Abstract
We investigated the genetic aspects of the large sex bias in the prevalence of autism spectrum disorder by monitoring changes in linkage when the family set for an affected sibling pair genome scan is subdivided on the basis of the sex of affected children. This produces a significant excess in the total number of linkage peaks (P=1.3×10-8) and identifies a major male-specific linkage peak at chromosome 17q11 (P<.01). These results suggest that sexual dichotomy is an important factor in the genetics of autism; the same strategy can be used to explore this possibility in other complex disorders that exhibit significant sex biases.
It is well established that male and female brains develop, are structured, and function differently. Indeed, a large body of research has accumulated that shows not only that males and females process input in different ways (Shaywitz et al. 1995; Rhodes and Rubin 1999; Wrase et al. 2003) but also that this sexual dichotomy extends to the macroscopic structures of the brain (Giedd et al. 1997; Wisniewski 1998). Genes located on the X chromosome and those genes directly related to the function of sex-related hormones are clear candidates. It is clear that sex hormones play many roles in normal male and female development. There is also increasing evidence that differentially regulated genes on the autosomes are integrally involved in normal sex-specific brain development (Reisert and Pilgrim 1991; Jin et al. 2001; Dewing et al. 2003). It remains unclear to what extent these regulatory effects may ultimately relate to hormones or X-chromosomal genes. Although the orchestration of all the factors involved in normal male and female development and functioning is complex and has only begun to be understood, it is evident that there are substantial differences between the sexes at the molecular level.
Many human diseases also show a striking level of sex bias in terms of disease prevalence and severity. One of the prime examples of such a disorder is autism spectrum disorder (MIM 209850) (Folstein and Rosen-Sheidley 2001). Autism is the most severe form of a set of complex neurobehavioral disorders that are marked by impaired language development and social reciprocity and by the stereotyped and repetitive behaviors. The male:female ratio of idiopathic autism is 4–10:1, and this ratio increases as the intelligence of the affected individuals increases (Folstein and Rosen-Sheidley 2001). There is a noticeable male bias in all of the pervasive developmental disorders, of which autism is the archetypal form. The possible exception is Rett syndrome (X-linked dominant), which affects predominantly females, primarily because most affected males are not viable. Other neurodevelopmental disorders, including dyslexia (Lambe 1999) and attention-deficit/hyperactivity disorder (Swanson et al. 1998), also show a male bias in prevalence. Although there is some evidence of X-chromosome contribution to aspects of complex social behavior (Skuse 1999), no association with other cognitive features—such as attention, executive functioning, or language—that underlie these neurodevelopmental disorders has been identified that can account for the strong sex bias. We reasoned that, since sex bias is a prominent and consistent feature of many neurodevelopmental disorders, it should be incorporated into the search for susceptibility alleles.
To conduct a genomewide survey to search for potential sex-related loci in autism, we started with linkage-scan data from a large affected sibling pair (ASP) data set (Autism Genetic Resource Exchange [AGRE]) and then subdivided the families for further scans that were intended to highlight the sex differences. The subdivision was based on the rationale that, under the hypothesis that at least some of the major genetic risk factors for autism differ between males and females, an affected female child is positive evidence that a family carries the relevant female risk factors, whereas families that have (so far) produced only affected male children are enriched for carrying the male risk factors. Thus, splitting the family set on the basis of the presence of an affected female child should stratify the cohort with autism into groups that are more homogeneous for sex-sensitive risk loci.
An essential addition to this basic strategy is a procedure for interpretation of the results in the presence of confounding biases. Specifically, the linkage scans performed on these sex-split sets are biased to have the linkage peaks present in the original unsplit set. In fact, the expected behavior would be that the LOD scores from the original genome scan would simply be reduced in proportion to the reduction in sample size in each subset. To account for this, we devised the following statistical methodology: recall that, for assessment of the significance of the original scan, the null hypothesis is that the autism cohort does not differ from the general population or, more specifically that the ASP identical-by-descent (IBD)–sharing probabilities (z0,z1,z2) do not differ from the Mendelian values of (0.25,0.50,0.25). In contrast, the null hypothesis for the splitting study is either that the split sets are a random division of the original cohort or that their splitting statistics would be obtained by a random sampling from the (z0,z1,z2) fractions that exist in the cohort. Thus, we propose to evaluate the primary significance of the splitting by comparing it with empirical results that are based on many matched random splittings. However, once we accept the split scans as meaningful subdivisions of the original data set, it is also interesting to assess them as genome scans in their own right—that is, relative to the null hypothesis of Mendelian sharing. For comparison of any scan (original or split) with this null hypothesis, we compare the observed scan with the corresponding random simulation scan data generated by genome scans of ASP sets created by the random generation of offspring-genotype data from the parental genotype data.
In the present study, genotype and phenotype data from 257 nuclear families with two or more affected children were obtained from AGRE, with the exclusion of MZ twins and individuals with nonidiopathic autism (Yonan et al. 2003). Affected children include those who have received a diagnosis of full autism, not quite autism (NQA), and broad-spectrum disorder. Sample details, diagnostic criteria, and genotyping procedures are described elsewhere (Geschwind et al. 2001; Yonan et al. 2003). We typed 408 markers at an average density of 8 cM, as described elsewhere, and the raw data have been made publicly available on the AGRE Web site (Yonan et al. 2003). Sibships were subdivided into two groups—the male-only (MO) group and the female-containing (FC) group—on the basis of the presence of at least one affected female within the family. The stratification of the data resulted in two comparably and reasonably sized genome-scan sets, with the MO group consisting of 148 (58%) independent nuclear families and the FC group consisting of 109 (42%) families (table 1). Within a family, each possible ASP was counted as one ASP. Therefore, a family with only two affected children contributed one ASP to the total number of ASPs of that group, whereas a family with three affected children contributed three possible ASPs to the total. The majority (77% in the MO group; 79% in the FC group) of affected children in both groups received a diagnosis of full autism.
Table 1.
Sample Characteristics
|
Sample |
||
| Characteristic | MO | FC |
| No. of families | 148 | 109 |
| No. of autistic individuals | 243 | 189 |
| No. of NQA-affected individualsa | 27 | 10 |
| No. of broad spectrum–affected individuals | 44 | 39 |
| Total no. of affected individuals | 314 | 238 |
| Total no. of ASPs | 170 | 145 |
See Geschwind et al. (2001) for more information.
Complete genome scans for linkage were performed on the unsplit family set, on the MO and FC sex-split family sets, and on various random simulation data sets described below. The scans were performed using Genehunter (version 1.3), with the nonparametric IBD-sharing maximum-likelihood score (MLS) for the autosomes, and Genehunter-Plus (version 1.2) with the IBD-sharing NPL score for the X chromosome (X-NPL). The scans were performed using the Haldane mapping function, with all ASPs weighted equally; sharing LOD scores were computed using the possible triangle method; and no assumptions were made about the mode of inheritance.
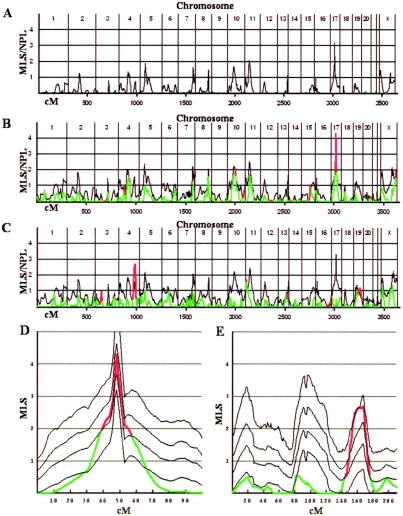
Across the genome, we calculated two different P value curves on the basis of the corresponding LOD score curves. The first is the local empiric P value for the local LOD score, which measures the significance of LOD scores relative to those from samples with no biases in sharing. This was obtained—for the original, MO, and FC scans—by performance of 1,000 randomly simulated genome scans in which offspring genotypes for each simulation were randomly generated from parental genotypes by assignment of random parental haplotypes and by use of appropriate intermarker recombination frequencies. These data provide the empirical probability of observing a LOD score of any given size at any given location throughout the genome, relative to the null hypothesis of Mendelian sharing. The second P value is the local sex-stratification P value, which is based on the LOD score in the original scan and which measures the significance of the enhancement in LOD score achieved by the splitting. To obtain this, we generated 1,000 random divisions of the 257 families analyzed in the original genome scan into two size-matched groups (consisting of 109 and 148 families) and performed linkage analysis for each sample subset. This provided the empirical probability of observing any LOD score at any given location throughout the genome, in either split scan, given the LOD scores observed in the given scan. The original genome scan LOD score is plotted in full (fig. 1A), and LOD scores obtained from the observed MO and FC families are also shown in full (fig. 1B and 1C), with the sex-stratification–based 95% CI LOD curves and regions of significant enhancement highlighted for reference. Close-up figures with 99% and 99.9% CI curves (based on 10,000 splitting simulations) are also shown for chromosomes of special interest (fig. 1D and 1E).
Figure 1.
Sex-stratified genome scans, with CIs and highlighted regions. A, Original genome scan based on all families. B and C, The genome scan for MO and FC families, respectively, shown as multicolored lines. The black line represents the sex-stratification–based 95% CI curve; the red sections of the scan curve denote regions where the LOD score exceeds the 95% CI (sex-stratification P<.05), whereas the green portions are within the CI. D, Close-up of the MO family scan in panel B for chromosome 17, with the addition of black lines that show the original genome scan (lowest curve) and the 99% and 99.9% sex-stratification CI curves (upper two curves). E, Close-up of the FC family scan in panel C for chromosome 4, with the addition of black lines that show the original genome scan (lowest curve) and the 99% and 99.9% sex-stratification CI curves (upper two curves).
A total of 25 distinct regions of the genome showed locally significant enhancement of linkage scores in our sex-split strategy (sex-stratification P<.05) (fig. 1B and 1C); however, the LOD scores involved are usually low. The only peak that reached genomewide significance was at chromosome 17q11 in the MO families (original MLS=3.2; MO MLS=4.3; sex-stratification P<.01; MO empiric P=.008) (fig. 1D). This LOD peak is located at 53cM, according to the Marshfield map (Center for Medical Genetics), flanked by markers D17S1294 and D17S798. The next largest and most significantly enhanced peak was at 4q32.3-35.1 in the FC families, although the peak does not reach genomewide significance (original MLS=0.76; FC MLS=2.7; sex-stratification P<.003; FC empiric P=.15) (fig. 1E). Two additional peaks on chromosomes 10q (original MLS=1.7; MO MLS=2.1; sex-stratification P<.03) and Xq (original X-NPL=0.87; MO X-NPL=2.0; sex-stratification P<.02) were detected in the MO families, but neither LOD score approaches genomewide significance.
It is important to assess whether such local peak enhancements remain significant when we correct for the multiple testing at many regions of the genome. There is some ambiguity as to how to perform this correction most appropriately, but a reasonable approach would be as follows. We can condition on the peaks “clearly” identified in the original scan as given. The overall significance of a particular enhancement at the peaks “clearly identified” in the original scan should be Bonferroni corrected for independent multiple testing at the total number of “comparable” peaks in the original scan. By extension, for peaks that appear essentially de novo in the split scans, the significances (both degree of enhancement and non-Mendelian sharing) should be corrected for multiple testing across the entire genome, as judged by the genomewide frequency of similar local results in the corresponding simulations, which is the most conservative form of multiple-testing correction. Under this approach, the enhancement of the dominant peak in the original scan at 17q11 requires no multiple-testing correction since it was uniquely identified. At most, it would require a factor of 2–4 for the number of comparable major peaks in the original scan. Either way, this still suggests genomewide-significant sex-related enhancement at 17q11. Similarly, the 4q32 region identified in the FC scan had a local empiric P value of .06 (MLS=0.76) in the original scan. The region contained 18 peaks of equal or greater local significance in the original scan, so its local sex-stratification enhancement of 0.003 should be corrected by this factor for multiple testing—which yields P=.054, a value that is marginally significant after correction. However, it would not be significant if it were instead considered to appear de novo in the FC scan and were given a full genomewide correction. The other two major enhanced peaks, at 10q and Xq, do not remain significant after similar correction for multiple testing of comparable peaks in the original scan.
Another important part of the splitting analysis is assessment of whether the splitting has in fact produced a global, significant enhancement of linkage scores. For this purpose, we introduced a global measure of how many “interesting LOD score peaks” are identified by the splitting. For the specific statistic, we count the number of disjoint regions in the two split scans in which both (a) the splitting produced a significant enhancement of the LOD score (sex-stratification P⩽.05) and (b) the resultant LOD score was nominally significant (local empirical P⩽.05); note that the corresponding LOD score threshold varies across the genome but has a median of 0.70 for the MLS and 1.1 for the X-NPL score. The sex-split strategy produced a count of 16 peaks (8 from MO; 8 from FC), whereas 1,000 randomly generated splittings (sets matched by size with the sex-split sets) resulted in an average count of 3.8 peaks (SD 1.5 peaks). The 1,000 simulations never produced >10 peaks; thus, the simulation-based bound on the sex-splitting significance is P<.001. Moreover, the simulated-peak counts quite accurately fit a Poisson distribution (as expected for random counting data), which predicts that P=1.3×10-8. This demonstrates that the specific separation into MO and FC groups has a highly nonrandom relation to the underlying genetics of the autistic cohort and strongly supports the hypothesis that the sex-split groups are more homogeneous for a variety of sex-related genetic risk factors. The simulation data also suggest that 12 of the 16 peaks (95% CI 9–15) are independent of random splitting effects. This provides an estimate for the total number of sex-related risk loci detected.
In contrast, similar peak-counting statistics in the original genome scan do not produce strong evidence that the autistic cohort is distinct from the general population. For example, examination of disjoint regions in the original scan in which the local empiric P⩽.05 resulted in a peak count of 16—which is not significant on the basis of the simulations on random-mating data, which show an expected peak count of 14.1 (SD 4.0; empirical P=.16). Even a survey of all other possible peak-defining criteria of local empiric P⩽P, for 0<P<1, does not show a highly significant excess of peak counts at any particular threshold. The most significant excess occurs for regions where the local empiric P⩽P=.10 (corresponding LOD thresholds vary across the genome, but median values are MLS>0.47 or X-NPL>0.95), where the observed peak count was 31, whereas the expected count was 19.9 (SD 5.7; empirical P=.04). Thus, the original scan gave little statistical evidence that a genetically distinct cohort had been isolated and was, at best, suggestive of a number of (mostly minor) linkage peaks interspersed with even greater numbers of false-positive results; this suggests that the genetic heterogeneity of the disorder has undermined the power of the study. In contrast, the sex-splitting procedure provides strong evidence for identification of a real genetic distinction and clearly identifies a number of interesting peaks, with relatively few false-positive results.
It is striking that the region of genomewide-significant linkage found in the MO set corresponds to the region at 17q11 that was identified as suggestive in the original AGRE genome scan. However, the MO MLS score of 4.3 achieves genomewide significance (empiric P=.008). Moreover, since there is little of the original linkage signal remaining at this region within the FC families (MLS=0.50; local empiric P=.10), the excess IBD sharing in the MO families is sufficient to account for the excess sharing originally detected in the whole-family set. These observations suggest that this locus corresponds to a genetic risk allele that is specific for the MO set (fig. 1D).
The risk factor harbored at the 17q11 locus is unknown and offers an important target for future research. One highly studied candidate gene, the serotonin transporter (SLC6A4), is located directly under this peak. Recently, other groups have published nominal associations with polymorphisms located in this gene (Conroy et al. 2004; McCauley et al. 2004). In the present study, the closest microsatellite to the peak (D17S1294) is 142 kb away from this gene, and no alleles show evidence for association with autism.
It is tempting to simply interpret MO-specific and FC-specific linkage peaks as male-specific and female-specific risk loci, but this simple conclusion is erroneous. Proper biological interpretation requires careful reasoning and, in any case, remains uncertain, even if we assume that complicating factors, such as sex-related ascertainment bias, can be ignored. In the present study, a reasonable interpretation is that MO-specific peaks represent male-specific genetic risk factors, but FC-specific peaks pose risk to both females and males. For clarity, note that by “male-specific genetic risk factors” we mean that if those genetic factors were imposed equally on a male and a female embryo with otherwise similar genetic backgrounds, the male would be substantially more likely to develop autism; likewise, “female-specific genetic risk factors” would be more likely to affect females. Under these definitions, an MO-specific risk locus—such as 17q11—can reasonably be interpreted as a male-specific risk locus, since, if the constellation of factors including it conferred similar risk in females, there would be comparable numbers of male-male and male-female ASPs linked to these factors and the latter ASPs would produce a strong signal in the FC scan, contrary to what is observed. In contrast, peaks unique to the FC scan—such as 4q32—do not suggest female-specific risk factors. This is because, in most FC families, one of the affected siblings is male (families with only affected females are rare, just 6.5% of families in the present study); hence, the FC-specific susceptibility alleles that result in affected females apparently also confer substantial risk to males in those families. Indeed, mere comparable risk would suggest that the FC set should contain comparable numbers of female-male and female-female ASPs, whereas, in reality, there is a significant 5:1 (female-male:female-female) bias. Thus, the alleles would seem to pose an even greater risk to males, but perhaps a lower overall prevalence renders the MO or whole-family genome scans underpowered to detect these factors. Conversely, the rarity of female-female ASPs means we lack the power to detect true female-specific risk factors analogous to 17q11.
One additional inference is that, whereas the 17q11 risk factor appears specific for males with autism, it is not highly essential, since males with autism are abundantly represented in the FC set, yet there is little evidence of linkage to 17q11. This highlights a potentially interesting molecular dichotomy in males: the (more common) form of autism that is linked to 17q11 and the (less common) form that is not. This distinction is not directly observable in an individual at present, but an observable approximation that is further justified by the general validity of our sex-splitting data would be to divide the male patients with autism into two groups (similar to the FC and MO set, in the present study). This dichotomy may provide a useful basis for defining more-consistent subphenotypes, risk groups, or therapeutic response groups.
It is notable that other classes of complex diseases with a strong genetic component show a high female:male ratio. In addition to a number of neurobehavioral and neuropsychiatric disorders, various autoimmune disorders have high heritability, obvious genetic complexity, and strong sex biases in prevalence or severity. There is evidence that hormone levels play a role in autoimmune diseases, since symptoms are known to increase or decrease with pregnancy and menopause. Although some studies have linked autoimmune disorders to the X chromosome (Barbesino et al. 1998), other studies have implicated autosomal regions (Tomer et al. 2003), which makes it necessary for researchers to examine autosomal-based sex differences to explain the significant clinical sex biases. Techniques similar to those employed in the present study may be useful for such investigations.
Motivated by the observation of the strong sex bias in the prevalence of autism, we developed a strategy for using sex status to decrease the sample heterogeneity in ASP studies and we devised a statistical methodology for analyzing the results. When applied to the AGRE ASP sample, stratification of families on the basis of the presence of an affected female sibling produced a highly significant enhancement of the linkage scan data, highlighted a number of otherwise minor linkage peaks from the original scan, and clearly identified a male-specific region of linkage on chromosome 17q11. This strategy is suitable for large ASP studies of other complex genetic disorders that manifest a strong sex bias, and the analytical method can be generalized to include stratifications based on other phenotypic categories as well.
Acknowledgments
We thank the families (from the AGRE program) who participated in this study, the Cure Autism Now (CAN) Foundation for supporting the AGRE program, and the scientists who have provided oversight to the AGRE Consortium (listed below). Thanks to Maricela Alarcon and Matthew Ogdie for insightful conversations and technical assistance. This work was supported by National Institutes of Health grant MH64547 and by a Graduate Assistance in Areas of National Need predoctoral training fellowship (to J.L.S.). Members of AGRE Consortium are Daniel H. Geschwind, University of California, Los Angeles; Maja Bucan, University of Pennsylvania, Philadelphia; W. Ted Brown, New York State Institute for Basic Research in Developmental Disabilities, New York; Joseph D. Buxbaum, Mt. Sinai School of Medicine, New York; T. Conrad Gilliam, Columbia Genome Center, New York; David A. Greenberg, Mt. Sinai School of Medicine, New York; David H. Ledbetter, Emory University, Atlanta; Stanley F. Nelson, University of California, David Geffen School of Medicine, Los Angeles; Jonathan Pevsner, Kennedy Kreiger Institute, Baltimore; Gerard D. Schellenberg, University of Washington and Veterans Affairs Medical Center, Seattle; Carole Samango-Sprouse, Children’s National Medical Center, Baltimore; and Rudolph E. Tanzi, Massachusetts General Hospital, Boston.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Autism Genetic Resource Exchange, http://www.agre.org/
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/ (for the Marshfield map)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for autism spectrum disorder)
References
- Barbesino G, Tomer Y, Concepcion ES, Davies TF, Greenberg DA, International Consortium for the Genetics of Autoimmune Thyroid Disease (1998) Linkage analysis of candidate genes in autoimmune thyroid disease. II. Selected gender-related genes and the X-chromosome. J Clin Endocrinol Metab 83:3290–3295 10.1210/jc.83.9.3290 [DOI] [PubMed] [Google Scholar]
- Conroy J, Meally E, Kearney G, Fitzgerald M, Gill M, Gallagher L (2004) Serotonin transporter gene and autism: a haplotype analysis in an Irish autistic population. Mol Psychiatry 9:587–593 10.1038/sj.mp.4001459 [DOI] [PubMed] [Google Scholar]
- Dewing P, Shi T, Horvath S, Vilain E (2003) Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Brain Res Mol Brain Res 118:82–90 [DOI] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B (2001) Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet 2:943–955 10.1038/35103559 [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Sowinski J, Lord C, Iversen P, Shestack J, Jones P, Ducat L, Spence SJ, AGRE Steering Committee (2001) The Autism Genetic Resource Exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet 69:463–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Castellanos FX, Rajapakse JC, Vaituzis AC, Rapoport JL (1997) Sexual dimorphism of the developing human brain. Prog Neuropsychopharmacol Biol Psychiatry 21:1185–1201 10.1016/S0278-5846(97)00158-9 [DOI] [PubMed] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G (2001) The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet 29:389–395 10.1038/ng766 [DOI] [PubMed] [Google Scholar]
- Lambe EK (1999) Dyslexia, gender, and brain imaging. Neuropsychologia 37:521–536 10.1016/S0028-3932(98)00146-8 [DOI] [PubMed] [Google Scholar]
- McCauley JL, Olson LM, Dowd M, Amin T, Steele A, Blakely RD, Folstein SE, Haines JL, Sutcliffe JS (2004) Linkage and association analysis at the serotonin transporter (SLC6A4) locus in a rigid-compulsive subset of autism. Am J Med Genet 127B:104–112 [DOI] [PubMed] [Google Scholar]
- Reisert I, Pilgrim C (1991) Sexual differentiation of monoaminergic neurons—genetic or epigenetic? Trends Neurosci 14:468–473 10.1016/0166-2236(91)90047-X [DOI] [PubMed] [Google Scholar]
- Rhodes ME, Rubin RT (1999) Functional sex differences (“sexual diergism”) of central nervous system cholinergic systems, vasopressin, and hypothalamic-pituitary-adrenal axis activity in mammals: a selective review. Brain Res Brain Res Rev 30:135–152 [DOI] [PubMed] [Google Scholar]
- Shaywitz BA, Shaywitz SE, Pugh KR, Constable RT, Skudlarski P, Fulbright RK, Bronen RA, Fletcher JM, Shankweiler DP, Katz L, Gore JC (1995) Sex differences in the functional organization of the brain for language. Nature 373:607–609 10.1038/373607a0 [DOI] [PubMed] [Google Scholar]
- Skuse DH (1999) Genomic imprinting of the X chromosome: a novel mechanism for the evolution of sexual dimorphism. J Lab Clin Med 133:23–32 10.1053/lc.1999.v133.a94575 [DOI] [PubMed] [Google Scholar]
- Swanson JM, Sergeant JA, Taylor E, Sonuga-Barke EJ, Jensen PS, Cantwell DP (1998) Attention-deficit hyperactivity disorder and hyperkinetic disorder. Lancet 351:429–433 10.1016/S0140-6736(97)11450-7 [DOI] [PubMed] [Google Scholar]
- Tomer Y, Ban Y, Concepcion E, Barbesino G, Villanueva R, Greenberg DA, Davies TF (2003) Common and unique susceptibility loci in Graves and Hashimoto diseases: results of whole-genome screening in a data set of 102 multiplex families. Am J Hum Genet 73:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski AB (1998) Sexually-dimorphic patterns of cortical asymmetry, and the role for sex steroid hormones in determining cortical patterns of lateralization. Psychoneuroendocrinology 23:519–547 10.1016/S0306-4530(98)00019-5 [DOI] [PubMed] [Google Scholar]
- Wrase J, Klein S, Gruesser SM, Hermann D, Flor H, Mann K, Braus DF, Heinz A (2003) Gender differences in the processing of standardized emotional visual stimuli in humans: a functional magnetic resonance imaging study. Neurosci Lett 348:41–45 10.1016/S0304-3940(03)00565-2 [DOI] [PubMed] [Google Scholar]
- Yonan AL, Alarcón M, Cheng R, Magnusson PKE, Spence SJ, Palmer AA, Grunn A, Juo S-HH, Terwilliger JD, Liu J, Cantor RM, Geschwind DH, Gilliam TC (2003) A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet 73:886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]