Abstract
Several quantitative trait loci (QTLs) that influence developmental dyslexia (reading disability [RD]) have been mapped to chromosome regions by linkage analysis. The most consistently replicated area of linkage is on chromosome 6p23-21.3. We used association analysis in 223 siblings from the United Kingdom to identify an underlying QTL on 6p22.2. Our association study implicates a 77-kb region spanning the gene TTRAP and the first four exons of the neighboring uncharacterized gene KIAA0319. The region of association is also directly upstream of a third gene, THEM2. We found evidence of these associations in a second sample of siblings from the United Kingdom, as well as in an independent sample of twin-based sibships from Colorado. One main RD risk haplotype that has a frequency of ∼12% was found in both the U.K. and U.S. samples. The haplotype is not distinguished by any protein-coding polymorphisms, and, therefore, the functional variation may relate to gene expression. The QTL influences a broad range of reading-related cognitive abilities but has no significant impact on general cognitive performance in these samples. In addition, the QTL effect may be largely limited to the severe range of reading disability.
Introduction
Developmental dyslexia (reading disability [RD] [MIM 600202]) is a specific difficulty learning to read, in the absence of obvious causes, such as low general intelligence or lack of educational opportunity (Smith et al. 1996). The prevalence is estimated at ∼5% in school-aged children (Smith et al. 1996). Dyslexia is highly familial and heritable, but the genetic background is complex and heterogeneous (DeFries et al. 1987; Fisher and DeFries 2002; Francks et al. 2002). Linkage studies have identified several genomic regions that may contain QTLs for dyslexia susceptibility, including regions on chromosomes 2, 3, 6, 15, and 18 (Fisher and DeFries 2002; Fisher et al. 2002). A candidate gene for dyslexia susceptibility (DYX1C1), located on 15q21, was recently reported to be disrupted by a balanced translocation in subjects with RD from one family (Taipale et al. 2003). Suggestive evidence was also reported of associations, at a more general population level, between RD and putative functional SNPs within this gene (Taipale et al. 2003). However, our analysis of DYX1C1 in 264 families with RD from the United Kingdom (Scerri et al., in press) and in 148 families from Canada (Wigg et al., in press) casts doubt on the functional significance of these SNPs, since, in these samples, RD appears to be associated with SNP alleles opposite to those implicated by Taipale et al. (2003). Therefore, the DYX1C1 associations await convincing replication.
The most consistently replicated linkage to RD is on 6p23-21.3; this linkage has been found in five separate samples from the United Kingdom and the United States (Cardon et al. 1994, 1995; Grigorenko et al. 1997; Fisher et al. 1999, 2002; Gayan et al. 1999; Grigorenko et al. 2000; Fisher and DeFries 2002). The 6p23-21.3 region showed one of the two strongest linkage signals in our previous genomewide screen of one of these samples (223 siblings in 89 unrelated nuclear families from the United Kingdom, each with at least one proband with RD) (Fisher et al. 2002; Marlow et al. 2003). However, linkage and association studies have failed, so far, to refine the candidate interval on 6p to smaller than ∼16 Mb (Fisher and DeFries 2002; Kaplan et al. 2002; Turic et al. 2003; Deffenbacher et al. 2004).
Developmental dyslexia is often characterized by impairments in several conceptually distinct cognitive skills that are related to reading or language processing (Castles and Coltheart 1993; Gayan and Olson 2001). In our studies, we have assessed probands and their siblings for a range of reading and language-related measures, such as single-word reading ability, spelling ability, and measures of phonological and orthographic coding (sounding out unfamiliar words and recognizing the spelling of familiar words, respectively [see the “Material and Methods” section]). These measures are strongly intercorrelated in clinical and epidemiological samples (Castles and Coltheart 1993; Gayan and Olson 2003) (e.g., correlations 0.41–0.76 in 223 U.K. siblings [Marlow et al. 2001]). The measures also show significant but weaker correlations with verbal/nonverbal intelligence (correlations 0.14–0.40 in 223 U.K. siblings [Marlow et al. 2001]). Twin studies have shown that the different reading-related traits may be influenced by both overlapping and trait-specific genetic factors (Gayan and Olson 2003). Likewise, twin studies also indicate that variabilities in reading-related measures and general intelligence have both overlapping and independent genetic components (Gayan and Olson 2003).
We previously used multivariate linkage analysis to show that the QTL on 6p23-21.3 appeared to influence trait variability that was shared between all reading-related cognitive measures but that was not shared with measures of general intelligence (IQ) (Marlow et al. 2003). We therefore reasoned that shared variance between reading-related measures and IQ could be considered as noise in the specific context of further linkage and association analysis of the 6p23-21.3 QTL. In the present study, we refined our linkage mapping of the QTL to a 5.8-Mb interval by removing the variance in reading-related measures that is shared with IQ. We then selected brain-expressed genes within the new candidate interval for association analysis with SNPs. We detected significant association with IQ-adjusted RD measures in our sample of 223 U.K. siblings, within a 77-kb region of strong intermarker linkage disequilibrium (LD) that spans the gene for TTRAP (tumor necrosis factor [TNF], tumor receptor-associated factor [TRAF], and TNF receptor-associated protein), and the first four exons of the neighboring uncharacterized gene KIAA0319. We replicated this QTL effect in two large, independent samples of sibships collected in the United Kingdom and the United States; these samples were selected to include probands who were severely affected with RD. We found that one main risk haplotype, with a frequency of ∼12%, was associated with RD in both the U.K. and U.S. samples. This is the first identification of a QTL that has been shown, by repeated replication, to be of relevance to many individuals with RD.
Material and Methods
The U.K. Family Sample (Samples 1 and 2)
We identified 264 unrelated nuclear families at the dyslexia clinic at the Royal Berkshire Hospital in Reading, United Kingdom. The majority (>90%) of the first 192 families were recruited on the basis of having (1) at least one proband whose single-word reading ability was >2 SDs below that predicted by tests of verbal or nonverbal reasoning (Marlow et al. 2001; Fisher et al. 2002) (see below for specific tests used) and (2) evidence of RD in one or more siblings of the proband with dyslexia (e.g., on the basis of parental reports or school history) (Marlow et al. 2001; Fisher et al. 2002). Many of the probands in these families had above-average verbal/nonverbal reasoning and roughly normal reading-related abilities (Marlow et al. 2003). To increase the representation of probands with poor reading in the sample, the majority (∼80%) of the remaining 72 families were recruited through probands that were required to have single-word reading ability ⩾1 SD below that predicted for their age, with a minimum IQ of 90 (again, see below for the specific tests). These families were recruited through a minimum of just one proband with RD, with no requirement for RD in siblings of the probands. All of the remaining families in both recruitment phases were collected via referral to the clinic of at least one reading-disabled proband by a qualified clinician. In practice, all of the sibships that were collected under the second recruitment scheme included a proband that met criteria for the first scheme (and 51% of first-scheme sibships included a proband that met second-scheme criteria). A battery of reading-related and cognitive tests was administered to all U.K. probands and siblings, regardless of recruitment, for the purpose of quantitative genetic analysis (see below). Subjects were given a full description of the experimental procedures, plus the option to ask questions or to withdraw at any time. Written informed consent was then obtained.
Blood samples or buccal swabs were donated by all available children and parents for the purpose of genomic DNA extraction. The 264 families comprised a total of 630 siblings for whom psychometric test scores and genomic DNA were available. Sibship sizes used in our analyses ranged from 2 to 5. Sibling ages ranged from 5.6 years to 30.6 years (median 11.7 years). The sample consisted uniformly of families of European origin.
We refer to the first 89 U.K. families (all from the first recruitment scheme) as “sample 1”; these were used in our previous genomewide linkage screen (Fisher et al. 2002) and also in the linkage analysis and initial association analysis that we report in the present study. Of these families, 82 were originally analyzed by Fisher et al. (1999) in a targeted linkage study of chromosome 6p. The remaining 175 U.K. families (“sample 2,” a mixture of both recruitment schemes) were used in the present study to test for replication of the associations found in sample 1.
The Colorado Family Sample (Sample 3)
The Colorado sample (sample 3) comprised 159 families drawn from the Colorado twin study of RD (DeFries et al. 1987; Gayan and Olson 2003). We identified twin pairs from the records of 27 Colorado school districts and selected those in which at least one member had a positive school history of reading difficulty (Gayan and Olson 2003). Each proband and all siblings were then administered a battery of psychometric tests (see below) for the purpose of quantitative genetic analysis. The sample investigated in the present study also included nontwin siblings in a significant proportion of families, which yielded 369 total siblings. Sibling ages ranged from 8.0 years to 18.9 years (median 10.8 years). We did not use pairs of MZ twins in this study. Genomic DNA samples were extracted from all siblings and available parents, and some samples were preamplified using the GenomiPhi kit (Amersham). Of the 159 families analyzed, 109 were used for our previous genome screen (Fisher et al. 2002). All families were of European descent. We used sample 3 for a further test of replication of our association findings.
Phenotype Measures: U.K. Samples (Samples 1 and 2)
We administered a battery of psychometric tests to all probands and cosiblings in each family, and we age-adjusted and standardized their scores against a normative control data set, as described elsewhere (Marlow et al. 2001; Fisher et al. 2002). These included measures of single-word reading ability (READ), spelling ability (SPELL), phonological decoding ability (PD) (use of rule-like letter-sound relations to derive the pronunciation of pseudowords), phonemic awareness (PA) (awareness of the phonemic structure of language), orthographic coding (OC-irreg) (use of word-specific spelling patterns to recognize real irregular words), orthographic coding assessed by a forced word choice test (OC-choice), and tests of verbal (SIM) and nonverbal (MAT) reasoning. Each measure was based on the percentage accuracy for an individual psychometric test (Marlow et al. 2001). All of these measures are significantly intercorrelated and familial (Marlow et al. 2001; Fisher et al. 2002). We used the sum of SIM and MAT as a proxy measure of IQ (Thompson 1982), and we linearly regressed reading-related scores on this IQ measure, for the total combined U.K. sample (samples 1 and 2), to obtain IQ-adjusted Z scores (mean 0 [1 SD]).
Phenotype Measures: U.S. Sample (Sample 3)
A battery of psychometric tests was administered to all twins and cosiblings in the U.S. sample. These tests were aimed at tapping the same range of cognitive skills as in the U.K. sample, although the actual tests differed in all but one case (OC-choice was common to both samples). We therefore had conceptually equivalent measures for sample 3 as in the U.K. samples, for the measures READ, SPELL, PD, and PA, together with the same measure of OC-choice as in the U.K. samples. The tests have been described elsewhere (Fisher et al. 2002; Gayan and Olson 2003). Unlike the U.K. measures, some of the Colorado measures were derived as composites from more than one test and/or from accuracy combined with latency data (Gayan and Olson 2003). Briefly, READ was derived as a composite of accuracy on two word recognition tests (Gayan and Olson 2003). PD was a composite of accuracy and latency on one- and two-syllable nonword reading. PA was a composite of accuracy on a phoneme-deletion task and weighted percentage correct on a phoneme-segmentation and -transposition task. SPELL and OC-choice were based on accuracy for individual tests of spelling and orthographic choice tasks, respectively. The reading-related measures were age-adjusted and standardized against an appropriate normative twin-based population (Gayan and Olson 2003). All of these measures are significantly intercorrelated and familial (Fisher et al. 2002; Gayan and Olson 2003). A standardized measure of full-scale IQ (Gayan and Olson 2003) was used for linear IQ adjustment of reading-related measures, in the same way as for the U.K. samples.
Genotyping
We had microsatellite marker genotype data for the families in sample 1 and sample 3 that were included in our previous genomewide screens (Fisher et al. 2002). Nineteen additional polymorphic microsatellite markers were genotyped as part of the present study in sample 1, in accordance with standard protocols (Fisher et al. 2002). Between 6 and 16 microsatellite markers were also genotyped for families in sample 2. Microsatellite genotype data were used only for linkage analysis or to increase the accuracy of identity-by-descent–sharing estimates for the modeling of residual linkage during association analysis (see below). Because of their high number of alleles and higher mutability compared with SNPs, microsatellites were not tested for trait-marker association.
All SNP genotyping was performed using the Sequenom system, in accordance with the manufacturer’s instructions. PCR primers and primer-extension probes were designed with the SpectroDESIGNER software, with the exception of the probes for three insertion/deletion polymorphisms, which were designed manually. All primer and probe sequences are available on request. In total, 57 SNPs were genotyped in sample 1, of which 48 spanned a 225-kb interval that surrounded our initial positive-association findings (see the “Results” section), with an average intermarker distance of 5 kb. Twenty of these SNPs—those that showed association in sample 1 or that were included in the same assay multiplexes—were genotyped in samples 2 and 3. We identified and eliminated Mendelian errors in our genotype data and used MERLIN (Abecasis et al. 2002) to identify and eliminate genotypes that indicated unlikely recombination events. We also used MERLIN to infer missing genotypes from relatives in unambiguous situations. No SNPs deviated significantly from Hardy-Weinberg equilibrium (data not shown).
Linkage Analysis
Quantitative linkage analysis of IQ-adjusted reading-related measures was performed using the 223 siblings from the first 89 U.K. families (sample 1), using polymorphic microsatellite genotype data from our previous genomewide linkage screen (Fisher et al. 2002), supplemented with data from 19 new microsatellite markers, and using the deCODE genetic map (Kong et al. 2002). Linkage analysis was performed within the variance-components (VC) framework (see below), as implemented in the package MERLIN (Abecasis et al. 2002), with no dominance variance and with the estimation of a single-trait mean. VC linkage analysis within this sample has been validated elsewhere by simulation (Fisher et al. 2002).
Intermarker LD Analysis and Haplotypes
Pairwise intermarker LD statistics for SNPs were calculated using HAPLOXT from the GOLD package (Abecasis and Cookson 2000), by use of founder-haplotype estimates derived from MERLIN (Abecasis et al. 2002). The GoldSurfer package was used to evaluate simultaneously pairs of LD features with a three-dimensional graphical interface (Pettersson et al., in press). To gauge the haplotype diversity within this region, SNP-based haplotypes that spanned LD region B (see the “Results” section) were estimated using MERLIN. Three SNPs (rs4504469, rs2038137, and rs2143340) within LD region B were identified, which, together, distinguished all of the observed haplotypes spanning region B with frequencies >3% (data not shown). We next used genotype data for these three SNPs as input for the haplotype-estimating method of Rohde and Fürst (2001), which uses a combination of an expectation-maximization algorithm and nuclear-family data to assign the most likely haplotypes to each individual, regardless of sporadic missing data. The best-estimate haplotypes from this method were used in marker-trait association analysis. Marker rs2143340, which distinguishes the risk haplotype from all other common haplotypes, was also genotyped in a panel of 190 random human control individuals from the European Collection of Cell Cultures (ECACC).
Marker-Trait Association Analysis
Marker-trait association was assessed using the “total-association” option of the package QTDT (Abecasis et al. 2000). In this analysis, maximum-likelihood modeling is first used to fit a null VC model to quantitative-trait data in sibships in which variance is partitioned into unshared environmental, shared environmental/polygenic, and QTL-specific components, with a single-trait mean. Subsequently, a full-association model is fitted that includes the same VC as the null model but includes the mean effects of the SNP alleles to model association (Abecasis et al. 2000). Residual familial and linkage effects are therefore explicitly modeled within this framework and do not bias the association test. The likelihood-ratio test statistic is distributed asymptotically as a χ2 with 1 df. This “total-association” test can be biased by population stratification, so we verified our results by using the QTDT package to perform a less powerful within-sibship orthogonal association test that is robust to stratification (the “orthogonal” test) (Abecasis et al. 2000). The orthogonal test produced significant associations within the same genomic interval as the total-association test, despite having lower power than the total-association test (data not shown). We performed a genotype-permutation procedure within QTDT to verify the results from the orthogonal test and to correct for any biasing effects of trait nonnormality; the significance levels derived from these permutations were similar to the asymptotic predictions for this test (data not shown). This suggested that trait nonnormality and/or phenotypic selection had not substantially biased the orthogonal test. There is no comparable way to obtain empirical significance levels for the total-association test, but we reasoned that the relatively unbiased behavior of the orthogonal test indicated that the total test should also be robust in our samples, since both tests are similar in design (Abecasis et al. 2000). We present the total-association results without allowing for dominance effects; when we allowed for dominance, the results remained effectively unchanged (data not shown).
Selection for RD Severity
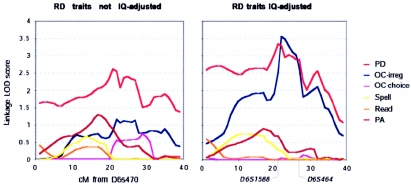
For both the U.K. and Colorado data sets, we aimed to increase our power to detect the QTL, in association analysis, by selecting only sibships that included individuals scoring below a threshold that was derived from those measures that showed the best evidence of linkage across 6p23-21.3 in each sample. Our multivariate linkage analysis had previously suggested that the 6p21.3-23 QTL influenced variability that was shared between all RD traits but that was not shared with IQ (Marlow et al. 2003). We therefore hypothesized that differences in the strengths of univariate linkages with the different RD traits were largely stochastic in nature and were influenced by trait- and sample-specific variations in sample sizes and patterns of missing genotype/phenotype data. Nonetheless, we reasoned that, within each sample, the relative strengths of univariate linkages could be used to indicate which specific phenotypes should be employed for severity selection, to maximize the power to detect association effects within any given sample. In the U.K. samples (samples 1 and 2), we used a threshold of −0.5 on a score calculated as the mean of individuals’ IQ-adjusted PD and OC-irreg (see linkage data for sample 1 in fig. 1; linkage in sample 2 was not significant [data not shown]). In the U.S. sample, we used a threshold of 0 on the mean of individuals’ IQ-adjusted READ and PD (these measures showed the strongest evidence of linkage in this sample [data not shown]). The threshold of 0 does not correspond to the unselected population mean, since the IQ adjustment was performed within the sample (see the “Material and Methods” section). The choice of a threshold value is necessarily arbitrary. Furthermore, the U.K. and Colorado samples could not be compared directly, since they were ascertained, tested, and standardized differently. In our choice of thresholds, we aimed to balance the increased potential power from phenotype selection against the decreased power from reduced sample size, as well as to have comparable sample sizes in the U.K. and Colorado selected samples (table 3). The phenotype selection yielded 126 families that included 313 siblings from the combined U.K. samples (samples 1 and 2) and 124 families that included 290 siblings from the U.S. sample (sample 3). The selected and total samples did not differ significantly for their IQs in either the U.K. or U.S. data sets. The P values that we present are not corrected for multiple hypothesis testing; a Bonferroni correction would be too conservative for multiple correlated traits and nonindependent genetic markers. Appropriate corrections for correlated phenotypes and genotypes are presently unknown.
Figure 1.
Linkage across 6p24.3-21.1 with reading-related measures in sample 1 (223 U.K. siblings, 195 total sibling pairs), before and after adjustment for IQ. IQ adjustment increased the strength of linkage and refined the QTL position.
Table 3.
Marker-Trait Association P Values in Two Independent Selected Samples[Note]
|
Selected Sibships from Samples 1 and 2, Combined (U.K.)(n = 313 Siblings [126 Families]) |
Selected Sibships from Sample 3 (U.S.)a(n = 290 Siblings [124 Families]) |
||||||||||||
|
P Value for Traitb |
P Value for Traitc |
||||||||||||
| Marker | LD Region | Risk Alleled | OC-irreg | OC-choice | PD | READ | SPELL | PA | Risk Alleled | PD | READ | SPELL | PA |
| rs699463 | A | Major | .0032 | .0231 | .0279 | .0153 | .0112 | ||||||
| rs4504469 | B | Major | .0011 | .0082 | .004 | .01 | |||||||
| rs2179515 | B | Major | .0012 | .0131 | .0004 | .0232 | |||||||
| rs761101 | B | Major | .0025 | .0057 | .0006 | .0325 | |||||||
| rs6456624 | B | Major | .0005 | .0045 | .0003 | .0157 | |||||||
| rs2328846 | B | Major | .0007 | .0017 | .0003 | .0155 | |||||||
| rs2235676 | B | Minor | .0023 | .0009 | .0041 | Minor | .0131 | ||||||
| rs2038137 | B | Major | .0013 | .0026 | .0002 | .0061 | |||||||
| k_pr_dele | B | Major | .0011 | .0032 | .0002 | .0086 | |||||||
| rs9467247 | B | Minor | .0006 | .0003 | .0373 | .0003 | .0016 | Minor | .0038 | .0421 | |||
| rs1555090 | B | Major | .001 | .0029 | .0003 | .0131 | |||||||
| rs3033236 | B | Minor | .0134 | .0104 | .0073 | Minor | .0421 | .0023 | .0145 | .0204 | |||
| rs2143340 | B | Minor | .01 | .0003 | .0115 | Minor | .0050 | .0273 | .0124 | ||||
| rs1061925 | B | Minor | .0009 | .0005 | .0008 | Minor | .0102 | .0340 | |||||
| tt_th_dele | B | Minor | .0181 | ||||||||||
| rs926529 | C | Major | .0132 | ||||||||||
| rs1885211 | C | ||||||||||||
| th_ex_3e | C | ||||||||||||
| rs3756814 | C | Minor | .0332 | ||||||||||
| rs6456632 | C | Major | .0415 | ||||||||||
Note.— All the reading-related measures were IQ-adjusted.
Single-tailed P values for testing the same marker allele as in the U.K. samples.
IQ showed no significant association in this analysis.
OC-choice and IQ showed no significant association in this analysis.
Only alleles that showed nominally significant marker-trait association (P<.05) are shown.
Polymorphisms identified by mutation screening analysis, which were not present in the public databases.
Polymorphism Screening
All of the known coding exons and predicted promoters of TTRAP, KIAA0319, THEM2, and ALDH5A1, as well as up to 289 bp of flanking sequence around each exon or promoter, were PCR amplified in 32 individuals with dyslexia from sample 1, who had the lowest mean scores derived from the IQ-adjusted measures PD and OC-irreg. The PCR products were screened using the WAVE DNA Fragment Analysis System (Transgenomic), which performs denaturing high-performance liquid chromatography (DHPLC) with hybridized amplimer duplexes across a range of temperatures. Any samples that showed heteroduplex formation were then resequenced. Fluorescence-based dideoxy sequencing was performed using BigDye Terminator sequencing kits (Applied Biosystems), followed by ABI 3700 capillary electrophoresis (Applied Biosystems). Sequencing primer details are available on request.
Results
Refinement of Linked Genomic Interval
Quantitative sib-pair linkage analysis, controlling for IQ as a linear covariate, provided increased evidence of linkage in our 223 U.K. siblings (sample 1), compared with analyses without IQ adjustment (fig. 1) (PD LOD 2.62 before IQ adjustment, LOD 3.34 after IQ adjustment; OC-irreg LOD 1.16 before IQ adjustment, LOD 3.48 after IQ adjustment). The adjustment for IQ focused our association mapping on the now clearly defined 1-LOD-unit interval obtained from linkage analysis of IQ-adjusted orthographic coding ability (OC-irreg [see the “Material and Methods” section]); this interval spanned 5.8 Mb from marker D6S1588 to marker D6S464 (fig. 1). IQ data were not available from 38 siblings in sample 1, and part of the striking increase in LOD score with IQ-adjusted OC-irreg, compared with analysis of the unadjusted trait, arises from the loss of these individuals. However, even compared with analysis of unadjusted traits in the 186 siblings with IQ data (not shown), the IQ adjustment resulted in an increase of 1.24 LOD units for OC-irreg and 0.77 LOD units for PD.
Association and LD Analysis
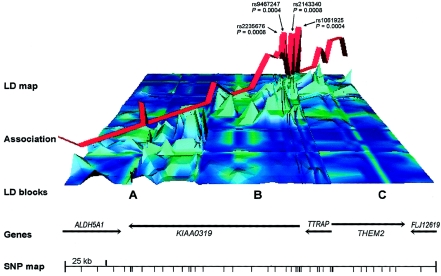
There were 80 known genes within the 5.8-Mb interval between D6S1588 and D6S464 in June 2002 database releases. Of these known genes, 40 encode histone proteins, which are not obvious functional candidates for a specific cognitive disorder. Of the remaining 40 genes, 8 were known to be expressed in brain tissue and were selected for association studies (distal to proximal: ALDH5A1, KIAA0319, TTRAP, THEM2, C6orf32, SCGN, BTN3A1, and BTN2A1 [LocusLink IDs: 7915, 9856, 51567, 55856, 9750, 10590, 11119, and 11120, respectively]). Summaries of the known functions of most of these genes are also available in the articles by Londin et al. (2003) and Deffenbacher et al. (2004). These eight genes fall within four distinct genomic clusters in the 5.8-Mb interval (University of California–Santa Cruz Genome Bioinformatics Web site); this facilitated association mapping, which relies on LD between markers that are in close proximity to one another (cluster 1: ALDH5A1, KIAA0319, TTRAP, and THEM2; cluster 2: C6orf32; cluster 3: SCGN; and cluster 4: BTN3A1 and BTN2A1).
After the analysis of 15 SNPs in sample 1 within these gene clusters, we obtained significant evidence of association of SNP rs1061925 with several IQ-adjusted measures (P=.0004 for OC-irreg; P=.0114 for READ; P=.0269 for OC-choice) (table A1 [online only]). We then analyzed 42 additional SNPs in sample 1 (including 4 biallelic insertion/deletions), within a 225-kb region surrounding rs1061925 (fig. 2 and table A1 [online only]). SNP associations with several reading-related measures were significant within a 77-kb region of strong intermarker LD (region B [fig. 2; online-only tables A1 and A3]) that spans TTRAP and the first four exons of the neighboring gene KIAA0319 (fig. 2). Our linkage analysis suggested that the QTL effect in sample 1 was most apparent in those sibships that included individuals who were severely impaired on IQ-adjusted OC-irreg and IQ-adjusted PD (fig. 1). Accordingly, we found that the significant marker-trait associations within LD region B (fig. 2) were derived predominantly from a subset of sibships that included at least one proband who scored <−0.5 SD on a mean score calculated from IQ-adjusted OC-irreg and IQ-adjusted PD (table 1).
Figure 2.
Representation of the 225-kb region surrounding the QTL in sample 1. The LD map shows a three-dimensional color-coded plot for pairwise intermarker LD statistics (48 biallelic markers). The colors represent D′ values: green indicates high LD, and blue indicates low LD. The height of the peaks represents the −log10 significance of pairwise LD; high peaks indicate significant LD. Three major LD regions are distinguishable: A, B, and C. The red ribbon represents the significance of SNP associations (−ln[P value]) with the IQ-adjusted orthographic coding (OC-irreg) phenotype (see also table A1 [online only]). The SNPs that showed the strongest associations (and the corresponding P values) are indicated by arrows. The genes within the region and the locations of the SNPs are shown at the bottom.
Table 1.
Marker-Trait Association P Values in Selected Siblings from Sample 1 (112 Siblings, 42 Families)[Note]
|
P Value for Trait |
|||||||||
| Marker | LD Region | Minor AlleleFrequencya (%) | Risk Alleleb | OC-irreg | OC-choice | PD | READ | SPELL | PA |
| rs2817220 | A | .23 | Major | .0326 | .0067 | .0004 | |||
| rs22252525 | A | .13 | |||||||
| rs807517 | A | .24 | Major | .0223 | .0017 | ||||
| rs1054899 | A | .34 | |||||||
| rs2760184 | A | .04 | |||||||
| rs807527 | A | .10 | |||||||
| rs699463 | A | .24 | Major | .0068 | |||||
| rs807530 | A | .41 | |||||||
| rs807535 | A | .10 | |||||||
| rs807540 | A | .08 | Major | .0203 | .0118 | .0085 | |||
| rs807545 | A | .06 | Major | .0244 | |||||
| rs807521 | A | .06 | Major | .0306 | |||||
| rs807525 | A | .06 | Major | .0308 | |||||
| rs807507 | A | .43 | |||||||
| rs807508 | A | .06 | Major | .0304 | |||||
| k_ex5 | A | .30 | Major | .044 | |||||
| rs4504469 | B | .47 | Major | .0003 | .0231 | .0005 | |||
| rs4576240 | B | .05 | |||||||
| rs4472344 | B | .03 | |||||||
| rs4236032 | B | .05 | |||||||
| rs2745333 | B | .45 | Major | .0002 | .0178 | .0001 | |||
| rs2179515 | B | .40 | Major | .0002 | .0021 | ||||
| rs761101 | B | .39 | Major | .0002 | .0255 | .0003 | |||
| rs6456624 | B | .44 | Major | .00006 | .036 | .0231 | .0007 | ||
| rs2328846 | B | .41 | Major | .00003 | .0001 | .0377 | |||
| rs2235676 | B | .14 | Minor | .0008 | .0442 | .0029 | |||
| rs2038137 | B | .43 | Major | .00006 | .0002 | .0285 | |||
| k_pr_del | B | .41 | Major | .00002 | .0001 | .0377 | |||
| rs9467247 | B | .19 | Minor | .0002 | .0167 | .0005 | |||
| rs1555090 | B | .40 | Major | .00003 | .0001 | .0377 | |||
| rs2294689 | B | .05 | |||||||
| rs3033236 | B | .17 | Minor | .0066 | .0288 | .0046 | |||
| rs3212232 | B | .26 | |||||||
| rs2143340 | B | .17 | Minor | .0012 | .0128 | .0011 | |||
| rs2056998 | B | .05 | |||||||
| rs1061925 | B | .11 | Minor | .00006 | .0213 | .0003 | |||
| th_ex1 | B | .11 | Minor | .0004 | .0214 | .0011 | |||
| tt_th_del | B | .06 | |||||||
| rs3181228 | B | .15 | |||||||
| rs2328847 | C | .01 | |||||||
| rs1555088 | C | .34 | Major | .0178 | |||||
| rs926529 | C | .36 | Major | .0177 | |||||
| rs1885211 | C | .46 | Major | .009 | |||||
| rs2092404 | C | .34 | Major | .0137 | .0405 | ||||
| th_ex3 | C | .04 | |||||||
| rs3756814 | C | .43 | Minor | .0178 | |||||
| rs6456632 | C | .15 | |||||||
| rs2067573 | C | .05 | |||||||
Note.— The selection included all the sibships with at least one sibling scoring <−0.5 SD on an average score of IQ-adjusted OC-irreg and PD. All reading-related measures are IQ-adjusted.
Minor allele frequency calculated in the founders only.
Only alleles with nominally significant marker-trait association (P<.05) are shown.
We then analyzed 20 of the polymorphisms in a second, independent set of 407 siblings from 175 unrelated U.K. nuclear families (sample 2). Each sibship again included at least one proband with RD. The evidence of marker-trait association within sample 2 was weak, and a combined analysis with sample 1 produced attenuated evidence of association (data not shown). However, we then analyzed the subset of sample 2 (201 siblings) that included at least one sib who met the −0.5 SD criterion for the mean of IQ-adjusted PD and OC-irreg, and again the evidence of association within LD region B was significant for many SNPs and for a range of IQ-adjusted RD measures (table 2). This replicated our initial findings in sample 1, and it confirmed that the effect is strongest in the lower tail of reading-related cognitive ability. Combined analysis of the subsets from sample 1 and sample 2 that met the −0.5 SD criterion (313 siblings in 126 families) yielded highly significant evidence of associations within LD region B with most reading-related measures (table 3) (e.g., rs9467247 and OC-irreg, P=.0006; rs9467247 and READ, P=.0003; rs1061925 and OC-choice, P=.0005; and rs1061925 and READ, P=.0008).
Table 2.
Marker-Trait Association P Values in Selected Siblings from Sample 2 (201 Siblings, 84 Families)[Note]
|
P Value for Traita |
|||||
| Marker | LD Region | OC-choice | PD | READ | SPELL |
| rs699463 | A | ||||
| rs4504469 | B | .0256 | .019 | ||
| rs2179515 | B | .0433 | .0283 | .0431 | |
| rs761101 | B | .0418 | |||
| rs6456624 | B | ||||
| rs2328846 | B | .0159 | .0464 | .0467 | |
| rs2235676 | B | .0037 | |||
| rs2038137 | B | .0128 | .021 | .0125 | |
| k_pr_del | B | .0269 | .0377 | .0207 | |
| rs9467247 | B | 0.0075 | .0396 | ||
| rs1555090 | B | .0248 | .0396 | .0277 | |
| rs3033236 | B | ||||
| rs2143340 | B | .0053 | |||
| rs1061925 | B | .0021 | |||
| tt_th_del | B | ||||
| rs926529 | C | .0326 | |||
| rs1885211 | C | ||||
| th_ex3 | C | .0399 | |||
| rs3756814 | C | ||||
| rs6456632 | C | ||||
Note.— The selection included all the sibships with at least one sibling scoring <−0.5 SD on an average score of IQ-adjusted OC-irreg and PD. All reading-related measures are IQ-adjusted.
OC-irreg and PA showed no significant association in this analysis.
We next analyzed 21 of the SNPs in an epidemiological sample of 369 siblings from 159 unrelated nuclear twin-based families from Colorado, each of which included at least one proband with RD (sample 3) (DeFries et al. 1987; Gayan and Olson 2003). Previous analyses of subsets of this sample, which used different markers from those described here, produced suggestive evidence of marker-trait associations within TTRAP and KIAA0319, although these studies also produced similarly suggestive evidence of association elsewhere within 6p23-21.3 (Kaplan et al. 2002; Deffenbacher et al. 2004). Our association analysis of sample 3 produced only weakly significant evidence of marker-trait association within the putative QTL (data not shown). However, we aimed once more to replicate the selection scheme used for the U.K. samples, by choosing families in which at least one sibling scored poorly on their average of IQ-adjusted READ and PD (see the “Material and Methods” section), since these measures showed the best evidence of linkage in this sample. The evidence of association was again significant for SNPs within the same genomic interval as that implicated in the U.K. samples (table 3) (e.g., rs9467247 and READ, P=.0038; rs9467247 and PA, P=.042; rs3033236 and READ, P=.0023; rs3033236 and SPELL, P=.015). We also tried other threshold-based selection methods, in addition to those described here, and found that all methods that yielded similar selected sample sizes also produced broadly comparable effects for the association data, but the associations were consistently strongest if the prior selection had been performed using the RD traits that showed the strongest linkage within each sample (data not shown). In addition, association analysis with the IQ-unadjusted RD phenotypes gave a pattern of results that was similar to that of the association analysis with IQ-adjusted traits, although the overall level of significance was lower.
Haplotype association analysis revealed one main risk haplotype that spanned the QTL (LD region B), that was common to the U.K. and U.S. populations, that had a frequency of ∼12% in all samples, and that was effectively distinguished from all other common haplotypes by the SNP rs2143340 (table 4). We genotyped rs2143340 in 190 control individuals of European ancestry (ECACC) and found the minor allele frequency to be effectively the same as in the parents from the U.K. and U.S. samples (15%–16%). This frequency is higher than the 12% frequency for the main risk haplotype, because the minor allele of rs2143340 occasionally falls on other haplotypes. We observed an increase in the minor allele frequency of rs2143340 (up to 28% [table 5]) only in U.K. and U.S. siblings that were selected for RD phenotypic severity (table 5). These sample selections were based on the same combinations of two phenotypic measures that showed the strongest evidence of linkage in either sample (see above and table 5).
Table 4.
Association P Values of Most Common rs4504469, rs2038137, and rs2143340 Haplotypes with Reading-Related Measures[Note]
|
Selected Subset from Samples 1 and 2, Combined (U.K.) |
Selected Subset from Sample 3 (U.S.) |
||||||||||||
|
P Value for Traita |
P Value for Traitb |
||||||||||||
| Haplotype | Frequency (%) | OC-irreg | OC-choice | PD | READ | SPELL | Mean Effectc | Frequency (%) | PD | READ | SPELL | PA | Mean Effectc |
| 111 | .41 | .0306 | −.10 | .46 | −.01 | ||||||||
| 221 | .36 | .0019 | .0314 | .0017 | .0004 | .0165 | .24 | .31 | −.01 | ||||
| 112 | .12 | .0045 | .00007 | .0024 | .0246 | −.34 | .12 | .017 | .0367 | .0183 | −.23 | ||
| 211 | .05 | .03 | .05 | .0187 | .0119 | .0243 | .30 | ||||||
| 121 | .04 | −.04 | .04 | .0245 | .0413 | .33 | |||||||
| Others | .02 | −.10 | .02 | −.07 | |||||||||
Note.— All reading-related measures are IQ-adjusted. 1 = major allele; 2 = minor allele.
PA showed no significant association in this analysis.
OC-choice showed no significant association in this analysis.
Mean phenotypic effect relative to all other haplotypes, calculated across the different measures.
Table 5.
Minor Allele Frequency for SNP rs2143340 within Phenotypic Subcategories in the U.K. (Samples 1 and 2) and U.S. (Sample 3) Siblings
|
Minor Allele Frequency (%) |
|||||
| PhenotypicGroup | <−1.5 | <−1 | <−0.5 | <0 | ALL |
| U.K.a | 28 | 19 | 16 | 16 | 16 |
| U.S.b | 23 | 16 | 16 | 15 | 15 |
The phenotypic groups were calculated on the average of IQ-adjusted OC-irreg and PD.
The phenotypic groups were calculated on the average of IQ-adjusted READ and PD.
Candidate-Gene Screening
The region of association spans the genomic extent of TTRAP but is also directly upstream of the proximal neighboring gene THEM2 (thioesterase superfamily member 2), and it encompasses the first four exons of the distal gene KIAA0319 (uncharacterized brain-expressed transcript) (fig. 2). We screened the exons and predicted promoters of TTRAP, KIAA0319, THEM2, and another neighboring gene (ALDH5A1 [fig. 2]) for mutations in 32 probands with severe RD. However, we did not find any variants that have obvious disruptive effects on these genes (table A2 [online only]). No coding polymorphisms were detected that tagged the main risk haplotype. The only SNP that we identified within the trait-associated region (LD region B [fig. 2]) that had an effect on protein sequence was rs4504469 (table A1 [online only]), within exon 4 of KIAA0319. However, the minor allele of this SNP has a frequency of 0.47 in our samples and therefore is not unique to the risk haplotype that we identified. Accordingly, the significances of phenotype associations for rs4504469 were less than those of other SNPs that distinguished the risk haplotype more effectively (e.g., rs1061925 [table 3]). The SNP rs2143340, which distinguishes the risk haplotype from all other common haplotypes, is within intron 2 of TTRAP and lies on an untranslated alternative transcript of this gene (Vega Human Genome Browser Web site).
Discussion
We have used quantitative-trait linkage and association analysis to identify a QTL that influences RD on 6p22.2, within the genomic region that has shown the most consistently replicated linkage to RD among studies internationally. We have refined this QTL to a 77-kb region of strong intermarker LD that spans the gene for TTRAP and part of the neighboring uncharacterized gene KIAA0319. The QTL is also directly upstream of a third uncharacterized gene, THEM2. We identified a single main risk haplotype that spanned the QTL, with a frequency of ∼12% in our RD samples, which were drawn from populations of European descent in the United Kingdom and United States. We did not detect any coding polymorphisms within KIAA0319, TTRAP, or THEM2 that have overt disruptive effects on these genes or that characterize the main risk haplotype. The variation at this QTL that is functionally relevant for RD may therefore influence the expression, splicing, or transcript stability of any of the KIAA0319, TTRAP, or THEM2 genes. The strong LD between all polymorphisms within the region of marker-trait association (fig. 2) means that the functionally relevant variation for RD may be difficult to identify by further genetic mapping, as has proven to be the case in studies of QTLs for other complex traits, including asthma and Crohn disease (Rioux et al. 2001; Zhang et al. 2003).
RT-PCR analysis and expression profiling show that TTRAP is expressed in most or all tissues (Londin et al. 2003) (GNF SymAtlas Web site). The TTRAP protein interacts with cytoplasmic TNF receptor–associated factors (TRAFs), as well as with the cytoplasmic domains of specific members of the TNF receptor superfamily (Pype et al. 2000). TTRAP may therefore be involved as a regulatory factor in TNF signal transduction. Overexpression of TTRAP inhibits activity of the TNF-responsive transcription factor NF-κB (Pype et al. 2000). TNF signaling via NF-κB regulates many normal physiological and pathogenic processes, including immune and autoimmune responses and tumorigenesis (Aggarwal 2003). NF-κB activity in neurons also responds to synaptic signaling and may bring about long-term changes in neuronal function that subserve learning (Meffert et al. 2003), whereas inhibition of NF-κB activity in forebrain neurons results in neurodegenerative-like phenotypes in mice (Fridmacher et al. 2003). Thus, TTRAP may potentially influence both normal and pathogenic neuronal processes via its effects on NF-κB activity. TTRAP may also interact with members of the ETS family of transcription factors (Pei et al. 2003) and can also be conjugated to the small ubiquitin-like modifier SUMO-1, which modifies protein-protein interactions, subcellular localization, and stability (Lee et al. 2003).
KIAA0319 is expressed most strongly in brain tissue, with weaker expression in a restricted number of other tissues (Londin et al. 2003) (GNF SymAtlas Web site). KIAA0319 is largely uncharacterized; the predicted protein contains a putative transmembrane domain and four polycystic kidney disease domains that have homology with extracellular domains of the polycystic kidney disease protein PKD1, in which they are involved in cell-adhesive functions (Streets et al. 2003). THEM2 is also largely uncharacterized. The predicted protein has homology with thioesterase enzymes, and the gene is expressed in most tissues (Londin et al. 2003).
Our association data show clearly that the 6p22 QTL influences a broad range of reading-related cognitive skills in our samples (table 3). Fluctuating levels of univariate linkage of 6p23-21.3 to different reading-related phenotypes within a given study sample have been interpreted as evidence that this QTL affects some reading-related cognitive processes but not others (Grigorenko et al. 1997). The attraction of this interpretation lay in the prospect of being able to “dissect” reading-related cognition by the assignment to distinct genetic loci of different cognitive components of reading. However, the specificity of the 6p QTL for distinct cognitive reading-related processes was not well replicated by linkage analysis as study sample sizes were increased or as new samples were analyzed (Fisher et al. 1999; Gayan et al. 1999; Grigorenko et al. 2000; Fisher and DeFries 2002; Francks et al. 2002). In our previous multivariate linkage analysis of 6p23-21.3 (performed in sample 1), we were unable to drop any single reading-related measure from the multivariate linkage model without significantly reducing the model fit, even though, in separate univariate tests, some of the measures showed no significant linkage at this locus (Marlow et al. 2003). Now, having performed association analysis directly at the presumed underlying QTL, using powerful association analysis based on modeling allelic mean effects, the true pattern of QTL pleiotropy across the full range of reading-related measures becomes apparent (tables 1–3).
It is worth noting that our association data, like previous linkage data, still apparently contain chance fluctuations in measured effect sizes for different RD traits; for example, the trait OC-choice shows highly significant association with the main risk haplotype in the selected U.K. sample, but no significant association in the selected U.S. sample, despite being based on the same phenotypic test (table 3). Also, there are many instances in which a single SNP may show significant association with certain RD measures in one sample and association with a different set of RD measures in a different sample (tables 1–3). Again, we interpret this as stochastic variation that is influenced by relatively small individual sample sizes and by sample-specific patterns of missing genotype/phenotype data. It is worth noting that when the selected samples 1 and 2 are combined (table 3), the associations within LD region B become highly significant across most of this LD region, as well as across most of the RD measures, as would be expected if the underlying effect is truly pleiotropic. Our prior multivariate linkage analysis, together with the nature of the LD landscape in this region, suggest that the associations that we have found within LD region B can be considered as replications in the different samples, even though the same SNP/phenotype combinations are not always significant in all of the samples.
The pleiotropic effect of this QTL on a range of reading-related measures shows that the development of these cognitive abilities are at least partly interdependent. Even if this QTL produces a primary core deficit that affects one specific aspect of reading-related cognition, such a primary deficit may cause a general inhibitory effect on the ability to learn all reading-related skills. It remains possible that other RD susceptibility loci may cause even more specific cognitive deficits than this QTL—deficits limited to just a single reading-related measure. However, any developmental interdependence between different reading-related cognitive skills may make such a simple trait-locus correspondence unlikely for any locus. Furthermore, the functional roles of individual proteins in CNS development are manifold, multiregional, multitemporal, and interaction-dependent. Thus, it may be naive to postulate that a single genetic locus can somehow encode a single cognitive phenotype (Pennington 1997; Fisher, in press). In addition, a subject's performance on an individual cognitive test is unlikely to correspond to activity in any unitary system within the brain (Goldberg and Weinberger 2004). Therefore, the kind of relatively specific cognitive disruptions that can be associated with brain lesions in adults may not provide good models for understanding the developmental genetics of reading-related cognition (Paterson et al. 1999). However, it may become possible, through association analysis of this QTL in large epidemiological samples, to reliably detect subtle differences in the relative mean effects of the locus on different RD measures and thereby to identify aspects of reading-related cognition that may be influenced particularly strongly by this QTL.
In our association models, the main “risk” haplotype that spanned the QTL had an average effect of −0.34 SD on IQ-adjusted reading-related measures within the phenotypically selected U.K. subset; the haplotype had an average effect of −0.23 SD in the Colorado subset, when compared with all other haplotypes (table 4). However, these effect-size estimates apply only to phenotypically selected samples. To obtain a precise estimate of the contribution of this QTL to RD susceptibility, it will be important to analyze its effect in other large clinical and epidemiological samples and also in relation to other QTLs for RD, as these are identified, since multilocus interactions are a possibility in RD etiology.
We detected the 6p22 QTL most strongly after selection of sibships for phenotypic severity. Consistent with this, the minor allele for SNP rs2143340, which distinguishes the “risk” haplotype from all other common haplotypes, was only substantially increased in frequency (up to 28%) in the probands who were most severely affected with RD (table 5). This allele was present at a base frequency of ∼16%, even in siblings in the upper range of reading ability. We genotyped rs2143340 in a panel of 190 control individuals and again found a 16% frequency for the minor allele. There are a number of possible explanations for these findings. Under the simplest QTL model, which involves only additive effects that are assumed to be constant across the full range of ability, we calculate that an allele with 12% frequency (the risk haplotype frequency) and a phenotypic effect of −0.3 SD would be expected to have only a moderately elevated frequency (18%) in individuals with average phenotypic values 2 SD below the mean. This may explain why phenotypic selection was necessary to detect the QTL effect in our samples. Alternatively, the phenotypic effect of the risk haplotype may be contingent either on the genotype at another unidentified locus (i.e., epistasis) or on environmental influences, such that only a subset of risk haplotype carriers experience a detectable phenotypic effect. No other replicated QTLs for RD have yet been identified that can be analyzed in a joint epistatic association model. A third possibility is that the risk haplotype that we have identified carries functionally relevant variants only on a subset of its overall diversity.
In contrast to the pleiotropic effect of this QTL for many reading-related measures, there is no association of the QTL with IQ in either the U.K. or U.S. samples (table 3). Our data therefore show directly, for the first time, that reading-related cognition can be influenced by at least one relatively specific genetic-developmental deficit, while other cognitive abilities are left intact. The QTL may influence a pathogenic process that is mostly restricted to regions of the brain involved in reading and language ability (Paulesu et al. 2001). Alternatively, the pathogenic process may occur more broadly throughout the developing brain, but the resulting damage may have its most detrimental effect on certain neuronal circuits—or a class of neuronal operations—that are necessary for reading-related cognition.
This QTL is the first identified genetic influence to be shown by repeated replication to be of relevance to many individuals with specific RD. The functions of TTRAP, KIAA0319, and THEM2 must now be studied in detail to determine which gene is responsible for the QTL effect and to identify the mechanism of pathogenesis.
Acknowledgments
We are grateful to all the families who participated in this study. We thank K. Taylor, for assistance with handling the U.K. phenotype data, and K. King, for helping with mutation screening. The Colorado Twin Study was supported in part by program project and center grants from National Institute of Child Health and Human Development (HD-11681 and HD-27802). I.L.M. was funded by the British Council and Natural Sciences and Research Council, S.E.F. is a Royal Society Research Fellow, and L.R.C. and A.P.M. are Wellcome Trust Principal Research Fellows. This work was funded by the Wellcome Trust.
Appendix A: Supplementary Tables
Table A1.
Marker-Trait Association P Values in the Complete (Unselected) Sample 1 (224 U.K. Siblings, 89 Families)[Note]
|
P Value for Trait |
||||||||
| Marker | Gene | Cumulative Distance(bp) | OC-irreg | OC-choice | PD | READ | SPELL | PA |
| rs2817220 | ALDH5A1 | 1 | .0459 | .0226 | ||||
| rs22252525 | ALDH5A1 | 9,349 | ||||||
| rs807517 | ALDH5A1 | 19,484 | .0155 | |||||
| rs1054899 | ALDH5A1 | 28,973 | .0207 | |||||
| rs2760184 | 35,971 | |||||||
| rs807527 | KIAA0319 | 39,255 | ||||||
| rs699463 | KIAA0319 | 39,683 | .0232 | .0153 | .0021 | .0064 | ||
| rs807530 | KIAA0319 | 40,719 | ||||||
| rs807535 | KIAA0319 | 46,509 | .0151 | .0289 | ||||
| rs807540 | KIAA0319 | 53,809 | ||||||
| rs807545 | KIAA0319 | 58,909 | ||||||
| rs807521 | KIAA0319 | 65,211 | .0486 | |||||
| rs807525 | KIAA0319 | 70,817 | .038 | |||||
| rs807507 | KIAA0319 | 74,647 | ||||||
| rs807508 | KIAA0319 | 76,063 | .0192 | .0213 | ||||
| k_ex5 | KIAA0319 | 78,584 | .0369 | .0096 | .0341 | |||
| rs4504469 | KIAA0319 | 83,664 | .0391 | |||||
| rs4576240 | KIAA0319 | 91,258 | ||||||
| rs4472344 | KIAA0319 | 96,020 | ||||||
| rs4236032 | KIAA0319 | 101,507 | ||||||
| rs2745333 | KIAA0319 | 112,977 | .0136 | .0267 | ||||
| rs2179515 | KIAA0319 | 122,983 | .0076 | |||||
| rs761101 | KIAA0319 | 127,312 | .018 | |||||
| rs6456624 | KIAA0319 | 134,003 | .0084 | |||||
| rs2328846 | KIAA0319 | 139,975 | .01 | |||||
| rs2235676 | KIAA0319 | 140,477 | .0008 | .0466 | .0149 | |||
| rs2038137 | KIAA0319 | 140,723 | .0292 | |||||
| k_pr_del | KIAA0319 | 141,944 | .008 | |||||
| k_pr_1 | KIAA0319 | 141,999 | .0004 | .0057 | .0114 | |||
| rs1555090 | 142,887 | .01 | ||||||
| rs2294689 | TTRAP | 148,326 | .0308 | |||||
| rs3033236 | TTRAP | 152,635 | .0038 | .0068 | .0157 | |||
| rs3212232 | TTRAP | 152,677 | ||||||
| rs2143340 | TTRAP | 153,851 | .0008 | .0037 | .0393 | .0089 | ||
| rs2056998 | TTRAP | 156,379 | .0289 | |||||
| rs1061925 | TTRAP | 160,787 | .0004 | .0269 | .0106 | |||
| rs3835233 | 161,986 | |||||||
| th_ex1 | THEM2 | 162,104 | .0017 | .0078 | .0259 | |||
| tt_th_del | THEM2 | 162,189 | ||||||
| rs3181228 | THEM2 | 162,442 | ||||||
| rs2328847 | THEM2 | 174,708 | ||||||
| rs1555088 | THEM2 | 179,576 | ||||||
| rs926529 | THEM2 | 182,545 | ||||||
| rs1885211 | THEM2 | 186,959 | .0364 | |||||
| rs2092404 | THEM2 | 192,168 | ||||||
| th_ex3 | THEM2 | 196,722 | ||||||
| rs3756814 | FLJ12619 | 200,615 | .0157 | .0339 | ||||
| rs6456632 | FLJ12619 | 216,077 | .0076 | .0096 | .0112 | .0046 | ||
| rs2067573 | 225,273 | |||||||
| rs1884160 | C6orf32 | 365,065 | ||||||
| rs2207401 | 377,761 | |||||||
| rs1406927 | SCGN | 1,148,622 | ||||||
| rs2072846 | SCGN | 1,149,884 | ||||||
| rs2072847 | SCGN | 1,156,667 | ||||||
| rs2049968 | SCGN | 1,176,533 | .0336 | |||||
| rs10105 | SCGN | 1,196,683 | ||||||
| rs14232 | BTN3A2 | 1,872,721 | ||||||
| rs2076030 | BTN2A3 | 1,921,636 | ||||||
Note.— All reading-related measures are IQ-adjusted.
Table A2.
List of Fragments Analyzed for Mutation Screening
| Gene and Fragment | PCR Forward Primer | PCR Reverse Primer | Size (bp) | DNA Variation | Positiona | IDb | Location/Effect | No. of Heterozygotesc | Typed?d |
| ALDH5A1: | |||||||||
| al_promoter_a | GGGTAGCTAGTATGACTTTGAACG | GTCCATAGATGCGCTGGTG | 395 | T →G | 119 | al_pr_1 | Promoter | 7 | |
| al_promoter_b | CGATCGCTCCCAATCAATAC | AAGCAAGAGGAGCGAGGAG | 459 | C→G | 230 | rs2744575 | Promoter | 12 | |
| … | … | … | … | C→T | 281 | rs4646828 | Promoter | 8 | |
| … | … | … | … | C→G | 356 | rs4646830 | Promoter | 13 | |
| al_promoter_ex_1 | TCTGTGGCTCTGCAACCTTC | GCATCCGGGCTCTCTCAC | 519 | None | |||||
| al_ex_2 | CACTCACGGACATACCCAGA | TTCTTCAGCTACAGCCAGCA | 386 | None | |||||
| al_ex_3 | AGGAACACAGAGCCATGCTT | ACACACTTCCACATCCAGCA | 389 | C→T | 166 | rs2760118 | His→Tyr | 10 | |
| … | … | … | … | C→T | 173 | rs3765310 | Pro→Leu | 1 | |
| al_ex_4 | AACATGCCTTCCTTTGCACT | TATGGCCTGCAGATGTCAGA | 400 | G→A | 195 | rs2744584 | Intronic | 8 | |
| … | … | … | … | G→A | 297 | rs2817220 | Intronic | 8 | Yes |
| al_ex_4b | TTGCTGAGGCTTTCCAGAAC | CCTAGGAAAAGGGGGAGCTA | 447 | None | |||||
| al_ex_5 | TTGGCACATGTTTGCTGTTT | TTCTGTGAGGGGGTTTCATC | 426 | None | |||||
| al_ex_6 | CACTACCACACCCAGCCTCT | ACGCAAACACACATCCTGAA | 396 | C→T | 166 | al_ex6_1 | Intronic | 3 | |
| … | … | … | … | G→A | 185 | al_ex6_2 | Intronic | 3 | |
| al_ex_7 | ATGGCAGTTTGAGCACATGA | TGTCAACCCACCCCTCTCTA | 402 | None | |||||
| al_ex_8 | TCTGCAAATGTGGTTCCTTCT | CAGCCCAGGAATCTCTTTCA | 414 | None | |||||
| al_ex_9 | TTCCTTTCCTCTCCCCCTTA | TGGGACTTTGAGATGTGTGC | 399 | None | |||||
| al_ex_10a | TGCCATATATGTCCTTTTATCCTG | GGCAGTGATCAAGCCTTTGT | 577 | None | |||||
| al_ex_10b | GCAGACTCCCAGAGAACCAG | TCAGGAATAGGGAGAGGTGCT | 400 | A→C | 48 | rs1054899 | 3′ UTR | 4 | Yes |
| al_ex_10c | AATTGTCCGTGCTTCTGTGA | CGGTGCTGCTCGTAACAAT | 417 | None | |||||
| KIAA0319: | |||||||||
| k_ promoter_1 | TGAGACGGAGTCTTGCTCTG | CCACAGAAAACTTGGGTGGA | 611 | G→A | 403 | rs1883592 | Promoter | 7 | |
| … | … | … | … | G→T | 413 | rs9467247 | Promoter | 9 | Yes |
| … | … | … | … | T→A | 441 | rs1883593 | Promoter | 1 | |
| … | … | … | … | C→G | 448 | rs1883594 | Promoter | 7 | |
| … | … | … | … | del T | 478 | k_pr_del_1 | Promoter | 8 | Yes |
| k_promoter_2 | CCTAGCACAGGTTCCCAGTC | AGTGATTCAGCGCCTTTCC | 519 | G→A | 129 | rs3756821 | Promoter | 12 | |
| 26bp del | 404 | k_pr_del_2 | Promoter | 11 | |||||
| k_ex_1_1 | GGAAAGGCGCTGAATCACT | CACGTTCACACCCTCGCG | 317 | G→A | 234 | k_ex1_1 | 5′ UTR | 3 | |
| k_ex_1_2 | GTGTGTAAGACCTGCGATG | AGGGCGTCTCGATTCTAC | 209 | C→T | 66 | k_ex1_2 | 5′ UTR | 2 | |
| … | … | … | … | A→C | 108 | rs2038135 | 5′ UTR | 2 | |
| … | … | … | … | C→G | 128 | rs2038136 | 5′ UTR | 4 | |
| … | … | … | … | A→C | 132 | rs2038137 | 5′ UTR | 10 | Yes |
| k_ex_2 | GCTCTGTGATGTGACCAGGA | AAACCAACAGCGTCTTCACC | 379 | C→T | 318 | rs4472344 | Intronic | 2 | Yes |
| k_ex_3a | TGCCTCTCTCCCTCATTAGAA | TTCTGATATCCTCAGGTGAGTCC | 397 | None | |||||
| k_ex_3b | TATGGGGACATGATGCTGAA | CCAAGCACATCCTGAACACA | 481 | A→C | 79 | rs4576240 | Tyr→Pro | 2 | Yes |
| k_ex_4 | TTGAGTTGCTTGAAGAGTTGGA | CCGTTTCTGGCACATGATAG | 391 | G→A | 198 | rs4504469 | Ala→Tyr | 12 | Yes |
| k_ex_5 | GAGGAAAAACGTGCCTTAGC | GTGAAAAAGAATCGGCGTTG | 349 | A→T | 282 | k_ex5 | Intronic | 10 | Yes |
| k_ex_6 | CAAGAGTGAGGGGATAAAAACAA | GCTCTATGCCGTTACGCTGT | 340 | None | |||||
| k_ex_7 | CCTCTGCAGCGAAGCTAAAG | GTCATCCAGGCACCAAAAAT | 379 | C→T | 183 | rs807508 | Intronic | 3 | Yes |
| k_ex_8 | TGTCCAGACAAGCTCCTGAA | TCCTCCTAGCTTTGATGCAGA | 374 | G→C | 160 | rs2744564 | Intronic | 1 | |
| … | … | … | … | ins T | 187 | rs3215493 | Intronic | 6 | |
| k_ex_9 | AGTGGGAACAGCCTCATTCA | ATAAAGGCAGGGACCCACTT | 381 | None | |||||
| k_ex_10 | AGCCTGGCATATTCTTCCAA | AGAGCCAATTGGTTGCTGTC | 498 | G→A | 239 | rs2744559 | Gly→Ser | 4 | |
| k_ex_11 | GCCTGGCCAAACACATTTCT | AAGTGTGGCATCTCCAAACC | 351 | None | |||||
| k_ex_12 | CACTCCAGGATTCCAAAGGA | TGCGCAAGCTGATTGTTTAC | 371 | T→C | 62 | rs2817195 | Intronic | 2 | |
| k_ex_13 | CAAATCGCATGCCTGTCATA | GCATCTGCTGCGAACAAATA | 339 | None | |||||
| k_ex_14 | TGCAAGGTGGAAGAGTTGTG | ATATTGTTTGCAGCCCAAGC | 328 | None | |||||
| k_ex_15 | GGATCAACAGAAGCCCAGAA | CATCCTCAAAGTGGGTCCAG | 347 | T→C | 60 | rs2760163 | Intronic | 4 | |
| … | … | … | … | C→T | 66 | rs2760164 | Intronic | 4 | |
| … | … | … | … | A→G | 95 | rs2744550 | Ser→Val | 4 | |
| … | … | … | … | T→C | 99 | rs2817191 | Val→Val | 4 | |
| k_ex_16 | CCTGTGCAAATGTCCAAGTG | GGCCATACTACTGCCCTTTG | 370 | A→G | 242 | k_ex16 | Intronic | 2 | |
| k_ex_17 | TCAATTCCTTGGCTCACAGA | TCCATGATTGGCAAGTCTGA | 350 | T→C | 108 | rs3903801 | Intronic | 9 | |
| … | … | … | … | T→C | 260 | rs807541 | Ala→Ala | 7 | |
| k_ex_18 | AGCCCTACACACCTCTTTGC | TGGACCTCCTCTCAAGCAAT | 364 | G→A | 116 | k_ex18_1 | Arg→His | 1 | |
| … | … | … | … | C→T | 305 | k_ex18_2 | Intronic | 4 | |
| … | … | … | … | G→T | 330 | k_ex18_3 | Intronic | 1 | |
| k_ex_19 | CAGGTGGCCTTTCCTTGTAG | TTTGTGGCAGAAAGTGCATTA | 332 | G→A | 209 | rs2243831 | Intronic | 13 | |
| … | … | … | … | T→C | 299 | k_ex19 | Intronic | 7 | |
| k_ex_20 | CTGACCTGGCCTTGTGTATG | GCTGACTCCAGCAAATCGTT | 351 | A→G | 180 | rs807535 | Lys→Lys | 5 | Yes |
| … | … | … | … | A→G | 245 | rs807534 | Tyr→Cys | 5 | |
| … | … | … | … | T→C | 285 | k_ex_20 | Intronic | 13 | |
| k_ex_21 | CAACCACAACGACTGACAGC | TGGGGAAGAAGGTCAATGAA | 359 | C→T | 37 | rs2817245 | Intronic | 12 | |
| … | … | … | … | A→G | 292 | k_ex21 | 3′ UTR | 2 | |
| Region that spanned TTRAP and THEM2: | |||||||||
| tt_th_1 | ACTTTTTAAGTGCAGGAACTG | GATACCAGGTAGCCCTGC | 446 | ins A | 67 | tt_ex1_2 | Intronic | 15 | |
| tt_th_2 | TCTGCCTCAGCATCGTCC | GCCTCCTGTTCCGCTTAAA | 491 | C→T | 456 | tt_ex1_1 | 5′ UTR | 1 | |
| tt_th_3 | GCTTTTTCACCTCAGGCTC | CTTTGGGCTTTGCGCAAG | 456 | ins CC | 188 | rs3835233 | 5′ UTR | 10 | Yes |
| … | … | … | … | C→T | 306 | th_ex1 | 5′ UTR | 5 | Yes |
| … | … | … | … | del CCT | 391 | tt_th_del | 5′ UTR | 6 | Yes |
| tt_th_4 | CACTAACTTCTGGACTTTCC | TTCGACCCAGCATATTTAG | 337 | A→G | 170 | th_ex1_2 | Intronic | 6 | |
| … | … | … | … | C→T | 246 | rs3181228 | Intronic | 3 | Yes |
| TTRAP: | |||||||||
| tt_ex_3 | GGGGATGCAAGTCATCAGTA | GCCCACAACCTTGAACTGTTA | 504 | T→C | 149 | rs2143340 | Intronic | 11 | Yes |
| tt_ex_4 | TGTTGTGGGATTTCCTTCATT | TCCTGAAAAATCATCCATTT | 391 | A→C | 250 | rs3212231 | Intronic | 13 | |
| … | … | … | … | T→C | 317 | rs3212232 | Intronic | 12 | Yes |
| … | … | … | … | del ATTAAT | 353 | rs3033236 | Intronic | 7 | Yes |
| tt_ex_5 | AAATATCTGTTACCTGTTTGC | TAAAGGCAATGGTTTTAAAAAG | 317 | None | |||||
| tt_ex_6 | TAGTGCTGATTTTACAGTTTC | GAGTGACTAAAGGTCCTA | 403 | T→C | 134 | rs1129644 | Asn→Asn | 8 | |
| … | … | … | … | A→G | 227 | tt_ex6 | Leu→Leu | 2 | |
| tt_ex_7 | CCCCTCTGGCTGTCTTGATA | AAAACCCACACTTGAAAAGCA | 502 | C→T | 107 | rs3181244 | Intronic | 14 | |
| … | … | … | … | G→C | 175 | rs3181245 | Intronic | 20 | |
| THEM2: | |||||||||
| th_ex_2 | TTCAATCCATTGTTCTTCATGG | GGTGCAGTTAAGGAGATAAGCA | 394 | None | |||||
| th_ex_3 | GTAAAATAAGTTACATAGGAACAC | GAAAACAAGATGCTTAAAGTG | 441 | A→G | 349 | th_ex3 | 3′ UTR | 1 | Yes |
Distance in bp from the first base of the forward primer.
Reference numbers (rs#) from dbSNP are used, when available.
Number of heterozygotes detected in 32 individuals with dyslexia.
Markers genotyped for the association analysis.
Table A3.
Raw Data for Intermarker LD Statistics Plotted in Figure 2
|
Standardized Disequilibrium Coefficient (D′) for Marker Pair |
|||||||||||||||||||||||||||||||||||||||||||||||
| rs22252525 | rs807517 | rs1054899 | rs2760184 | rs807527 | rs699463 | rs807530 | rs807535 | rs807540 | rs807545 | rs807521 | rs807525 | rs807507 | rs807508 | k_ex5 | rs4504469 | rs4576240 | rs4472344 | rs4236032 | rs2745333 | rs2179515 | rs761101 | rs6456624 | rs2328846 | rs2235676 | rs2038137 | k_pr_del | k_pr_1 | rs1555090 | rs2294689 | rs3033236 | rs3212232 | rs2143340 | rs2056998 | rs1061925 | th_ex1 | tt_th_del | rs3181228 | rs2328847 | rs1555088 | rs926529 | rs1885211 | rs2092404 | th_ex3 | rs3756814 | rs6456632 | rs2067573 | |
| rs2817220 | .592 | .7668 | .2745 | 1 | .6483 | .7184 | .7007 | .8449 | .4432 | .8234 | .7908 | .7982 | .5333 | .7685 | .0293 | .2776 | .553 | .714 | .4337 | .3912 | .2108 | .2684 | .2298 | .2205 | .1507 | .1953 | .2142 | .0079 | .2149 | .4122 | .0694 | .0378 | .0286 | .4045 | .2649 | .4061 | .1182 | .4907 | 1 | .0957 | .063 | .023 | .0674 | 1 | .0038 | .5721 | .5074 |
| rs22252525 | .9186 | .176 | 1 | .6709 | .2231 | .0247 | .7033 | .5419 | 1 | .4231 | .4394 | .8681 | 1 | .5247 | .3279 | 1 | .7124 | .6533 | .3972 | .2976 | .356 | .3224 | .1503 | .3161 | .1774 | .2045 | .1206 | .1464 | .6856 | .1322 | .106 | .0989 | .6868 | .1794 | .1612 | .2206 | .8543 | .6147 | .7222 | .7147 | .0096 | .0874 | 1 | .1229 | .0944 | .011 | |
| rs807517 | .2515 | 1 | .4907 | .5645 | .5066 | .7235 | .2349 | .9365 | .7785 | .7868 | .465 | .8744 | .0128 | .5043 | .6752 | .4034 | .8462 | .3214 | .4245 | .256 | .3569 | .4179 | .1701 | .398 | .4127 | .0311 | .4133 | .6615 | .4093 | .0575 | .0277 | .6615 | .2077 | .4033 | .0478 | .4468 | 1 | .1781 | .1619 | .0543 | .1379 | 1 | .0175 | .529 | .7248 | ||
| rs1054899 | 1 | .8164 | .3922 | .9236 | .9071 | .5081 | .9324 | .9401 | .9417 | .7669 | .9335 | .1376 | .216 | .3069 | .8636 | .4452 | .2035 | .2263 | .2059 | .2462 | .255 | .0301 | .2908 | .2684 | .0672 | .265 | .3571 | .1015 | .2572 | .1137 | .3614 | .1432 | .042 | .1984 | .0366 | .1473 | .4003 | .3693 | .3049 | .1742 | 1 | .128 | .1391 | 1 | |||
| rs2760184 | 1 | .233 | 1 | .4992 | 1 | 1 | 1 | 1 | 1 | 1 | .2197 | .1982 | .0062 | 1 | 1 | .2285 | .1308 | .0675 | .3475 | .0966 | .4802 | .1287 | .0241 | .646 | .0966 | .0076 | 1 | .4694 | .5764 | .0078 | .3534 | 1 | 1 | 1 | .3101 | .1221 | .1096 | .0476 | .1243 | 1 | .1897 | .5782 | 1 | ||||
| rs807527 | .7633 | 1 | 1 | .8733 | 1 | 1 | 1 | .7424 | 1 | .736 | .2658 | 1 | 1 | 1 | .2698 | .2703 | .132 | .2762 | .3142 | .2611 | .2997 | .2585 | .1787 | .3162 | .0551 | .1404 | .2123 | .1719 | .0524 | .6957 | 1 | .4012 | 1 | .6364 | .7793 | .7701 | .5105 | .6647 | 1 | .1263 | 1 | .0565 | |||||
| rs699463 | .9235 | .7812 | .5459 | .7639 | .7879 | .796 | .7528 | .7661 | .1899 | .1026 | .5691 | .7628 | .4551 | .1632 | .1025 | .1346 | .0838 | .1204 | .1114 | .093 | .1137 | .1097 | .1145 | .5006 | .0164 | .0714 | .1574 | .5033 | .0886 | .2824 | .0188 | .6905 | 1 | .0472 | .0099 | .0979 | .1556 | 1 | .0346 | .1214 | .5512 | ||||||
| rs807530 | .8176 | .6981 | .7411 | .7619 | .7626 | .7325 | .7339 | .0885 | .0606 | .2711 | .8272 | .3104 | .044 | .0641 | .0045 | .0967 | .0486 | .1679 | .0881 | .0566 | .0826 | .0541 | .4778 | .1618 | .1339 | .2202 | .4806 | .2033 | .0872 | .3408 | .199 | .287 | .105 | .1383 | .0911 | .0615 | 1 | .0482 | .0585 | .7189 | |||||||
| rs807535 | .7227 | .8457 | .8629 | .8633 | .6673 | .8986 | .25 | .232 | .3956 | .4456 | .4431 | .2829 | .1786 | .0907 | .2245 | .1023 | .0193 | .0651 | .0889 | .0306 | .0881 | .4541 | .0723 | .228 | .0759 | .4509 | .0029 | .5787 | .1523 | .2199 | 1 | .332 | .2465 | .4565 | .2446 | 1 | .2168 | .4056 | 1 | ||||||||
| rs807540 | .4147 | .4432 | .4848 | .5597 | .363 | .4438 | .4032 | 1 | .0574 | 1 | .2962 | .2105 | .0519 | .2157 | .3061 | .6633 | .3713 | .2341 | .0388 | .3084 | .0524 | .2005 | .3059 | .1244 | .0493 | .6229 | 1 | .1846 | .6925 | .4594 | .5936 | .5592 | .2762 | .5923 | .0418 | .2009 | .4563 | .0138 | |||||||||
| rs807545 | 1 | 1 | .6061 | 1 | .3783 | .3432 | 1 | .032 | 1 | .5465 | .1333 | .5193 | .2061 | .2526 | .1808 | .2719 | .2536 | .2613 | .249 | .3208 | .0614 | .3989 | .1395 | .3167 | .0984 | .0433 | 1 | .1741 | 1 | .091 | .1453 | .4469 | .0063 | 1 | .1232 | .0406 | 1 | ||||||||||
| rs807521 | 1 | .6336 | 1 | .4856 | .2549 | 1 | .0092 | 1 | .4418 | .0913 | .3786 | .1614 | .0701 | .1544 | .0987 | .1582 | .1679 | .066 | .4412 | .1189 | .4552 | .0736 | .4181 | .0856 | .0334 | 1 | .16 | 1 | .0638 | .0516 | .353 | .0527 | 1 | .0764 | .0017 | 1 | |||||||||||
| rs807525 | .6444 | 1 | .5002 | .2752 | 1 | .0067 | 1 | .4589 | .1254 | .3971 | .1988 | .0956 | .1387 | .127 | .1835 | .1935 | .0956 | .4565 | .1576 | .4757 | .099 | .4348 | .0733 | .0301 | 1 | .161 | 1 | .0481 | .0354 | .3725 | .0371 | 1 | .1196 | .029 | 1 | ||||||||||||
| rs807507 | .6065 | .3526 | .0111 | .6051 | 1 | .5453 | .0323 | .0059 | .0581 | .0299 | .0295 | .1145 | .0573 | .033 | .0783 | .0335 | .7249 | .0293 | .2263 | .009 | .728 | .1209 | .0914 | .5475 | .1691 | 1 | .1593 | .1836 | .2076 | .0893 | .4762 | .0341 | .0595 | .1794 | |||||||||||||
| rs807508 | .4142 | .3368 | 1 | .0221 | 1 | .5428 | .1932 | .5343 | .276 | .2417 | .1852 | .2665 | .3477 | .2604 | .238 | .3333 | .0575 | .4014 | .1419 | .3294 | .1021 | .0564 | 1 | .1698 | 1 | .0778 | .0285 | .4362 | .0764 | 1 | .0555 | .035 | 1 | ||||||||||||||
| k_ex5 | .1007 | .126 | .7612 | .0278 | .0954 | .203 | .2459 | .0146 | .195 | .01 | .1122 | .1731 | .2024 | .2093 | .6272 | .2201 | .0238 | .2363 | .6276 | .0622 | .0347 | .6473 | .562 | 1 | .3804 | .2752 | .1694 | .2252 | .8412 | .12 | .0565 | .6715 | |||||||||||||||
| rs4504469 | .8039 | 1 | .9069 | .8415 | .8177 | .7183 | .6836 | .8265 | .8599 | .7673 | .8095 | .5503 | .8257 | 1 | .5677 | .0174 | .5352 | 1 | .8272 | .9291 | .1483 | .1728 | 1 | .5134 | .4881 | .107 | .3904 | .5022 | .2459 | .3015 | .473 | ||||||||||||||||
| rs4576240 | 1 | .9027 | .7041 | .8671 | .761 | .8375 | .8899 | .3984 | .8933 | .8895 | .7918 | .8894 | 1 | .2403 | .1973 | .2439 | 1 | .2565 | .1455 | 1 | .5237 | 1 | .8672 | .7184 | .4291 | .87 | 1 | .2189 | .5208 | 1 | |||||||||||||||||
| rs4472344 | 1 | 1 | .7356 | 1 | .7297 | .7898 | 1 | .7965 | .7891 | .5942 | .7888 | .9108 | .5119 | 1 | .5144 | .9108 | 1 | .1545 | 1 | 1 | 1 | .7398 | .7375 | 1 | .8761 | 1 | 1 | .7961 | 1 | ||||||||||||||||||
| rs4236032 | .8171 | 1 | .8852 | 1 | 1 | .3936 | 1 | 1 | .7929 | 1 | 1 | .2522 | .2262 | .2434 | 1 | .2434 | .1652 | .0362 | .5456 | 1 | 1 | .8667 | .5433 | 1 | 1 | .1434 | .4889 | .0034 | |||||||||||||||||||
| rs2745333 | .7805 | .8837 | .6977 | .7677 | .8597 | .7212 | .7637 | .7731 | .7677 | 1 | .4225 | .0142 | .5443 | 1 | .943 | .9309 | .2298 | .2575 | 1 | .5398 | .4967 | .2153 | .4284 | .5173 | .2496 | .4191 | .4929 | ||||||||||||||||||||
| rs2179515 | .8578 | .9488 | .9399 | .8759 | .9106 | .9393 | .7861 | .9396 | .8462 | .7345 | .0188 | .7954 | .8465 | .8419 | .8186 | .8249 | .5571 | 1 | .6337 | .6204 | .2782 | .5484 | 1 | .1966 | .7158 | .7048 | |||||||||||||||||||||
| rs761101 | .8755 | .8486 | .8918 | .8448 | .8593 | .9235 | .8486 | 1 | .6756 | .0122 | .8167 | 1 | .933 | .9144 | .6224 | .7978 | 1 | .6107 | .5556 | .4434 | .5117 | 1 | .2944 | .8294 | .6493 | ||||||||||||||||||||||
| rs6456624 | .9319 | .9318 | .9341 | .9315 | .7744 | .9315 | 1 | .7374 | .034 | .837 | 1 | .9146 | .8104 | .8207 | .6523 | 1 | .7479 | .7288 | .3398 | .5313 | 1 | .1181 | .8418 | .8459 | |||||||||||||||||||||||
| rs2328846 | .8458 | 1 | .9759 | .7866 | 1 | 1 | .8273 | .0359 | .7863 | 1 | .8109 | .9192 | .8281 | .6776 | 1 | .6779 | .6178 | .3315 | .5813 | 1 | .2763 | .8431 | .6577 | ||||||||||||||||||||||||
| rs2235676 | .8512 | .8925 | .8218 | .8458 | 1 | .523 | .6959 | .7075 | 1 | .9404 | .7263 | .5992 | .6076 | 1 | .6172 | .5487 | .5906 | .617 | 1 | .5649 | .6638 | 1 | |||||||||||||||||||||||||
| rs2038137 | .9876 | .7932 | 1 | 1 | .8327 | .0824 | .7928 | 1 | .8167 | .922 | .8343 | .6878 | 1 | .7458 | .6865 | .362 | .5723 | .0197 | .2538 | .8479 | .105 | ||||||||||||||||||||||||||
| k_pr_del | .7823 | .9758 | 1 | .8238 | .029 | .8225 | 1 | .8669 | .9163 | .8277 | .6766 | 1 | .664 | .6034 | .3424 | .5534 | .7463 | .2802 | .8426 | .4848 | |||||||||||||||||||||||||||
| k_pr_1 | .7857 | .0721 | .5908 | .8939 | .83 | .0728 | .8433 | .7978 | .6786 | .6416 | .583 | .5958 | .542 | .5907 | .4706 | 1 | .152 | .6127 | .6559 | ||||||||||||||||||||||||||||
| rs1555090 | 1 | .8266 | .0485 | .7854 | 1 | .8101 | .9188 | .8281 | .6761 | 1 | .6788 | .6178 | .3385 | .5825 | 1 | .2863 | .8424 | .6577 | |||||||||||||||||||||||||||||
| rs2294689 | .4655 | 1 | .7413 | 1 | 1 | .3958 | 1 | 1 | .6443 | .8665 | .8598 | .9204 | .6087 | 1 | 1 | .4984 | .0025 | ||||||||||||||||||||||||||||||
| rs3033236 | .8078 | .7255 | .4639 | .7466 | .6708 | .7696 | .7836 | 1 | .8925 | .837 | .7287 | .7321 | 1 | .3593 | .7184 | 1 | |||||||||||||||||||||||||||||||
| rs3212232 | .8744 | 1 | .8179 | .7663 | 1 | .9345 | 1 | .9341 | .8559 | .4464 | .9345 | 1 | .8809 | .8276 | 1 | ||||||||||||||||||||||||||||||||
| rs2143340 | .7406 | .97 | .8036 | .8472 | .6727 | 1 | 1 | .9455 | .8481 | .8941 | 1 | .3563 | .6303 | 1 | |||||||||||||||||||||||||||||||||
| rs2056998 | 1 | .3958 | 1 | 1 | .6444 | .8649 | .858 | .9206 | .611 | 1 | 1 | .4987 | .0027 | ||||||||||||||||||||||||||||||||||
| rs1061925 | .9174 | 1 | .6811 | 1 | 1 | .9224 | .9435 | 1 | 1 | .8656 | .7273 | 1 | |||||||||||||||||||||||||||||||||||
| th_ex1 | 1 | .3793 | 1 | 1 | .9004 | .9265 | .8932 | 1 | .7072 | .2914 | 1 | ||||||||||||||||||||||||||||||||||||
| tt_th_del | 1 | 1 | 1 | 1 | .8653 | 1 | 1 | 1 | 1 | .0296 | |||||||||||||||||||||||||||||||||||||
| rs3181228 | 1 | .8319 | .8782 | .9589 | .8338 | 1 | .861 | .7097 | 1 | ||||||||||||||||||||||||||||||||||||||
| rs2328847 | 1 | 1 | .4004 | 1 | 1 | 1 | .3879 | 1 | |||||||||||||||||||||||||||||||||||||||
| rs1555088 | .9435 | .886 | .791 | 1 | .6812 | .8564 | .8021 | ||||||||||||||||||||||||||||||||||||||||
| rs926529 | .8287 | .7254 | 1 | .5868 | .8478 | .8045 | |||||||||||||||||||||||||||||||||||||||||
| rs1885211 | .8538 | 1 | .4605 | .4011 | .755 | ||||||||||||||||||||||||||||||||||||||||||
| rs2092404 | .6982 | .8638 | .299 | .5714 | |||||||||||||||||||||||||||||||||||||||||||
| th_ex3 | .6102 | 1 | 1 | ||||||||||||||||||||||||||||||||||||||||||||
| rs3756814 | .8769 | .3285 | |||||||||||||||||||||||||||||||||||||||||||||
| rs6456632 | 1 | ||||||||||||||||||||||||||||||||||||||||||||||
| χ2 P for Marker Pair |
|||||||||||||||||||||||||||||||||||||||||||||||
| rs22252525 | rs807517 | rs1054899 | rs2760184 | rs807527 | rs699463 | rs807530 | rs807535 | rs807540 | rs807545 | rs807521 | rs807525 | rs807507 | rs807508 | k_ex5 | rs4504469 | rs4576240 | rs4472344 | rs4236032 | rs2745333 | rs2179515 | rs761101 | rs6456624 | rs2328846 | rs2235676 | rs2038137 | k_pr_del | k_pr_1 | rs1555090 | rs2294689 | rs3033236 | rs3212232 | rs2143340 | rs2056998 | rs1061925 | th_ex1 | tt_th_del | rs3181228 | rs2328847 | rs1555088 | rs926529 | rs1885211 | rs2092404 | th_ex3 | rs3756814 | rs6456632 | rs2067573 | |
| rs2817220 | .0084 | 0 | .0001 | .0138 | .0185 | 0 | 0 | 0 | .1324 | 0 | 0 | 0 | 0 | 0 | .6031 | .0003 | .0775 | .106 | .1544 | 0 | .0053 | .0001 | .008 | .0012 | .4898 | .0056 | .0017 | .9103 | .0017 | .1844 | .7169 | .5027 | .88 | .1969 | .2706 | .135 | .4478 | .0115 | .2613 | .1056 | .3024 | .8066 | .2749 | .0499 | .9597 | .0011 | .215 |
| rs22252525 | 0 | .317 | .1406 | 0 | .0146 | .872 | .0327 | 0 | .0703 | .378 | .3527 | 0 | .0632 | 0 | .037 | .0674 | 0 | .2179 | .0094 | .1463 | .0429 | .122 | .3812 | .3779 | .288 | .234 | .0845 | .3951 | 0 | .0392 | .6379 | .1167 | 0 | .6522 | .7304 | .0129 | .0088 | .0067 | .0003 | .0005 | .9496 | .3886 | .2399 | .5081 | .1542 | .9147 | |
| rs807517 | .0004 | .026 | .0901 | 0 | 0 | 0 | .4523 | 0 | 0 | 0 | 0 | 0 | .83 | 0 | .0407 | .375 | .0082 | .0001 | 0 | .0005 | .0001 | 0 | .4562 | 0 | 0 | .6584 | 0 | .0499 | .0386 | .3043 | .8883 | .0499 | .4095 | .141 | .7508 | .0289 | .2742 | .0041 | .0109 | .5771 | .0325 | .0719 | .825 | .0044 | .0966 | ||
| rs1054899 | 0 | .0003 | 0 | 0 | 0 | .0581 | 0 | 0 | 0 | 0 | 0 | .041 | .0039 | .2305 | .0001 | .076 | .0052 | .0183 | .0189 | .0087 | .0021 | .8585 | .0003 | .0015 | .6191 | .0015 | .027 | .5009 | .0002 | .4491 | .0241 | .4487 | .8481 | .5143 | .718 | .8373 | 0 | .0002 | 0 | .0711 | .0103 | .0377 | .1346 | .0024 | |||
| rs2760184 | .2555 | .5741 | .0001 | .434 | .3427 | .2751 | .2802 | .2555 | .0055 | .3029 | .6005 | .4973 | .9292 | .4945 | .2915 | .4176 | .7148 | .839 | .3181 | .7653 | .4639 | .682 | .943 | .2147 | .7653 | .9119 | .1133 | .0051 | .3214 | .9095 | .6343 | .2786 | .3736 | .0891 | .0038 | .7459 | .7732 | .8693 | .7408 | .5079 | .4215 | .3155 | .4432 | ||||
| rs807527 | .0056 | 0 | .0188 | 0 | .1623 | .1101 | .0984 | .0004 | .1266 | 0 | .1082 | .146 | .2891 | .1389 | .1079 | .1118 | .3542 | .1404 | .0294 | .5536 | .0429 | .0722 | .046 | .0278 | .3536 | .0858 | .0568 | .031 | .386 | .1621 | .0981 | 0 | .0156 | .0002 | .0029 | .004 | .0118 | .0113 | .3433 | .4777 | .0089 | .4519 | |||||
| rs699463 | 0 | 0 | .079 | 0 | 0 | 0 | 0 | 0 | .0008 | .159 | .0625 | 0 | .1247 | .0301 | .1697 | .0419 | .334 | .067 | .583 | .1708 | .084 | .4967 | .0823 | .0002 | .8331 | .6063 | .3832 | .0001 | .6955 | .2564 | .9058 | .0003 | .2591 | .4116 | .9358 | .1921 | .0099 | .0334 | .7346 | .4786 | .1558 | ||||||
| rs807530 | 0 | .0017 | .0001 | 0 | 0 | 0 | .0002 | .2774 | .3392 | .1977 | .0033 | .1267 | .4756 | .4288 | .9463 | .2217 | .4889 | .2483 | .1992 | .4286 | .4753 | .4447 | .0154 | .2091 | .1107 | .0933 | .0142 | .2128 | .6261 | .183 | .1354 | .6399 | .2055 | .1056 | .1461 | .4113 | .002 | .4393 | .597 | .0068 | |||||||
| rs807535 | .1178 | 0 | 0 | 0 | 0 | 0 | .0026 | .0437 | .4178 | .5083 | .3434 | .0168 | .117 | .3778 | .0919 | .3173 | .7375 | .5398 | .3872 | .6208 | .3928 | .3262 | .1985 | .0045 | .1611 | .3314 | .9934 | .1454 | .1762 | .4305 | .4424 | .0744 | .1933 | .001 | .1748 | .1655 | .0556 | .1188 | .0894 | ||||||||
| rs807540 | .5573 | .5197 | .4627 | .0179 | .6235 | .001 | .0359 | .2024 | .5532 | .1821 | .1357 | .2431 | .7608 | .2766 | .0741 | .201 | .0342 | .1685 | .7021 | .0708 | .4135 | .6577 | .0168 | .1798 | .4524 | .2617 | .1705 | .0053 | .1587 | .0221 | .04 | .0626 | .2255 | .0408 | .6488 | .2906 | .3194 | .8589 | |||||||||
| rs807545 | 0 | 0 | .0072 | 0 | .24 | .1182 | .1847 | .7087 | .1671 | .0103 | .6158 | .0488 | .4409 | .3038 | .0327 | .2579 | .3012 | .01 | .3126 | .6763 | .8845 | .2083 | .1256 | .6813 | .1792 | .5699 | .2262 | .0452 | .6832 | .75 | .6282 | .0388 | .9631 | .3895 | .5877 | .6536 | .2937 | ||||||||||
| rs807521 | 0 | .0037 | 0 | .092 | .2143 | .1515 | .9094 | .1362 | .0258 | .7375 | .1185 | .5574 | .7578 | .039 | .6557 | .4884 | .0683 | .7722 | .5247 | .7722 | .132 | .3626 | .5555 | .1851 | .6167 | .22 | .0489 | .6047 | .6208 | .6987 | .079 | .6875 | .3497 | .7444 | .9964 | .2454 | |||||||||||
| rs807525 | .0028 | 0 | .0785 | .175 | .1453 | .9349 | .1305 | .0189 | .6395 | .0969 | .4608 | .6702 | .063 | .5601 | .4145 | .0343 | .6702 | .5042 | .694 | .1081 | .2238 | .5335 | .2591 | .6578 | .2125 | .0469 | .5993 | .7016 | .7853 | .0601 | .7711 | .3425 | .6 | .9394 | .2385 | ||||||||||||
| rs807507 | .0071 | .0006 | .8807 | .0006 | .0094 | .0016 | .5956 | .9359 | .3609 | .6669 | .6281 | .3387 | .3341 | .5951 | .5357 | .5847 | .0079 | .7838 | .0274 | .9329 | .0071 | .362 | .5488 | .0523 | .131 | .127 | .0263 | .0128 | .0003 | .2322 | .0764 | .6415 | .5721 | .4135 | |||||||||||||
| rs807508 | .1803 | .1291 | .172 | .7861 | .1549 | .0115 | .4901 | .0401 | .3299 | .3315 | .0262 | .2711 | .1635 | .0105 | .3406 | .6617 | .8922 | .2048 | .1168 | .6666 | .1568 | .4629 | .2337 | .0548 | .6208 | .7879 | .9235 | .0477 | .7916 | .3717 | .8135 | .7039 | .2677 | ||||||||||||||
| k_ex5 | .2905 | .6827 | 0 | .9263 | .3064 | .1041 | .0308 | .8618 | .0655 | .9103 | .2791 | .1082 | .0042 | .0504 | 0 | .0063 | .8635 | .003 | 0 | .5328 | .7743 | 0 | .0033 | .0075 | .0024 | .0277 | .0679 | .0617 | .0001 | .1327 | .4498 | .0001 | |||||||||||||||
| rs4504469 | .0004 | .0017 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .8618 | .0001 | 0 | 0 | 0 | .4671 | .2118 | .1212 | 0 | 0 | .0529 | 0 | .1705 | .0008 | .0161 | .1248 | ||||||||||||||||
| rs4576240 | .3389 | 0 | .0016 | .0018 | .0038 | .0053 | .0004 | .4159 | .0002 | .0004 | .0433 | .0004 | .1914 | .5866 | .5501 | .5802 | .1817 | .6429 | .8063 | .2768 | 0 | .6392 | .0026 | .0159 | .0505 | .0022 | .3978 | .3702 | 0 | .3093 | |||||||||||||||||
| rs4472344 | .3464 | .0012 | .0646 | .0055 | .0626 | .024 | .1541 | .0193 | .0244 | .2866 | .0247 | 0 | .4124 | .0301 | .4089 | 0 | .2027 | .856 | .4103 | .1206 | .7316 | .0732 | .0756 | .0001 | 0 | .536 | .0053 | 0 | .4413 | ||||||||||||||||||
| rs4236032 | .0001 | .0002 | .0006 | .0005 | 0 | .4247 | 0 | 0 | .0433 | 0 | .1831 | .5656 | .4853 | .5823 | .1739 | .6636 | .7782 | .9687 | 0 | .6333 | .0004 | .0027 | .0115 | .0003 | .3802 | .5492 | 0 | .9616 | |||||||||||||||||||
| rs2745333 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .0013 | .862 | 0 | 0 | 0 | 0 | .2784 | .0511 | .1116 | 0 | 0 | .0001 | 0 | .1462 | .0004 | .0006 | .0976 | ||||||||||||||||||||
| rs2179515 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .0046 | 0 | .7955 | 0 | .0046 | .0001 | .0002 | .0111 | .0009 | .2674 | 0 | 0 | 0 | 0 | .0518 | .0253 | 0 | .095 | |||||||||||||||||||||
| rs761101 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | .0001 | 0 | .8642 | 0 | .0001 | 0 | 0 | .0673 | 0 | .177 | 0 | 0 | 0 | 0 | .0117 | .0005 | 0 | .0438 | ||||||||||||||||||||||
| rs6456624 | 0 | 0 | 0 | 0 | 0 | 0 | .0012 | 0 | .7687 | 0 | .0011 | 0 | .0002 | .01 | .0001 | .2333 | 0 | 0 | 0 | 0 | .0007 | .1547 | 0 | .0006 | |||||||||||||||||||||||
| rs2328846 | 0 | 0 | 0 | 0 | 0 | .0001 | 0 | .7408 | 0 | .0001 | 0 | 0 | .01 | 0 | .1617 | 0 | 0 | 0 | 0 | .0095 | .0006 | 0 | .0375 | ||||||||||||||||||||||||
| rs2235676 | 0 | 0 | 0 | 0 | .0445 | 0 | .0014 | 0 | .0449 | 0 | 0 | .2895 | .0479 | .4806 | .0013 | .0045 | .0001 | .0013 | .1978 | 0 | .018 | .1053 | |||||||||||||||||||||||||
| rs2038137 | 0 | 0 | 0 | 0 | 0 | .4387 | 0 | 0 | 0 | 0 | .0076 | 0 | .1503 | 0 | 0 | 0 | 0 | .9581 | .0014 | 0 | .6134 | ||||||||||||||||||||||||||
| k_pr_del | 0 | 0 | .0001 | 0 | .7937 | 0 | .0001 | 0 | 0 | .0102 | 0 | .1628 | 0 | 0 | 0 | 0 | .0534 | .0006 | 0 | .126 | |||||||||||||||||||||||||||
| k_pr_1 | 0 | .479 | 0 | 0 | 0 | .4741 | 0 | 0 | 0 | .0087 | .0429 | .0001 | .0006 | 0 | .0021 | .1067 | .1029 | .0064 | .1996 | ||||||||||||||||||||||||||||
| rs1555090 | .0001 | 0 | .6587 | 0 | .0001 | 0 | 0 | .01 | 0 | .1632 | 0 | 0 | 0 | 0 | .0097 | .0004 | 0 | .0375 | |||||||||||||||||||||||||||||
| rs2294689 | .3068 | .0025 | .0983 | 0 | .0752 | .58 | .2814 | .0338 | 0 | .0028 | .0038 | 0 | 0 | .3926 | .0001 | 0 | .9699 | ||||||||||||||||||||||||||||||
| rs3033236 | 0 | 0 | .3093 | 0 | 0 | 0 | .0043 | .5153 | 0 | 0 | 0 | 0 | .149 | .0005 | .0044 | .0693 | |||||||||||||||||||||||||||||||
| rs3212232 | 0 | .0022 | .0007 | .0051 | .0199 | 0 | .2856 | 0 | 0 | 0 | 0 | .0482 | 0 | 0 | .0162 | ||||||||||||||||||||||||||||||||
| rs2143340 | .0992 | 0 | 0 | 0 | .0149 | .4268 | 0 | 0 | 0 | 0 | .152 | .0006 | .0118 | .0716 | |||||||||||||||||||||||||||||||||
| rs2056998 | .0757 | .58 | .2814 | .0335 | 0 | .0031 | .0042 | 0 | 0 | .3933 | .0001 | 0 | .9674 | ||||||||||||||||||||||||||||||||||
| rs1061925 | 0 | .1208 | .0479 | .5284 | 0 | 0 | 0 | 0 | .2551 | 0 | .0205 | .1508 | |||||||||||||||||||||||||||||||||||
| th_ex1 | .1477 | .3348 | .6274 | 0 | .0002 | 0 | .0004 | .3006 | 0 | .4169 | .2219 | ||||||||||||||||||||||||||||||||||||
| tt_th_del | .0663 | .7297 | .0058 | .0051 | .0009 | .0044 | .4656 | .0005 | .0515 | .6652 | |||||||||||||||||||||||||||||||||||||
| rs3181228 | .4441 | 0 | 0 | 0 | 0 | .157 | 0 | 0 | .0852 | ||||||||||||||||||||||||||||||||||||||
| rs2328847 | .2303 | .2307 | .4358 | .2261 | .7591 | .22 | .2453 | .7137 | |||||||||||||||||||||||||||||||||||||||
| rs1555088 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||||||||||||||||||||||||||||||||||
| rs926529 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||||||||||||||||||||||||||||||||||
| rs1885211 | 0 | .0004 | 0 | .0014 | .0011 | ||||||||||||||||||||||||||||||||||||||||||
| rs2092404 | .1146 | 0 | .0595 | .1223 | |||||||||||||||||||||||||||||||||||||||||||
| th_ex3 | .0325 | .1293 | 0 | ||||||||||||||||||||||||||||||||||||||||||||
| rs3756814 | 0 | .1613 | |||||||||||||||||||||||||||||||||||||||||||||
| rs6456632 | .0614 | ||||||||||||||||||||||||||||||||||||||||||||||
Electronic-Database Information
The URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/
- GNF SymAtlas, http://symatlas.gnf.org/SymAtlas/
- LocusLink, http://www.ncbi.nlm.nih.gov/LocusLink/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RD and the 6p23-21.3 region)
- University of California–Santa Cruz Genome Bioinformatics, http://genome.ucsc.edu/
- Vega Human Genome Browser, http://vega.sanger.ac.uk/Homo_sapiens/
References
- Abecasis GR, Cardon LR, Cookson WO (2000) A general test of association for quantitative traits in nuclear families. Am J Hum Genet 66:279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 [DOI] [PubMed] [Google Scholar]
- Aggarwal BB (2003) Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol 3:745–756 [DOI] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC (1994) Quantitative trait locus for reading disability on chromosome 6. Science 266:276–279 [DOI] [PubMed] [Google Scholar]
- ——— (1995) Quantitative trait locus for reading disability: correction. Science 268:1553 [DOI] [PubMed] [Google Scholar]
- Castles A, Coltheart M (1993) Varieties of developmental dyslexia. Cognition 47:149–180 [DOI] [PubMed] [Google Scholar]
- Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, Smith SD (2004) Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: linkage and association analyses. Hum Genet 115:128–138 [DOI] [PubMed] [Google Scholar]
- DeFries JC, Fulker DW, LaBuda MC (1987) Evidence for a genetic aetiology in reading disability of twins. Nature 329:537–539 [DOI] [PubMed] [Google Scholar]
- Fisher SE. Tangled webs: tracing the connections between genes and cognition. Cognition (in press) [DOI] [PubMed] [Google Scholar]
- Fisher SE, DeFries JC (2002) Developmental dyslexia: genetic dissection of a complex cognitive trait. Nat Rev Neurosci 3:767–780 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Francks C, Marlow AJ, MacPhie IL, Newbury DF, Cardon LR, Ishikawa-Brush Y, Richardson AJ, Talcott JB, Gayan J, Olson RK, Pennington BF, Smith SD, DeFries JC, Stein JF, Monaco AP (2002) Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat Genet 30:86–91 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP (1999) A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet 64:146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francks C, MacPhie IL, Monaco AP (2002) The genetic basis of dyslexia. Lancet Neurol 1:483–490 [DOI] [PubMed] [Google Scholar]
- Fridmacher V, Kaltschmidt B, Goudeau B, Ndiaye D, Rossi FM, Pfeiffer J, Kaltschmidt C, Israel A, Memet S (2003) Forebrain-specific neuronal inhibition of nuclear factor-κB activity leads to loss of neuroprotection. J Neurosci 23:9403–9408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayan J, Olson RK (2001) Genetic and environmental influences on orthographic and phonological skills in children with reading disabilities. Dev Neuropsychol 20:483–507 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Genetic and environmental influences on individual differences in printed word recognition. J Exp Child Psychol 84:97–123 [DOI] [PubMed] [Google Scholar]
- Gayan J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, Pennington BF, DeFries JC (1999) Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet 64:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg TE, Weinberger DR (2004) Genes and the parsing of cognitive processes. Trends Cogn Sci 8:325–335 [DOI] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Hart LA, Speed WC, Shuster A, Pauls DL (1997) Susceptibility loci for distinct components of developmental dyslexia on chromosomes 6 and 15. Am J Hum Genet 60:27–39 [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls DL (2000) Chromosome 6p influences on different dyslexia-related cognitive processes: further confirmation. Am J Hum Genet 66:715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DE, Gayan J, Ahn J, Won TW, Pauls D, Olson RK, DeFries JC, Wood F, Pennington BF, Page GP, Smith SD, Gruen JR (2002) Evidence for linkage and association with reading disability on 6p21.3-22. Am J Hum Genet 70:1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Lee BH, Yoshimatsu K, Maeda A, Ochiai K, Morimatsu M, Araki K, Ogino M, Morikawa S, Arikawa J (2003) Association of the nucleocapsid protein of the Seoul and Hantaan hantaviruses with small ubiquitin-like modifier-1-related molecules. Virus Res 98:83–91 [DOI] [PubMed] [Google Scholar]
- Londin ER, Meng H, Gruen JR (2003) A transcription map of the 6p22.3 reading disability locus identifying candidate genes. BMC Genomics 4:25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow AJ, Fisher SE, Francks C, MacPhie IL, Cherny SS, Richardson AJ, Talcott JB, Stein JF, Monaco AP, Cardon LR (2003) Use of multivariate linkage analysis for dissection of a complex cognitive trait. Am J Hum Genet 72:561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlow AJ, Fisher SE, Richardson AJ, Francks C, Talcott JB, Monaco AP, Stein JF, Cardon LR (2001) Investigation of quantitative measures related to reading disability in a large sample of sib-pairs from the UK. Behav Genet 31:219–230 [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D (2003) NF-κB functions in synaptic signaling and behavior. Nat Neurosci 6:1072–1078 [DOI] [PubMed] [Google Scholar]
- Paterson SJ, Brown JH, Gsodl MK, Johnson MH, Karmiloff-Smith A (1999) Cognitive modularity and genetic disorders. Science 286:2355–2358 [DOI] [PubMed] [Google Scholar]
- Paulesu E, Demonet JF, Fazio F, McCrory E, Chanoine V, Brunswick N, Cappa SF, Cossu G, Habib M, Frith CD, Frith U (2001) Dyslexia: cultural diversity and biological unity. Science 291:2165–2167 [DOI] [PubMed] [Google Scholar]
- Pei H, Yordy JS, Leng Q, Zhao Q, Watson DK, Li R (2003) EAPII interacts with ETS1 and modulates its transcriptional function. Oncogene 22:2699–2709 [DOI] [PubMed] [Google Scholar]
- Pennington BF (1997) Using genetics to dissect cognition. Am J Hum Genet 60:27–39 [PMC free article] [PubMed] [Google Scholar]
- Pettersson F, Jonsson O, Cardon LR. GOLDsurfer: three dimensional display of linkage disequilibrium. Bioinformatics (in press) [DOI] [PubMed] [Google Scholar]
- Pype S, Declercq W, Ibrahimi A, Michiels C, Van Rietschoten JG, Dewulf N, de Boer M, Vandenabeele P, Huylebroeck D, Remacle JE (2000) TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-κB activation. J Biol Chem 275:18586–18593 [DOI] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, et al (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet 29:223–228 [DOI] [PubMed] [Google Scholar]
- Rohde K, Fürst R (2001) Haplotyping and estimation of haplotype frequencies for closely linked biallelic multilocus genetic phenotypes including nuclear family information. Hum Mutat 17:289–295 [DOI] [PubMed] [Google Scholar]
- Scerri TS, Fisher SE, Francks C, MacPhie IL, Paracchini S, Richardson AJ, Stein JF, Monaco AP. Putative functional alleles of DYX1C1 are not associated with dyslexia susceptibility in a large sample of sibling pairs from the UK. J Med Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Gilger JW, Pennington BF (1996) Dyslexia and other specific learning disorders. Churchill Livingston, New York [Google Scholar]
- Streets AJ, Newby LJ, O’Hare MJ, Bukanov NO, Ibraghimov-Beskrovnaya O, Ong AC (2003) Functional analysis of PKD1 transgenic lines reveals a direct role for polycystin-1 in mediating cell-cell adhesion. J Am Soc Nephrol 14:1804–1815 [DOI] [PubMed] [Google Scholar]
- Taipale M, Kaminen N, Nopola-Hemmi J, Haltia T, Myllyluoma B, Lyytinen H, Muller K, Kaaranen M, Lindsberg PJ, Hannula-Jouppi K, Kere J (2003) A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci USA 100:11553–11558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson ME (1982) Assessment of children with specific reading difficulties using the British Ability Scales. Brit J Psychol 73:461–478 [DOI] [PubMed] [Google Scholar]
- Turic D, Robinson L, Duke M, Morris DW, Webb V, Hamshere M, Milham C, Hopkin E, Pound K, Fernando S, Grierson A, Easton M, Williams N, Van Den Bree M, Chowdhury R, Gruen J, Stevenson J, Krawczak M, Owen MJ, O’Donovan MC, Williams J (2003) Linkage disequilibrium mapping provides further evidence of a gene for reading disability on chromosome 6p21.3-22. Mol Psychiatry 8:176–185 [DOI] [PubMed] [Google Scholar]
- Wigg KG, Couto JM, Feng Y, Anderson B, Cate-Carter TD, Macciardi F, Tannock R, Lovett MW, Humphries TW, Barr CL (2004) Support for EKN1 as the susceptibility locus for dyslexia on 15q21. Mol Psychiatry (in press) [DOI] [PubMed] [Google Scholar]
- Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, Edser P, Bhattacharyya S, Dunham A, Adcock IM, Pulleyn L, Barnes PJ, Harper JI, Abecasis G, Cardon L, White M, Burton J, Matthews L, Mott R, Ross M, Cox R, Moffatt MF, Cookson WO (2003) Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet 34:181–186 [DOI] [PubMed] [Google Scholar]