Abstract
Linkage and association studies have recently implicated dystrobrevin-binding protein 1 (DTNBP1) in the etiology of schizophrenia. We analyzed seven previously tested DTNBP1 single-nucleotide polymorphisms (SNPs) in a cohort of 524 individuals with schizophrenia or schizoaffective disorder and 573 control subjects. The minor alleles of three SNPs (P1578, P1763, and P1765) were positively associated with the diagnosis of schizophrenia or schizoaffective disorder in the white subset of the study cohort (258 cases, 467 controls), with P1578 showing the most significant association (odds ratio 1.76, P = .0026). The same three SNPs were also associated in a smaller Hispanic subset (51 cases, 32 controls). No association was observed in the African American subset (215 cases, 74 controls). A stratified analysis of the white and Hispanic subsets showed association with the minor alleles of four SNPs (P1578, P1763, P1320, and P1765). Again, the most significant association was observed for P1578 (P = .0006). Haplotype analysis supported these findings, with a single risk haplotype significantly overrepresented in the white sample (P = .005). Our study provides further evidence for a role of the DTNBP1 gene in the genetic etiology of schizophrenia.
Schizophrenia (MIM #181500) is a common, complex disorder affecting ∼1% of the population worldwide. Its high heritability (∼80%) and elevated sibling recurrence risk (λS≈10) indicate a strong genetic component (McGuffin et al. 1995; Cardno and Gottesman 2000; Levinson and Mowry 2000). Current data suggest that several loci with low individual effects contribute to the genetic risk (Risch 1990). The identification of susceptibility genes for common, complex disorders is challenging, since large numbers of well-diagnosed cases are needed to detect weak effects of individual susceptibility genes (e.g., Altshuler et al. 2000; reviewed by Risch 2000). An added obstacle in the study of the genetic basis of psychiatric disorders like schizophrenia is the lack of objective biomarkers to aid in determination of phenotypes. Genomewide scans can be used to pinpoint the chromosomal location of susceptibility genes. However, because of the low marker resolution, the linked intervals are large—requiring additional approaches, such as linkage disequilibrium (LD) mapping, to identify disease-associated genes under linkage peaks.
Despite the difficulties in standardizing phenotypic information, genomewide scans have now reliably implicated several loci in the etiology of schizophrenia, including a region on chromosome 6p (6p24-22) (Lewis et al. [2003] and references within). Straub et al. (2002) subsequently narrowed the interval on chromosome 6p by using LD mapping to analyze a sample of 270 Irish pedigrees who were multiply affected with schizophrenia, finding significant association of several variants in the DTNBP1 gene on chromosome 6p22.3. Reanalysis of these data identified a haplotype block, which spanned 30 kb and contained a single high-risk haplotype of relatively recent evolutionary origin (van den Oord et al. 2003). Several replication studies in different populations have been reported since then, and most have confirmed an association with DTNBP1 (Morris et al. 2003; Schwab et al. 2003; Tang et al. 2003; Van Den Bogaert et al. 2003; Kirov et al. 2004; Williams et al. 2004). However, despite the overall confirmation of an association with DTNBP1, there was considerable heterogeneity among the associated alleles and haplotypes, suggesting allelic heterogeneity at this locus. The majority of studies have employed a family-based approach (Straub et al. 2002; Schwab et al. 2003; Tang et al. 2003; van den Oord et al. 2003; Kirov et al. 2004). Van Den Bogaert et al. (2003), who observed increased significance in positive-family-history cases in a Swedish case-control sample, hypothesized that DTNBP1 is particularly involved in the etiology of schizophrenia in cases with familial loading.
DTNBP1 is an evolutionarily conserved protein, which binds α- and β-dystrobrevin—components of the dystrophin-associated protein complex (DPC) in muscle and brain (Benson et al. 2001). In muscle, DTNBP1 colocalizes with α-dystrobrevin at the sarcolemma, and disruption of the DPC results in Duchenne and Becker muscular dystrophies (DMD and BMD) (Straub and Campbell 1997; Muntoni et al. 2003). It is interesting that DMD and schizophrenia share a number of phenotypes: both disorders have neurological findings, including ocular-control abnormalities characteristic of cerebellar problems (Lui et al. 2001; Anderson et al. 2002; Avila et al. 2002). Furthermore, recent research has highlighted the presence of cognitive impairments in DMD and BMD, which may relate to the now well-established cognitive deficits observed in schizophrenia (North et al. 1996; Hinton et al. 2000; reviewed by Mehler 2000). These findings suggest that the DPC may be involved in the etiology of both muscular dystrophy and schizophrenia. However, Talbot et al. (2004) recently reported presynaptic DTNBP1 reductions at hippocampal formation sites in patients with schizophrenia. These sites lack neuronal dystrobrevin, indicating that the role of DTNBP1 in schizophrenia may be independent of the DPC. In addition, Talbot et al. (2004) showed that the same patients with schizophrenia exhibited an increase in vesicular glutamate transporter 1 (VGluT-1), providing evidence of a potential role of DTNBP1 in glutamate neurotransmission, which has been implicated in the pathogenesis of schizophrenia (Collier and Li 2003).
Despite intensive resequencing efforts, no DTNBP1 coding variants have yet been identified (dbSNP build 120 [see dbSNP Home Page]) (Williams et al. 2004). Bray et al. (2003), however, have recently detected strong allele-specific expression of the DTNBP1 gene in the brain, raising the possibility that cis-acting variation may contribute to the role of DTNBP1 in the etiology of schizophrenia.
We present here an independent replication of the association of the DTNBP1 locus with schizophrenia. This is the first report of a positive association in a U.S. sample and the first study examining DTNBP1 variants in an African American population. Our sample consisted of 524 cases (patients with a diagnosis of either schizophrenia [n=421] or schizoaffective disorder [n=103]) and 573 controls of various ethnicities.
Cases were recruited from the inpatient and outpatient clinical services of the Zucker Hillside Hospital, a division of the North Shore–Long Island Jewish Health System, where patients are screened for potential recruitment into research studies by the Clinical Assessment and Training (CAT) unit of the National Institutes of Health–funded Hillside Hospital Intervention Research Center. The CAT unit monitors the inpatient and outpatient hospital census daily and conducts preliminary screening and recruitment functions. Inclusion criteria for screening for this study included a clinical diagnosis of a psychotic disorder, no active substance abuse, and the ability to provide informed consent. After providing written informed consent, each subject was assessed with the Structured Clinical Interview for DSM-IV Axis I disorders (SCID, version 2.0), which was administered by trained raters. Standardized diagnostic assessments were supplemented with clinical information obtained by a review of medical records and interviews with family informants when possible, and all diagnostic information was compiled into a narrative case summary. Information on the onset and course of Axis I illness, the presence of Axis II pathology, the presence of Axis III diagnoses, and a brief description of the subject’s psychosocial and occupational functioning during the course of illness was presented to a consensus diagnostic committee that consisted of a minimum of three senior faculty with DSM-IV diagnostic experience, as well as other faculty and trainees with SCID experience. All available information was used to arrive at a consensus DSM-IV diagnosis.
Healthy controls were recruited by the Zucker Hillside Hospital Normal Control Program and were ascertained in the same manner as the cases. Potential participants were recruited by use of local newspaper advertisements, flyers, and community Internet resources and underwent initial telephone screening to assess eligibility criteria. The nonpatient SCID (SCID-NP) was administered to subjects who met eligibility criteria, to rule out the presence of an Axis I psychiatric disorder; a urine toxicology screen for drug use and an assessment of the subject's family history of psychiatric disorders were also performed. Exclusion criteria included (current or past) Axis I psychiatric disorder, psychotropic drug treatment, substance abuse, a first-degree family member with an Axis I psychiatric disorder, or the inability to provide written informed consent. A subset of the control cohort was collected through the Massachusetts General Hospital Clinical Research Program, in conjunction with the Harvard Medical School–Partners Center for Genetics and Genomics in Boston. This subset comprised disease-free subjects >18 years of age. For the purposes of the study, “disease” was defined as current or past diagnoses made by a medical-care provider that required medication or other forms of treatment or therapy. Surgery was considered to be an exclusionary criterion, with the exception of orthopedic procedures, appendectomy, or surgeries that were related to trauma. Subjects who took prescription medications or regularly used over-the-counter medications were also excluded. Eligible subjects completed a structured family and medical questionnaire that detailed current and past history of psychiatric illness and pharmacological or psychotherapeutic psychiatric treatment. All responses to the self-report form were confirmed by physicians in clinical interviews with subjects. Physical exams were completed at the study visit. This study was approved by the institutional review boards of the North Shore–Long Island Jewish Health System and Harvard Medical School–Partners Center for Genetics and Genomics. The largest ethnic groups represented in the study were whites (258 cases [mean age 39.4 years; 35.7% female/64.3% male] and 467 controls [mean age 39.3 years; 51.8% female/48.2% male]), African Americans (215 cases [mean age 35.9 years; 36.3% female/63.7% male] and 74 controls [mean age 31.9 years; 59.5% female/40.5% male]), and Hispanics (51 cases [mean age 34.2 years; 35.3% female/64.7% male] and 32 controls [mean age 32.9 years; 40.6% female/59.4% male]).
We genotyped seven of the SNPs that were initially described by Straub et al. (2002). They included P1635, which had yielded the most significant results in the study by Straub et al. (2002); P1320, which was most significant in the studies by van den Oord et al. (2003) and Schwab et al. (2003); and the haplotype-tag SNPs P1578, P1763, and P1765, which were described by van den Oord et al. (2003). All SNPs are listed in table 1. Genotyping was performed by MALDI-ToF mass spectrometry by use of the Sequenom system. Tests of Hardy-Weinberg equilibrium (HWE) were performed at each SNP locus (Hill 1974). P1635 was the only SNP that was out of HWE across several ethnic groups in cases and controls and was therefore excluded from all analyses.
Table 1.
Allele Frequencies and P Values for Six DTNBP1 SNP Markers[Note]
|
Patients with Schizophreniaor Schizoaffective Disorder |
Patients with Schizophrenia |
||||||||||
| Minor-Allele Frequency |
Minor-Allele Frequency |
||||||||||
| Subset and SNP ID (dbSNP Number) | Alleles(Major/Minor) | Controlsa | Casesb | Allele P | Trend P | OR | Controlsa | Casesc | Allele P | Trend P | OR |
| White: | |||||||||||
| P1583 (rs909706) | C/T | .342 | .346 | .8795 | .8801 | 1.02 | .342 | .349 | .8051 | .8046 | 1.03 |
| P1578 (rs1018381) | C/T | .073 | .121 |
.0026 |
.0037 |
1.76 | .073 | .113 |
.0194 |
.0213 |
1.62 |
| P1763 (rs2619522) | A/C | .156 | .207 |
.0169 |
.0195 |
1.41 | .156 | .213 |
.0146 |
.0161 |
1.47 |
| P1320 (rs760761) | C/T | .163 | .197 | .1194 | .1276 | 1.26 | .163 | .207 | .0649 | .0676 | 1.34 |
| P1765 (rs2691528) | G/A | .157 | .208 |
.0171 |
.0195 |
1.41 | .157 | .213 |
.0162 |
.0176 |
1.46 |
| P1325 (rs1011313) | C/T | .070 | .060 | .4733 | .4786 | .85 | .070 | .058 | .4380 | .4397 | .82 |
| Hispanic: | |||||||||||
| P1583 (rs909706) | C/T | .345 | .330 | .8491 | .8496 | .94 | .345 | .325 | .8073 | .8150 | .91 |
| P1578 (rs1018381) | C/T | .060 | .177 | .0506 |
.0449 |
3.36 | .060 | .171 | .0654 |
.0460 |
3.23 |
| P1763 (rs2619522) | A/C | .154 | .324 |
.0243 |
.0319 |
2.63 | .154 | .305 |
.0480 |
.0565 | 2.41 |
| P1320 (rs760761) | C/T | .150 | .300 |
.0351 |
.0540 |
2.43 | .150 | .278 | .0775 | .1012 | 2.18 |
| P1765 (rs2691528) | G/A | .150 | .320 |
.0171 |
.0239 |
2.67 | .150 | .300 |
.0385 |
.0473 |
2.43 |
| P1325 (rs1011313) | C/T | .117 | .080 | .4411 | .4802 | .66 | .117 | .088 | .5692 | .6043 | .73 |
| African American: | |||||||||||
| P1583 (rs909706) | C/T | .221 | .234 | .7518 | .7567 | 1.08 | .221 | .236 | .72692 | .73135 | 1.09 |
| P1578 (rs1018381) | C/T | .317 | .301 | .7210 | .7270 | .93 | .317 | .301 | .72910 | .73492 | .93 |
| P1763 (rs2619522) | A/C | .390 | .357 | .4949 | .5043 | .87 | .390 | .356 | .49307 | .50293 | .87 |
| P1320 (rs760761) | C/T | .345 | .349 | .9286 | .9324 | 1.02 | .345 | .358 | .78009 | .79132 | 1.06 |
| P1765 (rs2691528) | G/A | .371 | .358 | .7715 | .7737 | .94 | .371 | .358 | .78441 | .78663 | .95 |
| P1325 (rs1011313) | C/T | .043 | .058 | .4819 | .4842 | 1.38 | .043 | .057 | .51561 | .52014 | 1.36 |
Note.— Significant P values are shown in bold, underlined italics. P values that approach significance are shown in bold italics.
Sample sizes were as follows: white (n=467), Hispanic (n=32), African American (n=74).
Sample sizes were as follows: white (n=258), Hispanic (n=51), African American (n=215).
Sample sizes were as follows: white (n=196), Hispanic (n=41), African American (n=184).
Statistical inference for single-SNP associations was based on χ2 test statistics. For each SNP, an allelic association test and the Cochran-Armitage trend test (test for additive allelic effects) were performed (Sasieni 1997). Odds ratios (ORs) associated with the minor allele were also produced for each SNP. To reduce the potential influence of population stratification, the χ2 tests were computed separately for each of the three largest ethnic groups: whites, African Americans, and Hispanics. A stratified Cochran-Armitage trend test was used in the analysis of association across ethnic groups. Haplotype associations were explored using score tests that account for linkage phase ambiguity (Schaid et al. 2002; Lake et al. 2003). The score tests, derived from generalized linear models, are used for global tests of association, as well as haplotype-specific tests. In addition, a permutation algorithm was applied in the testing framework to find the maximum of the haplotype-specific score statistics and the associated P value for this maximum. The Haplo Stats program, which implements the methods of Schaid et al. (2002), was used for these analyses. Haplotypes were imputed, and frequencies were estimated using the estimation facility in Haplo Stats, which is based on the expectation-maximization algorithm.
The results of the single-SNP analyses are presented in table 1. The minor alleles of three SNPs (P1578, P1763, and P1765) were positively associated with the diagnosis of schizophrenia or schizoaffective disorder in the white subset. The most significant P value was observed for P1578 (OR 1.76; allele test P=.0026; trend test P=.0037). All three SNPs remained significant when the analysis was restricted to patients with schizophrenia (n=196), and a fourth SNP, P1320, approached significance (OR 1.34; allele test P=.0649; trend test P=.0676). For the four associated SNPs, the observed associations occurred with the minor allele. Therefore, on the single-SNP level, our results agree with those obtained by Straub et al. (2002) and van den Oord et al. (2003), with the exception of P1578 (which did not reach significance in their analyses). Despite the small sample size, the same SNPs were associated in the Hispanic subgroup, but with higher estimated ORs. For example, P1578 was positively associated with the diagnosis of schizophrenia or schizoaffective disorder in the Hispanic subset (OR 3.36; allele test P=.0506; trend test P=.0449). When restricted to patients with schizophrenia, only P1763 and P1765 remained significant in the Hispanic cohort, which is likely due to the small sample size, as the estimated ORs suggest. When the data from the white and Hispanic subsets were analyzed jointly with a stratified Cochran-Armitage trend test, the results were more pronounced (see table 2). It is interesting that no association of the above SNPs was observed in the African American sample (215 cases, 74 controls), and the estimated ORs did not indicate any trend of association.
Table 2.
Combined Analysis of Whites and Hispanics with Schizophrenia or Schizoaffective Disorder[Note]
| SNP | Alleles(Major/Minor) | Test Statistic | P |
| P1583 | C/T | −.0814 | .9352 |
| P1578 | C/T | 3.4141 |
.0006 |
| P1763 | A/C | −2.9569 |
.0031 |
| P1320 | C/T | −2.1394 |
.0324 |
| P1765 | G/A | −3.0123 |
.0026 |
| P1325 | C/T | .9298 | .3525 |
Note.— Significant P values are shown in bold, underlined italics.
Although recent case-control studies have reported DTNBP1 associations with schizophrenia (Kirov et al. 2004; Williams et al. 2004), a previous study raised the possibility that DTNBP1 variation may be particularly involved in familial cases of schizophrenia (Van Den Bogaert et al. 2003). When we restricted our analysis to those cases with a positive family history (whites: n=58), none of the SNPs remained significant (data not shown); however, this should be interpreted with caution, since the sample size is very small.
LD between the selected SNPs we tested was assessed by use of Lewontin’s D′ statistic. We observed high LD from P1583 to P1765 in all ethnic groups (see table 3). This 11-kb interval generally showed very high LD in previous studies. In the white group, the marker pair P1325-P1320 showed the lowest D′ value, as was the case in the samples used in the studies by Straub et al. (2002) and Schwab et al. (2003). In the Hispanic group, the lowest D′ values were observed for marker pairs involving P1578 (P1325-P1578 and P1320-P1578). Similarly, van den Oord et al. (2003) found that marker pairs involving P1578 showed considerably lower LD than other marker pairs.
Table 3.
LD (D′) at the DTNBP1 Locus
|
LD (D′) for SNPs |
|||||
| Subsetand SNP | P1583 | P1578 | P1763 | P1320 | P1765 |
| White: | |||||
| P1578 | .93 | … | … | … | … |
| P1763 | .96 | 1.00 | … | … | … |
| P1320 | .87 | .96 | .97 | … | … |
| P1765 | .94 | 1.00 | 1.00 | .97 | … |
| P1325 | .97 | .78 | 1.00 | .56 | .94 |
| Hispanic: | |||||
| P1578 | .84 | … | … | … | … |
| P1763 | .94 | .99 | … | … | … |
| P1320 | 1.00 | .81 | 1.00 | … | … |
| P1765 | 1.00 | .99 | 1.00 | 1.00 | … |
| P1325 | 1.00 | .23 | 1.00 | 1.00 | 1.00 |
| African American: | |||||
| P1578 | 1.00 | … | … | … | … |
| P1763 | 1.00 | 1.00 | … | … | … |
| P1320 | 1.00 | .85 | .99 | … | … |
| P1765 | 1.00 | .99 | 1.00 | .97 | … |
| P1325 | .91 | 1.00 | 1.00 | 1.00 | .96 |
In whites, Hispanics, and African Americans, five haplotypes had estimated frequencies >0.025 and were included in the association testing. Together, these haplotypes accounted for 94%–98% of all haplotypes (table 4). One haplotype consisted of the rare (minor) allele for SNPs P1578, P1763, P1320, and P1765, and the common (major) allele for P1325 and P1583 was overrepresented in the white and Hispanic groups. However, significance was reached only in the white population (haplotype-specific P=.005; maximum haplotype-specific P=.024). The overrepresentation of the C-T-C-T-A-C haplotype in whites and Hispanics is consistent with the associations of the rare alleles for markers P1578, P1763, P1320, and P1765 on the single-SNP level and with the high LD we observed in this interval. In the African American group, this haplotype was present at almost equal frequencies in cases and controls, consistent with the negative associations on the single-SNP level. It is worth noting that P1578, which reached the most significant P values in the single-SNP analyses, is also the only SNP with the associated (minor) allele observed solely on the overrepresented haplotype. Our markers included two sets of haplotype-tag SNPs described by van den Oord et al. (2003) (P1578-P1763 and P1578-P1765). However, our risk haplotype differed from the one reported by this group, suggesting that these tag SNPs may not be generally applicable across different populations. Van den Oord et al. (2003) defined a haplotype block (P1792–P1635), in which a limited number of haplotypes accounted for the majority of all observed haplotypes. Since our data show high LD extending from P1765 to P1583 (the 5′-most marker we genotyped), we could not examine this feature in our sample.
Table 4.
DTNBP1 Haplotypes[Note]
|
Haplotypes |
Frequency in |
||||||||
| Subset | P1583 (C/T)a | P1578 (C/T)a | P1763 (A/C)a | P1320 (C/T)a | P1765 (G/A)a | P1325 (C/T)a | Casesb,c | Controlsd | Haplotype-Specific P |
| White: | |||||||||
| C | C | A | C | G | C | .4505 | .4999 | .0793 | |
| T | C | A | C | G | T | .0578 | .0648 | .6027 | |
| T | C | A | C | G | C | .2817 | .2662 | .5587 | |
| C | C | C | T | A | C | .0816 | .0818 | .9468 | |
| C | T | C | T | A | C | .1174 | .0714 |
.005 |
|
| Hispanic: | |||||||||
| C | C | A | C | G | C | .3482 | .4946 | .0959 | |
| T | C | A | C | G | T | .0787 | .1167 | .4188 | |
| T | C | A | C | G | C | .2496 | .2217 | .7146 | |
| C | C | C | T | A | C | .1471 | .0750 | .2095 | |
| C | T | C | T | A | C | .1471 | .0750 | .1602 | |
| African American: | |||||||||
| C | C | A | C | G | C | .3884 | .4444 | .5444 | |
| T | C | A | C | G | T | .0513 | .0158 | .8075 | |
| T | C | A | C | G | C | .1788 | .1699 | .7573 | |
| C | C | C | T | A | C | .0810 | .0617 | .4308 | |
| C | T | C | T | A | C | .2630 | .2525 | .9304 | |
Note.— The significant P value is shown in bold, underlined italics.
Major allele/minor allele.
Individuals with a diagnosis of schizophrenia or schizoaffective disorder.
Total frequency in cases: .9891 (whites), .9706 (Hispanics), .9624 (African Americans).
Total frequency in controls: .9841 (whites), .9830 (Hispanics), .9443 (African Americans).
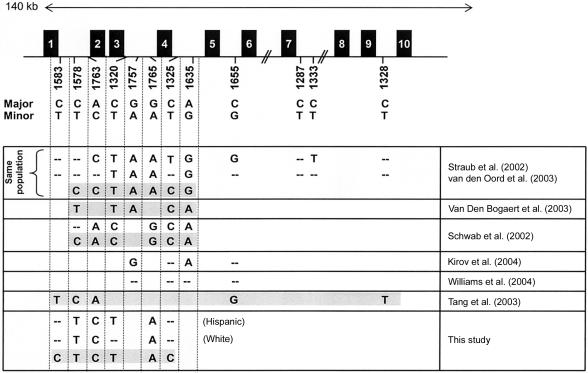
Figure 1 summarizes our findings in relation to previously reported associations for the DTNBP1 locus. Comparisons were restricted to the initial SNP set described by Straub et al. (2002). A growing number of studies, including ours, have now reported association of several DTNPB1 SNPs with schizophrenia. However, a precise comparison is hampered by the fact that the SNP sets overlapped but were not identical. Overall, there is considerable disagreement both within studies (differences between single-SNP associations and alleles present on associated haplotypes) and between studies. As described above, the samples in our study showed significant association with the minor alleles (T-C-T-A) for P1578, P1763, P1320, and P1765 in two different ethnic groups, and a single risk haplotype, which includes the same rare alleles plus the common alleles for P1325 and P1583, emerged in the white cohort. Five of these SNPs are present on the risk haplotype identified by van den Oord et al. (2003), and, for four of them, our risk haplotype contains the same alleles as the haplotype described by van den Oord et al. (2003).
Figure 1.
Comparison of DTNBP1 associations. The DTNBP1 gene is shown in the upper part of the figure (GenBank accession number NM_183041; variant 3). Exons are represented by boxes, and the SNPs described by Straub et al. (2002) are indicated below. Associated alleles and haplotypes, as reported elsewhere, are summarized in the lower part of the figure. Significant associations are indicated by the respective alleles. Two hyphens (“--”) represent a tested but nonsignificant marker. Single risk haplotypes are shaded. Complex haplotype patterns (such as those described by Straub et al. [2002] and Kirov et al. [2004]) or haplotypes including a significant number of novel SNPs (Williams et al. 2004) were omitted but are discussed in the text.
The only exception is P1578, for which our haplotype contained the rare allele (which we also found to be significantly associated on the single-SNP level but which did not show significant P values in the study by van den Oord et al. [2003]). Of interest, van den Oord et al. (2003) saw considerably lower LD with marker pairs involving P1578 than with most other pairs. It is conceivable that the haplotype we detected in our population is evolutionarily related to the haplotype described by van den Oord et al. (2003), as seems to be true for the risk haplotype identified in a Swedish population by Van Den Bogaert et al. (2003). In their sample, three of the five alleles present on the haplotype are identical to the ones on the haplotype identified by van den Oord et al. (2003). Again, P1578 is different, and, like our haplotype, it contains the rare allele (T). Schwab et al. (2003) observed positive associations for five of the SNPs described by Straub et al. (2002), however, with the common allele for each marker. Consistent with these results, one haplotype, consisting of the common allele for each SNP, was overtransmitted in their samples. Finally, the two most recent studies included a substantial number of novel SNPs, in addition to several of the SNPs described by Straub et al. (2002): Kirov et al. (2004) observed positive associations with the common allele for two of the SNPs (P1635 and P1757) described by Straub et al. (2002), but they were unable to assign a single risk haplotype, because of the large number of significant two-, three-, and four-marker haplotypes. Williams et al. (2004) genotyped four of the SNPs described by Straub et al. (2002) but found no association on the single-SNP or haplotype level. A highly significant haplotype was observed only after adding novel SNPs (Williams et al. 2004).
In summary, there is now little doubt that the DTNBP1 gene plays a significant role in the genetic etiology of schizophrenia, since eight studies (including ours) have reported significant associations with the same markers, all located in the 5′ portion of the gene. To our knowledge, this is the first study examining DTNBP1 variants in different U.S. populations. It is also the first study analyzing DTNBP1 variants in an African American population. We observed positive associations of DTNBP1 variants in both the white and Hispanic groups. No significant associations were observed in the African American population, which is most likely due to an insufficient sample size, as suggested by the ORs (table 1). It is possible that DTNBP1 may not contribute to schizophrenia in this ethnic group or that its contribution may be significantly smaller than in other populations.
We are aware of the potential danger of spurious results caused by hidden population substructure. Although this is a possible limitation of our study, empirical data obtained from European American, African American, and European populations suggest that carefully matched studies of moderate size are unlikely to contain stratification levels leading to false-positive associations (Ardlie et al. 2002; Pankow et al. 2002). A second possible limitation of our study is that, overall, our sample showed a sex imbalance. However, there is currently no evidence suggesting a sexually dimorphic effect of the DTNBP1 gene on the development of schizophrenia.
We observed an overrepresentation of the same haplotype in two ethnic groups, suggesting that the disease-causing variant may be identical in the Hispanic and white populations in our study. However, a comparison of the reported studies so far does not support a single disease-associated variant at the DTNBP1 locus, since association has been found with different haplotypes. It is therefore possible that the contribution of DTNBP1 to schizophrenia is defined by several susceptibility alleles that have arisen on different haplotypes.
A review of the disease-causing variants identified to date shows that most represent missense or nonsense mutations that affect the coding region (Botstein and Risch 2003). However, no DTNBP1 coding variants have yet been identified, and it is therefore possible that the functional variant(s) are not located within the protein-coding portion of the gene. A recent study has reported allelic differences in DTNBP1 expression in the brain, suggesting that regulatory variant(s) influence DTNBP1 expression levels (Bray et al. 2003). In this context, it is important to note that Williams et al. (2004) observed the strongest evidence of association for a haplotype that contained a putative regulatory SNP. A thorough analysis of the complete variation of the DTNBP1 locus, including all regulatory elements across multiple populations, will be needed to dissect the contribution of the functional variant(s) of the DTNBP1 gene to schizophrenia.
Acknowledgments
This project was supported by National Institutes of Health grants HD034980-09 (to R.K.) and K23MH001760 (to A.K.M.). We thank Dr. Marco Ramoni, for critical reading of the manuscript, and Dr. Bernice Morrow, for providing services to generate lymphoblastoid cell lines and DNA.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/index.html (for the SNPs listed in )
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for DTNBP1 [accession number NM_183041])
- Haplo Stats, http://www.mayo.edu/hsr/people/schaid.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for schizophrenia)
References
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES (2000) The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26:76–80 [DOI] [PubMed] [Google Scholar]
- Anderson JL, Head SI, Rae C, Morley JW (2002) Brain function in Duchenne muscular dystrophy. Brain 125:4–13 [DOI] [PubMed] [Google Scholar]
- Ardlie KG, Lunetta KL, Seielstad M (2002) Testing for population subdivision and association in four case-control studies. Am J Hum Genet 71:304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila MT, Hong E, Thaker GK (2002) Current progress in schizophrenia research. Eye movement abnormalities in schizophrenia: what is the nature of the deficit? J Nerv Ment Dis 190:479–480 [DOI] [PubMed] [Google Scholar]
- Benson MA, Newey SE, Martin-Rendon E, Hawkes R, Blake DJ (2001) Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J Biol Chem 276:24232–24241 [DOI] [PubMed] [Google Scholar]
- Botstein D, Risch N (2003) Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet Suppl 33:228–237 [DOI] [PubMed] [Google Scholar]
- Bray NJ, Buckland PR, Owen MJ, O’Donovan MC (2003) Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet 113:149–153 [DOI] [PubMed] [Google Scholar]
- Cardno AG, Gottesman II (2000) Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet 97:12–17 [PubMed] [Google Scholar]
- Collier DA, Li T (2003) The genetics of schizophrenia: glutamate not dopamine? Eur J Pharmacol 480:177–184 [DOI] [PubMed] [Google Scholar]
- Hill WG (1974) Estimation of linkage disequilibrium in randomly mating populations. Heredity 33:229–239 [DOI] [PubMed] [Google Scholar]
- Hinton VJ, De Vivo DC, Nereo NE, Goldstein E, Stern Y (2000) Poor verbal working memory across intellectual level in boys with Duchenne dystrophy. Neurology 54:2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Ivanov D, Williams NM, Preece A, Nikolov I, Milev R, Koleva S, Dimitrova A, Toncheva D, O'Donovan MC, Owen MJ (2004) Strong evidence for association between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia in 488 parent-offspring trios from Bulgaria. Biol Psychiatry 55:971–975 [DOI] [PubMed] [Google Scholar]
- Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, Schaid DJ (2003) Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Heredity 55:56–65 [DOI] [PubMed] [Google Scholar]
- Levinson DF, Mowry BJ (2000) Genetics of schizophrenia. In: Pfaff DW, Berrettini WH, Joh TH, Maxson SC (eds) Genetic influences on neural and behavioral functions. CRC Press, New York, pp 47–82 [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, et al (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet 73:34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui F, Fonda S, Merlini L, Corazza R (2001) Saccadic eye movements are impaired in Duchenne muscular dystrophy. Doc Ophthalmol 103:219–228 [DOI] [PubMed] [Google Scholar]
- McGuffin P, Owen MJ, Farmer AE (1995) Genetic basis of schizophrenia. Lancet 346:678–682 [DOI] [PubMed] [Google Scholar]
- Mehler MF (2000) Brain dystrophin, neurogenetics and mental retardation. Brain Res Brain Res Rev 32:277–307 [DOI] [PubMed] [Google Scholar]
- Morris DW, McGhee KA, Schwaiger S, Scully P, Quinn J, Meagher D, Waddington JL, Gill M, Corvin AP (2003) No evidence for association of the dysbindin gene (DTNBP1) with schizophrenia in an Irish population-based study. Schizophr Res 60:167–172 [DOI] [PubMed] [Google Scholar]
- Muntoni F, Torelli S, Ferlini A (2003) Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol 2:731–740 [DOI] [PubMed] [Google Scholar]
- North KN, Miller G, Iannaccone ST, Clemens PR, Chad DA, Bella I, Smith TW, Beggs AH, Specht LA (1996) Cognitive dysfunction as the major presenting feature of Becker’s muscular dystrophy. Neurology 46:461–465 [DOI] [PubMed] [Google Scholar]
- Pankow JS, Province MA, Hunt SC, Arnett DK (2002) Regarding “Testing for population subdivision and association in four case-control studies.” Am J Hum Genet 71:1478–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet 46:222–228 [PMC free article] [PubMed] [Google Scholar]
- Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 [DOI] [PubMed] [Google Scholar]
- Sasieni PD (1997) From genotypes to genes: doubling the sample size. Biometrics 53:1253–1261 [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA (2002) Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab SG, Knapp M, Mondabon S, Hallmayer J, Borrmann-Hassenbach M, Albus M, Lerer B, Rietschel M, Trixler M, Maier W, Wildenauer DB (2003) Support for association of schizophrenia with genetic variation in the 6p22.3 gene, dysbindin, in sib-pair families with linkage and in an additional sample of triad families. Am J Hum Genet 72:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O’Neill FA, Walsh D, Kendler KS (2002) Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet 71:337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub V, Campbell KP (1997) Muscular dystrophies and the dystrophin-glycoprotein complex. Curr Opin Neurol 10:168–175 [DOI] [PubMed] [Google Scholar]
- Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, Hahn CG, Siegel SJ, Trojanowski JQ, Gur RE, Blake DJ, Arnold SE (2004) Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest 113:1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JX, Zhou J, Fan JB, Li XW, Shi YY, Gu NF, Feng GY, Xing YL, Shi JG, He L (2003) Family-based association study of DTNBP1 in 6p22.3 and schizophrenia. Mol Psychiatry 8:717–718 [DOI] [PubMed] [Google Scholar]
- Van Den Bogaert A, Schumacher J, Schulze TG, Otte AC, Ohlraun S, Kovalenko S, Becker T, Freudenberg J, Jonsson EG, Mattila-Evenden M, Sedvall GC, Czerski PM, Kapelski P, Hauser J, Maier W, Rietschel M, Propping P, Nothen MM, Cichon S (2003) The DTNBP1 (dysbindin) gene contributes to schizophrenia, depending on family history of the disease. Am J Hum Genet 73:1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Oord EJ, Sullivan PF, Jiang Y, Walsh D, O’Neill FA, Kendler KS, Riley BP (2003) Identification of a high-risk haplotype for the dystrobrevin binding protein 1 (DTNBP1) gene in the Irish study of high-density schizophrenia families. Mol Psychiatry 8:499–510 [DOI] [PubMed] [Google Scholar]
- Williams NM, Preece A, Morris DW, Spurlock G, Bray NJ, Stephens M, Norton N, Williams H, Clement M, Dwyer S, Curran C, Wilkinson J, Moskvina V, Waddington JL, Gill M, Corvin AP, Zammit S, Kirov G, Owen MJ, O’Donovan MC (2004) Identification in 2 independent samples of a novel schizophrenia risk haplotype of the dystrobrevin binding protein gene (DTNBP1). Arch Gen Psychiatry 61:336–344 [DOI] [PubMed] [Google Scholar]