejtmancik JF, Plager DA, Trese MT (1997b) Evidence for genetic heterogeneity in X-linked familial exudative vitreoretinopathy. Genomics 44:247–248 10.1006/geno.1997.4863 [DOI] [PubMed] [Google Scholar]
- Shastry BS, Trese MT (1997) Familial exudative vitreoretinopathy: further evidence for genetic heterogeneity. Am J Med Genet 69:217–218 [DOI] [PubMed] [Google Scholar]
- Smith RJH, Holcomb JD, Daiger SP, Caskey CT, Pelias MZ, Alford BR, Fontenot DD, Hejtmancik JF (1989) Exclusion of Usher syndrome gene from much of chromosome 4. Cytogenet Cell Genet 50:102–106 [DOI] [PubMed] [Google Scholar]
- Toomes C, Bottomley HM, Jackson RM, Towns KV, Scott S, Mackey DA, Craig JE, Jiang L, Yang Z, Trembath R, Woodruff G, Gregory-Evans CY, Gregory-Evans K, Parker MJ, Black GCM, Downey LM, Zhang K, Inglehearn CF (2004) Mutations in LRP5 or FZD4 underlie the common familial exudative vitreoretinopathy locus on chromosome 11q. Am J Hum Genet 74:721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wesenbeeck L, Cleiren E, Gram J, Beals RK, Bénichou O, Scopelliti D, Key L, Renton T, Bartels C, Gong Y, Warman ML, de Vernejoul MC, Bollerslev J, Van Hul W (2003) Six novel missense mutations in the LDL receptor-related protein 5 (LRP5) gene in different conditions with an increased bone density. Am J Hum Genet 72:763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K, Nathans J (2004) Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell 116:883–895 10.1016/S0092-8674(04)00216-8 [DOI] [PubMed] [Google Scholar]