Abstract
Facioscapulohumeral muscular dystrophy (FSHD) is associated with contractions of the D4Z4 repeat in the subtelomere of chromosome 4q. Two allelic variants of chromosome 4q (4qA and 4qB) exist in the region distal to D4Z4. Although both variants are almost equally frequent in the population, FSHD is associated exclusively with the 4qA allele. We identified three families with FSHD in which each proband carries two FSHD-sized alleles and is heterozygous for the 4qA/4qB polymorphism. Segregation analysis demonstrated that FSHD-sized 4qB alleles are not associated with disease, since these were present in unaffected family members. Thus, in addition to a contraction of D4Z4, additional cis-acting elements on 4qA may be required for the development of FSHD. Alternatively, 4qB subtelomeres may contain elements that prevent FSHD pathogenesis.
Facioscapulohumeral muscular dystrophy (FSHD or FSHD1A [MIM 158900]) is a progressive myopathy that is mainly characterized by an often-asymmetric wasting and weakness of the facial, shoulder, and upper-arm muscles. FSHD displays a large clinical variability in expression and progression and can also present with nonmuscular features like hearing impairment and subclinical retinovasculopathy (Padberg 2004). In severe cases, CNS symptoms such as mental retardation and epileptic seizures can be present (Funakoshi et al. 1998; Miura et al. 1998).
In the vast majority of patients, autosomal dominant FSHD is associated with a contraction of the D4Z4 repeat in the subtelomere of chromosome 4q. The D4Z4 repeat normally consists of a polymorphic array of 11–100 KpnI units, each 3.3 kb in size. In patients with FSHD, the D4Z4 repeat is reduced to 1–10 units (Wijmenga et al. 1992; van Deutekom et al. 1993). A rough inverse relationship has been established between the severity and progression of the disease and the residual D4Z4 repeat size (Lunt et al. 1995; Tawil et al. 1996).
A homologous and equally polymorphic repeat is present on chromosome 10q, but contractions of this repeat have not been associated with disease (Bakker et al. 1995; Lemmers et al. 2001; Zhang et al. 2001). Moreover, completely or partially translocated repeats have been identified on chromosomes 4 and 10 in ∼20% of the population (Lemmers et al. 1998; van Overveld et al. 2000). Nevertheless, only small repeats on chromosome 4 have been reported in patients with FSHD.
The D4Z4 repeat can be visualized on EcoRI-HindIII–double-digested DNA that has been hybridized with probe p13E-11 (Wijmenga et al. 1992) after separation by pulsed field gel electrophoresis (PFGE). This probe recognizes the D4F104S1 locus that lies just proximal to D4Z4 on chromosomes 4 and 10. Thus, after hybridization, four fragments will be visualized, two from each chromosome. Small sequence variations between chromosome 4–derived and chromosome 10–derived repeat units allow differentiation between both chromosomes by the use of specific restriction enzymes: chromosome 4–derived units are resistant to BlnI and are sensitive to XapI, whereas chromosome 10–derived units have the opposite characteristics (Deidda et al. 1996; Lemmers et al. 2001).
There is increasing evidence that the contraction of D4Z4 causes a local chromatin alteration as well as the transcriptional deregulation of genes by cis- or trans-acting mechanisms (Gabellini et al. 2002; Jiang et al. 2003; Masny et al. 2004). A change in local chromatin conformation is corroborated by the observation that FSHD alleles are hypomethylated at D4Z4 (van Overveld et al. 2003).
Apparently, a contraction of the D4Z4 repeat on chromosome 4 is not sufficient to cause disease, since FSHD is uniquely associated with only one of the two variants of 4qter. The two 4qter alleles (4qA and 4qB) differ by sequence variations distal to D4Z4. Although both alleles are almost equally common in the population, FSHD alleles are always of the 4qA type (Lemmers et al. 2002; van Geel et al. 2002). Since, in the control population, both alleles have an equal mitotic propensity to rearrange, it was proposed that a functional difference between both chromosome ends must explain the unique association of FSHD with 4qA. However, since these studies were performed using samples from anonymous blood donors, we could not exclude the possibility that small D4Z4 repeats on 4qB chromosomes are pathogenic. By studying three families (∼1% of Dutch patients with FSHD) that were ascertained by the unusual observation that all probands were heterozygous for an FSHD-sized allele, we here provide strong evidence that 4qB chromosomes that carry FSHD-sized D4Z4 repeats do not cause disease. The probes used for detailed D4Z4 repeat analysis, as well as their positions in the restriction map of chromosomes 4 and 10, are depicted in figure 1.
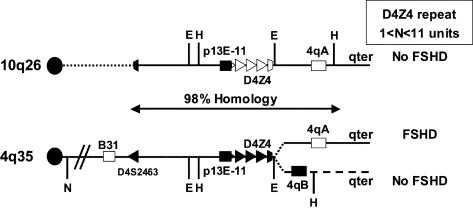
Figure 1.
Schematic presentation of the telomeric regions of chromosome 10q26 and the two variants of chromosome 4q35, 4qA and 4qB. The region distal to D4S2463 shows 98% homology between chromosomes 10q and 4q, except for the region distal to D4Z4 on the 4qB variant. Both 4q alleles are almost equally present in the population, but FSHD has been associated with only the 4qA variant. Chromosome 4–type D4Z4 repeats are depicted as blackened triangles, and the chromosome 10 D4Z4 repeats are shown as unblackened triangles. The locations of the EcoRI (E) and HindIII (H) restriction sites adjacent to D4Z4 are shown, as well as probes p13E-11 (blackened box), 4qB (blackened box), and 4qA (unblackened box). Chromosomal assignment depicted in figure 2G–2I makes use of the NotI restriction site (N) proximal to the chromosome 4–specific probe B31 (unblackened box).
In the first family (Rf215 [fig. 2A]), the proband (III-1) already had severe symptoms of FSHD at the age of 12 years. In addition, he presented with telecanthus, heterochromia iridis, and a white forelock, all features of Waardenburg syndrome (WS) (DeStefano et al. 1998). However, he had no hypertelorism. The father (II-1), paternal grandfather (I-1), and paternal uncle all had a slowly progressing form of FSHD; there was no history of muscle disease in the family of the mother (II-2) throughout three generations. Physical and neurological examinations of the mother, the maternal uncle (II-3), and the grandmother (I-4) were normal.
Figure 2.
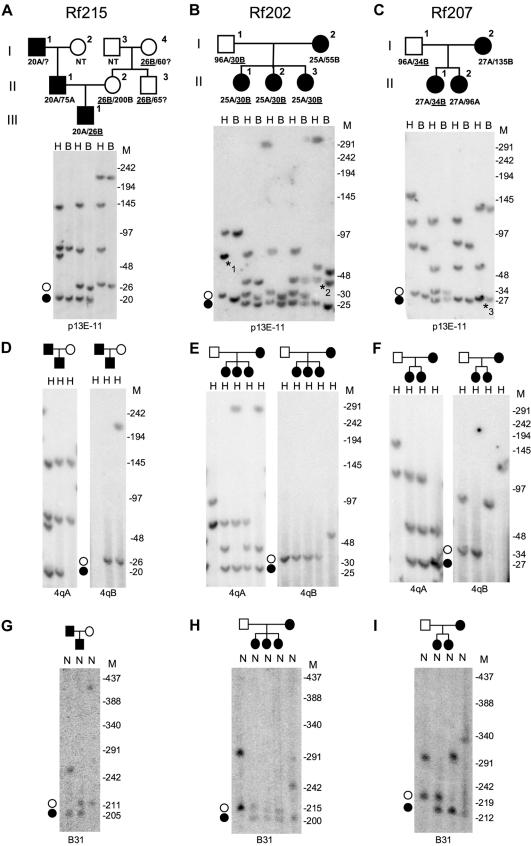
A–F, Southern blot analysis of samples from three families with FSHD (Rf215, Rf202, and Rf207). DNA was double-digested with EcoRI-HindIII (E) and EcoRI-BlnI (B), separated by PFGE, and hybridized with probe p13E-11. The FSHD-sized 4qB alleles (unblackened circles) of 26 kb, 30 kb, and 34 kb, respectively, are underlined (A–C). FSHD-causing 4qA alleles are indicated with blackened circles. For allelotyping (4qA/4qB), DNA was digested with HindIII, separated by PFGE, and subsequently hybridized with probes 4qA and 4qB (D–F). Marker sizes (in kb) are indicated on the right. A and D, In family Rf215, the affected proband (III-1) carried two chromosome 4–type FSHD-sized D4Z4 repeats. Allelotyping shows that the 20-kb D4Z4 repeat is of 4qA origin and was inherited from the affected father. The 26-kb D4Z4 repeat was inherited from the healthy mother and is of 4qB origin (D). Furthermore, the affected child carried chromosome 10 repeats of 70 kb and 145 kb. Segregation analysis showed that the FSHD-causing allele was inherited from the affected grandfather (I-1) and that the nonpathogenic short 4qB allele was inherited from the healthy maternal grandmother (I-4) and is also observed in a healthy uncle (II-3) of the affected proband (III-1) (A). B and E, In family Rf202, three affected sisters (II-1, II-2, and II-3) were identified as carriers of two FSHD-sized D4Z4 repeats. Allele-sizing and allelotyping identified a 25-kb 4qA allele and a 30-kb 4qB allele (B and E). The 30-kb 4qB allele was inherited from the healthy father (I-1), and the FSHD-causing 25-kb 4qA allele was inherited from the affected mother (I-2). Comigrating chromosome 10 alleles of 75 kb in the father are marked with “*1.” The mother carried a 45-kb chromosome 4–type allele (marked with “*2”), which originated from chromosome 10 (translocated chromosome 4–type repeat) and was passed to her daughters II-1 and II-3. C and F, In family Rf207, the oldest affected daughter (II-1) carried two FSHD-sized repeats. Allele-sizing and allelotyping identified a 27-kb 4qA allele and a 34-kb 4qB allele (C and F). The 34-kb 4qB allele was inherited from the healthy father (I-1), and the FSHD-causing 27-kb 4qA allele was inherited from the affected mother (I-2). Further analyses show that the FSHD allele in the mother comigrates with a chromosome 10 allele of 27 kb (marked with “*3”). G–I, Chromosome assignment of the D4Z4 repeats in three families with FSHD (Rf215, Rf202, and Rf207). NotI-digested DNA plugs and the chromosome 4–specific probe B31 were used. The NotI fragment is 185 kb larger than the EcoRI-HindIII fragment on chromosome 4. Marker sizes (in kb) are indicated on the right. G, The two chromosome 4–derived repeats of 20 kb and 26 kb in the compound heterozygous affected proband (III-1) in family Rf215 are visualized at 205 kb (20 kb + 185 kb) and 211 kb (26 kb + 185 kb). These results show that both FSHD-sized repeats do indeed reside on chromosome 4. Furthermore, the chromosomal assignment reveals that the chromosome 4–type 75-kb repeat in the father resides on chromosome 4 (75 kb + 185 kb = 260 kb), just like the 200-kb repeat in the mother (200 kb + 185 kb = 385 kb). H, Chromosomal assignment in family Rf202 shows that the two FSHD-sized alleles inherited by the three affected sisters (II-1, II-2, and II-3) are both located on chromosome 4. They are visualized at 210 kb (25 kb + 185 kb) and 215 kb (30 kb + 185 kb), corresponding to the paternal 25-kb repeat and the maternal 30-kb repeat, respectively. The other chromosome 4 alleles from the father and the mother are visualized at 281 kb (96 kb + 185 kb) and 240 kb (55 kb + 185 kb), respectively. I, The two FSHD-sized alleles in the affected daughter (II-1) of family Rf207 are located on chromosome 4 and are visible at 212 kb (27 kb + 185 kb) and 219 kb (34 kb + 185 kb).
PFGE analysis of the proband (III-1) demonstrated the presence of two BlnI-resistant D4Z4 repeats of 20 kb and 26 kb. In addition, chromosome 10–derived alleles of 70 kb and 145 kb were identified on the basis of their sensitivity to BlnI (fig. 2A).
The allelic origin of all D4Z4 fragments was determined on HindIII-digested DNA with the use of probes 4qA and 4qB. This showed that the 20-kb repeat was located on a 4qA chromosome, whereas the 26-kb fragment originated from a 4qB allele. As expected, both chromosome 10 alleles hybridized with probe 4qA (fig. 2D).
To identify the chromosomal origin of these fragments, NotI-digested DNA was hybridized with the chromosome 4–specific probe B31. The chromosome 4–specific probe B31 recognized the terminal NotI fragment on which D4Z4 resides; this yielded a fragment that was 185 kb larger than the observed D4Z4-containing EcoRI-HindIII fragment (Lemmers et al. 2003). Hybridization of probe B31 with NotI-digested DNA of the proband yielded chromosome 4–derived fragments of 205 kb (20 kb + 185 kb) and 211 kb (26 kb + 185 kb) (fig. 2G), which confirmed the chromosome 4 origin of both of the FSHD-sized D4Z4 repeats.
Segregation analysis demonstrated that the 20-kb 4qA allele was also present in the affected family members. Consequently, the 26-kb 4qB allele found in the proband was inherited from his healthy mother (II-2). This allele was also present in the unaffected grandmother (I-4) and in the unaffected uncle (II-3) of the proband (fig. 2A).
Since mutations in PAX3 have been associated with WS and since the first homeodomain of DUX4, a putative gene residing in each D4Z4 unit (Gabriels et al. 1999), is highly homologous to that of PAX3, we performed a mutation analysis for PAX3 in the proband by PCR amplification of all protein-encoding exons, including splice-donor and splice-acceptor sites, followed by direct sequence analysis (Pandya et al. 1996). No mutations were detected in PAX3 (data not shown).
In the second family (Rf202 [fig. 2B]), the proband (I-2) had been identified as having FSHD at the age of 33 years. At the age of 63 years, she presented with typical FSHD, including moderate, sometimes asymmetric, weakness and wasting of the facial, shoulder girdle, and upper-arm muscles. Her husband (I-1) was 64 years old and presented no abnormalities on clinical and neurological examination. All daughters (II-1, II-2, and II-3) showed a slowly progressing form of FSHD, with mild-to-moderate facial weakness, moderate-to-severe asymmetric shoulder girdle weakness, and moderate abdominal weakness. All three siblings also suffered from common but unrelated autoimmune diseases, including primary hypothyroidism and hypoparathyroidism (II-1); chronic aspecific respiratory affections and sarcoidosis (II-2); and ulcerative colitis, sclerotic cholangitis, and activated protein C resistance (II-3).
PFGE analysis revealed that all three sisters carried chromosome 4–derived D4Z4 repeats of 25 kb and 30 kb and a chromosome 10–derived D4Z4 repeat of 70 kb. In addition, individual II-2 carried a chromosome 10 allele of 300 kb, whereas individuals II-1 and II-3 carried a translocated chromosome 4–type D4Z4 repeat of 45 kb on chromosome 10 (fig. 2B).
Allelotyping performed using probes 4qA and 4qB showed that the 25-kb chromosome 4–derived repeat resides on 4qA, whereas the 30-kb repeat is located on a 4qB chromosome. The pathogenic 25-kb D4Z4 repeat was also identified in other affected family members (data not shown). The chromosome 10–derived repeats were recognized by probe 4qA, and the translocated 45-kb chromosome 4–derived repeat on chromosome 10 in individual I-2 was of 4qA origin (fig. 2E). As expected, the affected mother carried the 25-kb D4Z4 repeat on 4qA, whereas the healthy father carried the 30-kb 4qB-derived D4Z4 repeat.
After hybridization with probe B31, the chromosomal assignment revealed NotI fragments of 210 kb (25 kb + 185 kb) and 215 kb (30 kb + 185 kb) in all three sisters, which confirmed the presence of the 25-kb and 30-kb D4Z4 fragments on chromosome 4 (fig. 2H). In agreement with these findings, hybridization with probe p13E-11 revealed the additional chromosome 10 fragments and confirmed that the 45-kb chromosome 4–like allele in individuals I-2, II-1, and II-3 is translocated to chromosome 10 (data not shown).
Finally, in the third family (Rf207 [fig. 2C]), the mother (I-2) of the proband was identified as having FSHD at the age of 49 years. She showed mild facial weakness, moderate-to-severe shoulder girdle weakness, and bilateral foot extensor weakness. Her husband had no symptoms or signs of FSHD at examination. Her oldest daughter (II-1) had a medical history of Klippel-Trenaunay-Weber (KTW) syndrome and presented with mild facial weakness and asymmetric weakness and atrophy of the shoulder girdle and foot extensor muscles. The youngest daughter (II-2) presented with mild facial and shoulder paresis.
PFGE analysis revealed that patient II-1 carried the familial FSHD allele of 27 kb, which, as expected, resided on a 4qA chromosome. In addition, she carried a chromosome 4–derived repeat of 34 kb of 4qB origin and chromosome 10–derived repeats of 55 kb and 110 kb (fig. 2F). Again, all chromosome assignments were based on the sensitivity of the repeats for BlnI or XapI and were confirmed on NotI-digested DNA, which was hybridized with probe B31 (fig. 2C and 2I).
Patient II-1 inherited the disease allele from her affected mother, who also carried a 4qB allele of 135 kb and chromosome 10–derived repeats of 27 kb and 55 kb. Thus, the 34-kb repeat of 4qB origin was inherited from the healthy father.
Heterozygosity or homozygosity for two FSHD-sized D4Z4 alleles is very uncommon (∼1% in Dutch patients with FSHD). Recently, we described two families in which each proband was heterozygous for two D4Z4 repeats <38 kb and presented with a typical FSHD phenotype (Wohlgemuth et al. 2003). In addition, a patient was described who was homozygous for an FSHD allele because of consanguinity (Tonini et al. 2004a). All these alleles were on a 4qA allele/chromosome, the subtelomeric variant of 4q that is associated with the disease. It was suggested that a dose effect of both alleles might aggravate the disease, although this could not be confirmed in the homozygous patient.
In contrast to these patients, the heterozygous individuals reported here had one repeat that resided on a 4qA chromosome and another repeat that was located on a 4qB chromosome. In these cases, the repeat that comigrates with disease in these families resides on the 4qA chromosome, whereas the FSHD-sized repeat on the 4qB chromosome is always inherited from a healthy parent. Therefore, the presence of an FSHD-sized D4Z4 repeat on a 4qB chromosome in five healthy individuals without any myopathic features strongly suggests that contracted D4Z4 repeats on 4qB chromosomes are not associated with FSHD. Formerly, we could not rule out the possibility that D4Z4 repeats <38 kb on 4qB chromosomes might be associated with disease, since these data were derived from anonymous blood donors (Lemmers et al. 2002). However, in the present study, all 4qB repeats are well within the FSHD size range, and, despite the fact that nonpenetrance seems to be more prominent in the upper size range of the D4Z4 repeat (Lunt 2000; Tonini et al. 2004b), some myopathic features should have been noted in these individuals if D4Z4 repeats <38 kb on 4qB chromosomes cause FSHD. This nonpathogeneity of short D4Z4 repeats on 4qB chromosomes may also explain the rare finding of FSHD-sized D4Z4 repeats that do not cosegregate with disease in families with unrelated neuromuscular disorders (Auer-Grumbach et al. 2000).
It is interesting to note that all heterozygous patients described here showed additional diseases that are normally not associated with FSHD. At present, it is unclear whether or not these additional diseases are independent of the heterozygosity for FSHD-sized alleles on chromosomes 4qA and 4qB.
All three siblings in family Rf202 suffered from common but unrelated autoimmune diseases. A potential autoimmune mechanism in the muscle pathology of FSHD has long been debated. In at least 40% of biopsies, inflammatory infiltrations with a T-cell repertoire different from that in patients with myositis or other muscular dystrophies can be seen in the muscle of patients with FSHD (Arahata et al. 1995). In addition, a few cases have been reported in which the onset of FSHD seems to be related to the onset of a viral infection or vaccination (Tawil et al. 1993; Tupler et al. 1998).
The index patient in family Rf207 had KTW syndrome. KTW syndrome is commonly sporadic and might be a genetic condition. Putative loci, including the recently discovered susceptibility gene VG5Q (Tian et al. 2004), are not related to the FSHD locus. Nevertheless, it is interesting that both KTW syndrome and FSHD have been suggested to be caused by a similar disease model involving transcriptional derepression of essential disease genes (Gabellini et al. 2004).
Patient III-1 in family Rf215 showed several features common to WS, which is often caused by mutations in the transcription factor PAX3 (Baldwin et al. 1992; Tassabehji et al. 1992; DeStefano et al. 1998). Nevertheless, we did not find evidence of the presence of mutations in the PAX3 gene in this patient, although the mutation detection strategy does not exclude the presence of large exonic deletions, intronic mutations, or mutations affecting PAX3 gene transcription regulation. PAX3 and its orthologue, PAX7, are involved in muscle differentiation during embryogenesis and in adult muscle regeneration (Buckingham et al. 2003), and their homeodomains are closely related to the first homeodomain of the FSHD candidate gene DUX4. Although expression analyses of DUX4 have failed to identify its gene product, evidence of the DUX4 protein was recently found in myoblast cultures of patients with FSHD (Coppee et al. 2004). It is tempting to speculate that ectopic expression of DUX4 may have caused this WS-like phenotype, but this hypothesis cannot be pursued, since muscle biopsies from this patient are currently unavailable.
This study demonstrates that D4Z4 repeats <38 kb that reside on chromosome 4qB do not cause FSHD. Therefore, in addition to a contraction of D4Z4, additional cis-acting elements on 4qA may be required for the development of FSHD. Alternatively, 4qB subtelomeres may contain elements that prevent FSHD pathogenesis. Regardless of the exact mechanism, this study demonstrates that common subtelomeric polymorphisms can be of importance to the development of human disease. Since large variant alleles also exist for other chromosome ends (Wilkie et al. 1991; Macina et al. 1994; Trask et al. 1998; Mefford and Trask 2002), it will be interesting to study their role in common diseases that are often associated with subtelomeric rearrangements, such as idiopathic mental retardation (Flint et al. 1995).
Acknowledgments
This research was made possible by the Prinses Beatrix Fonds, the Stichting Spieren voor Spieren, the Muscular Dystrophy Association USA, the FSH Society, the Stichting FSHD, the Shaw family, and the National Institutes of Health.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FSHD1A) [PubMed]
References
- Arahata K, Ishihara T, Fukunaga H, Orimo S, Lee JH, Goto K, Nonaka I (1995) Inflammatory response in facioscapulohumeral muscular dystrophy (FSHD): immunocytochemical and genetic analyses. Muscle Nerve 2:S56–S66 [PubMed] [Google Scholar]
- Auer-Grumbach M, Wagner K, Strasser-Fuchs S, Loscher WN, Fazekas F, Millner M, Hartung HP (2000) Clinical predominance of proximal upper limb weakness in CMT1A syndrome. Muscle Nerve 23:1243–1249 [DOI] [PubMed] [Google Scholar]
- Bakker E, Wijmenga C, Vossen RH, Padberg GW, Hewitt J, van der Wielen M, Rasmussen K, Frants RR (1995) The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve 2:S39–S44 [PubMed] [Google Scholar]
- Baldwin CT, Hoth CF, Amos JA, Dasilva EO, Milunsky A (1992) An exonic mutation in the Hup2 paired domain gene causes Waardenburg syndrome. Nature 355:637–638 [DOI] [PubMed] [Google Scholar]
- Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F (2003) The formation of skeletal muscle: from somite to limb. J Anat 202:59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppee F, Mattéotti C, Ansseau E, Sauvage S, Leclercq I, Leroy A, Marcowycz A, Gerbaux C, Figlewicz D, Ding H, Belayew A (2004) The DUX gene family and FSHD. In: Upadhyaya M, Cooper DN (eds) Facioscapulohumeral muscular dystrophy: clinical medicine and molecular cell biology. Garland Science/BIOS Scientific Publishers, Oxon, United Kingdom [Google Scholar]
- Deidda G, Cacurri S, Piazzo N, Felicetti L (1996) Direct detection of 4q35 rearrangements implicated in facioscapulohumeral muscular dystrophy (FSHD). J Med Genet 33:361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano AL, Cupples LA, Arnos KS, Asher JH, Baldwin CT, Blanton S, Carey ML, da Silva EO, Friedman TB, Greenberg J, Lalwani AK, Milunsky A, Nance WE, Pandya A, Ramesar RS, Read AP, Tassabejhi M, Wilcox ER, Farrer LA (1998) Correlation between Waardenburg syndrome phenotype and genotype in a population of individuals with identified PAX3 mutations. Hum Genet 102:499–506 [DOI] [PubMed] [Google Scholar]
- Flint J, Wilkie AOM, Buckle VJ, Winter RM, Holland AJ, McDermid HE (1995) The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet 9:132–140 [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Goto K, Arahata K (1998) Epilepsy and mental retardation in a subset of early onset 4q35-facioscapulohumeral muscular dystrophy. Neurology 50:1791–1794 [DOI] [PubMed] [Google Scholar]
- Gabellini D, Green M, Tupler R (2002) Inappropriate gene activation in FSHD. A repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110:339–248 [DOI] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R (2004) When enough is enough: genetic diseases associated with transcriptional derepression. Curr Opin Genet Dev 14:301–307 [DOI] [PubMed] [Google Scholar]
- Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A (1999) Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236:25–32 [DOI] [PubMed] [Google Scholar]
- Jiang G, Yang F, van Overveld PG, Vedanarayanan V, van der Maarel S, Ehrlich M (2003) Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum Mol Genet 12:2909–2921 [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM (2002) Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet 32:235–236 [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, Osborn M, Haaf T, Rogers M, Frants RR, Padberg GW, Cooper DN, van der Maarel SM, Upadhyaya M (2003) D4F104S1 deletion in facioscapulohumeral muscular dystrophy: phenotype, size, and detection. Neurology 61:178–183 [DOI] [PubMed] [Google Scholar]
- Lemmers RJL, de Kievit P, van Geel M, van der Wielen MJ, Bakker E, Padberg GW, Frants RR, van der Maarel SM (2001) Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann Neurol 50:816–819 [DOI] [PubMed] [Google Scholar]
- Lemmers RJLF, van der Maarel SM, van Deutekom JCT, van der Wielen MJR, Deidda G, Dauwerse HG, Hewitt J, Hofker M, Bakker E, Padberg GW, Frants RR (1998) Inter- and intrachromosomal subtelomeric rearrangements on 4q35: implications for facioscapulohumeral muscular dystrophy (FSHD) aetiology and diagnosis. Hum Mol Genet 7:1207–1214 [DOI] [PubMed] [Google Scholar]
- Lunt P (2000) Facioscapulohumeral muscular dystrophy: diagnostic and molecular aspects. In: Deymeer F (ed) Neuromuscular diseases: from basic mechanisms to clinical management. Vol 18. Karger, Basel, Switzerland [Google Scholar]
- Lunt PW, Jardine PE, Koch MC, Maynard J, Osborn M, Williams M, Harper PS, Upadhyaya M (1995) Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD). Hum Mol Genet 4:951–958 [DOI] [PubMed] [Google Scholar]
- Macina RA, Negorev DG, Spais C, Ruthig LA, Hu XL, Riethman HC (1994) Sequence organization of the human chromosome 2q telomere. Hum Mol Genet 3:1847–1853 [DOI] [PubMed] [Google Scholar]
- Masny PS, Bengtsson U, Chung SA, Martin JH, Engelen BM, van der Maarel SM, Winokur S (2004) Localization of 4q35.2 to the nuclear periphery: is FSHD a nuclear envelope disease? Hum Mol Genet 13:1857–1871 [DOI] [PubMed] [Google Scholar]
- Mefford HC, Trask BJ (2002) The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet 3:91–102 [DOI] [PubMed] [Google Scholar]
- Miura K, Kumagai T, Matsumoto A, Iriyama E, Watanabe K, Goto K, Arahata K (1998) Two cases of chromosome 4q35-linked early onset facioscapulohumeral muscular dystrophy with mental retardation and epilepsy. Neuropediatrics 29:239–241 [DOI] [PubMed] [Google Scholar]
- Padberg GW (2004) Facioscapulohumeral muscular dystrophy: a clinician’s experience. In: Upadhyaya M, Cooper DN (eds) Facioscapulohumeral muscular dystrophy: clinical medicine and molecular cell biology. Garland Science/BIOS Scientific Publishers, Oxon, United Kingdom [Google Scholar]
- Pandya A, Xia XJ, Landa BL, Arnos KS, Israel J, Lloyd J, James AL, Diehl SR, Blanton SH, Nance WE (1996) Phenotypic variation in Waardenburg syndrome: mutational heterogeneity, modifier genes or polygenic background? Hum Mol Genet 5:497–502 [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Read AP, Newton VE, Harris R, Balling R, Gruss P, Strachan T (1992) Waardenburg syndrome patients have mutations in the human homolog of the Pax-3 paired box gene. Nature 355:635–636 [DOI] [PubMed] [Google Scholar]
- Tawil R, Forrester J, Griggs RC, Mendell J, Kissel J, McDermott M, King W, Weiffenbach B, Figlewicz D (1996) Evidence for anticipation and association of deletion size with severity in facioscapulohumeral muscular dystrophy. Ann Neurol 39:744–748 [DOI] [PubMed] [Google Scholar]
- Tawil R, Storvick D, Feasby TE, Weiffenbach B, Griggs RC (1993) Extreme variability of expression in monozygotic twins with FSH muscular dystrophy. Neurology 43:345–348 [DOI] [PubMed] [Google Scholar]
- Tian XL, Kadaba R, You SA, Liu MG, Timura AA, Yang L, Chen QY, Szafranski P, Rao SQ, Wu L, Housman DE, DiCorleto PE, Driscoll DJ, Borrow J, Wang Q (2004) Identification of an angiogenic factor that when mutated causes susceptibility to Klippel-Trenaunay syndrome. Nature 427:640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonini MM, Pavanello RC, Gurgel-Giannetti J, Lemmers RJ, van der Maarel SM, Frants RR, Zatz M (2004a) Homozygosity for autosomal dominant facioscapulohumeral muscular dystrophy (FSHD) does not result in a more severe phenotype. J Med Genet 41:e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonini MMO, Passos-Bueno MR, Cerqueira A, Matioli SR, Pavanello R, Zatz M (2004b) Asymptomatic carriers and gender differences in facioscapulohumeral muscular dystrophy (FSHD). Neuromusc Disorders 14:33–38 [DOI] [PubMed] [Google Scholar]
- Trask BJ, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, Kuo WL, Massa H, Morrish T, Naylor S, Nguyen OT, Rouquier S, Smith T, Wong DJ, Youngblom J, van den Engh G (1998) Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum Mol Genet 7:13–26 [DOI] [PubMed] [Google Scholar]
- Tupler R, Barbierato L, Memmi M, Sewry CA, De Grandis D, Maraschio P, Tiepolo L, Ferlini A (1998) Identical de novo mutation at the D4F104S1 locus in monozygotic male twins affected by facioscapulohumeral muscular dystrophy (FSHD) with different clinical expression. J Med Genet 35:778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR (1993) FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 2:2037–2042 [DOI] [PubMed] [Google Scholar]
- van Geel M, Dickson MC, Beck AF, Bolland DJ, Frants RR, van der Maarel SM, de Jong PJ, Hewitt JE (2002) Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics 79:210–217 [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Deidda G, Sandkuijl L, Padberg GW, Frants RR, van der Maarel SM (2000) Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum Mol Genet 9:2879–2884 [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM (2003) Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet 35:315–317 [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, van Ommen GJ, Padberg GW, Frants RR (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 2:26–30 [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Higgs DR, Rack KA, Buckle VJ, Spurr NK, Fischel-Ghodsian N, Ceccherini I, Brown WR, Harris PC (1991) Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell 64:595–606 [DOI] [PubMed] [Google Scholar]
- Wohlgemuth M, Lemmers RJ, van der Kooi EL, van der Wielen MJ, van Overveld PG, Dauwerse H, Bakker E, Frants RR, Padberg GW, van der Maarel SM (2003) Possible phenotypic dosage effect in patients compound heterozygous for FSHD-sized 4q35 alleles. Neurology 61:909–913 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Forner J, Fournet S, Jeanpierre M (2001) Improved characterization of FSHD mutations. Ann Genet 44:105–110 [DOI] [PubMed] [Google Scholar]