Introduction
Amyotrophic lateral sclerosis (ALS) is a common adult-onset neurodegenerative disease leading to paralysis and death typically within 2–5 years of diagnosis. Approximately 10% of ALS cases are inherited, with the remainder of cases being sporadic in origin. This distribution of familial and sporadic disease is similar to other adult-onset neurodegenerative diseases, such as Parkinson disease and Alzheimer disease (see, e.g., Przedborski et al. 2003). The annual incidence of ALS is 1–2 per 100,000, leading to a lifetime risk of developing ALS of 1 per 800 (Cleveland and Rothstein 2001). Except for atypical variants of ALS, the familial form of disease is clinically indistinguishable from the sporadic cases (see, e.g., Hand and Rouleau 2002). There is a slight male-to-female preponderance (1.3:1–1.6:1) that appears to be decreasing (Nelson 1995). Similarly, some epidemiologic studies suggest that the overall incidence of ALS is rising (Riggs and Schochet 1992).
Pathologic features of ALS include loss of motor neurons in the spinal ventral horns, most brainstem motor nuclei, and motor cortex. Interestingly, Onuf’s nucleus, which controls urethral and sphincter function, and motor neurons in the oculomotor, trochlear, and abducens cranial nerve nuclei are spared. Histopathological features include ubiquitinated inclusions in lower motor neurons and axonal swellings that are thought to contain disarrayed neurofilaments (Ince 2000). It is thought that the process of ALS begins with an initiation or triggering event, followed by the propagation of motor neuron demise up and down the spinal cord (see Armon [2003] for a detailed description of this hypothesis). The exact mechanisms underlying the selective motor neuron degeneration in ALS remain elusive, but experimental evidence implicates many potential factors, including oxidative damage, excitotoxicity, apoptosis, abnormal neurofilament function, defects in axonal transport, aberrant protein processing and degradation, increased inflammation, and mitochondrial dysfunction (Cleveland and Rothstein 2001; Bruijn et al. 2004). Although hypotheses abound, it is difficult to determine which of these processes is most important in triggering cell dysfunction and death and what determines the selective vulnerability of motor neurons.
The World Federation of Neurology diagnostic criteria for ALS include the presence of upper and lower motor neuron degeneration with a progressive phenotype in the absence of evidence that indicates other diseases (Revised Criteria for the Diagnosis of ALS Web site). Typically, there is no cognitive impairment or loss of sensory nerve function, although there are ALS variants that include these symptoms. Upper motor neuron signs include clonus and hyperreflexia, and lower motor neuron signs include atrophy, weakness, and fasciculations. ALS diagnoses are categorized as “clinically definite,” “clinically probable,” and “clinically possible,” on the basis of the number and location of the cardinal signs. The diagnosis of ALS is often one of excluding other diseases and waiting for the disease to progress to meet the full diagnostic criteria. A variety of studies are often performed to exclude syndromes that have symptoms that mimic ALS, including postpoliomyelitis syndrome; multifocal motor neuropathy; endocrinopathies, especially hyperparathyroid or hyperthyroid states; lead intoxication; infections; and paraneoplastic syndromes (Motor Syndromes Web site; Revised Criteria for the Diagnosis of ALS Web site).
Genetics of ALS
There is an obvious genetic component in only ∼10% of ALS cases; these cases exhibit significant phenotypic and genetic heterogeneity. At least 12 genetic loci, with dominant, recessive, and X-linked patterns of inheritance, have been associated with familial ALS and related ALS syndromes (table 1). Although specific genetic alterations do not appear to cause sporadic ALS, a number of potential susceptibility and modifier loci have been identified (table 2). I will discuss the genes and loci that have been implicated in the causation and/or susceptibility of both sporadic and familial ALS.
Table 1.
Familial ALS Loci
|
MIM Number |
|||||||||
| Disease | Type(s) ofInheritance | Disease | Gene | Gene | Onset | Chromosome | Interval | Size(Mb) | Reference(s) |
| ALS1 | Dominant and recessive (D90A) | 105400 | 147450 | SOD1 | Adult | 21q22.1 | Rosen et al. 1993; Al-Chalabi et al. 1998 | ||
| ALS2 | Recessive | 205100 | 606352 | Alsin (ALS2) | Juvenile | 2q33 | Hadano et al. 2001; Yang et al. 2001 | ||
| ALS3 | Dominant | 606640 | Adult | 18q21 | D18S846–D18S1109 | 8 | Hand et al. 2002; Sapp et al. 2003 | ||
| ALS4 | Dominant | 602433 | 608465 | SETX | Juvenile | 9q34 | Chen et al. 2004 | ||
| ALS5 | Recessive | 602099 | Juvenile | 15q15.1-q21.1 | Hentati et al. 1998 | ||||
| ALS6 | Dominant | 608030 | Adult | 16q12 | D16S339–D16S3032 | 4.2 | Abalkhail et al. 2003; Ruddy et al. 2003; Sapp et al. 2003 | ||
| ALS7 | Dominant | 608031 | Adult | 20ptel-p13 | Telomere–D20S199 | 1 | Sapp et al. 2003 | ||
| ALS8 | Dominant | 608627 | 605704 | VAPB | Adult | 20q13.33 | Nishimura et al. 2004a, 2004b | ||
| ALS-FTD | Dominant | 105550 | Adult | 9q21-22 | D9S301–D9S167 | 17 | Hosler et al. 2000 | ||
| ALS X | Dominant | Adult | Xp11-q12 | Siddique et al. 1998a | |||||
| ALS with dementia, Parkinsonism | Dominant | 600274 | 157140 | MAPT | Adult | 17q21 | Clark et al. 1998; Hutton et al. 1998 | ||
| Progressive LMN disease | Dominant | 601143 | 607641 | DCTN1 | Adult | 2p13 | Puls et al. 2003 | ||
Table 2.
Human Susceptibility and Modifier Loci
| Gene | MIM Number | Chromosome | Variant | Association | Reference |
| NEFH | 162230 | 22q12.1-q13.1 | KSP deletions | Sporadic | Al-Chalabi et al. 1999 |
| VEGF | 192240 | 6p12 | Promoter SNPs | Sporadic | Lambrechts et al. 2003 |
| SMN1 | 600354 | 5q12.2-q13.3 | Copy number | Sporadic | Corcia et al. 2002 |
| SMN2 | 601627 | 5q12.2-q13.3 | Copy number | Sporadic | Veldink et al. 2001 |
| CNTF | 118945 | 11q12.2 | Null allele | Familial | Giess et al. 2002 |
| ApoE ɛ4 | 107741 | 19q13.2 | ɛ4 genotype | Sporadic | Drory et al. 2001 |
| EAAT2 | 600300 | 11p13-p12 | Decreased expression | Familial, sporadic | Rothstein et al. 1995 |
| GluR2 | 138247 | 4q32-q33 | Altered RNA editing | Sporadic | Kawahara et al. 2004 |
Adult-Onset ALS Genes
There are at least six dominantly inherited, adult-onset ALS genes (table 1); however, only the gene for ALS1 (MIM 105400)—copper-zinc superoxide dismutase (SOD1 [MIM 147450]), on chromosome 21q22.1—has been identified. Mutations in SOD1 account for ∼20% of familial ALS (Rosen et al. 1993). Identifying the genes causing the remaining 80% of familial ALS cases will be challenging, since many of the remaining loci appear to segregate in individual families.
Cytoplasmic Copper-Zinc Superoxide Dismutase
The first ALS-associated gene to be identified was the SOD1 gene, on human chromosome 21 (Rosen et al. 1993). SOD1 is a 153–amino acid protein, containing one copper and one zinc, that is predominantly located in the cytoplasm as a homodimer. SOD1 detoxifies superoxide, creating oxygen and hydrogen peroxide, which can then be cleared by catalase and glutathione peroxidase. Copper is required for SOD1 activity, whereas zinc is thought to stabilize the protein structure. To date, >100 unique mutations in SOD1 have been identified (Andersen et al. 2003; alsod.org Web site). The majority of mutations in SOD1 are missense mutations, with a small percentage of deletion and insertion mutations that result in prematurely terminated SOD1 polypeptides. The expression of a mutant SOD1 polypeptide, with or without residual SOD1 activity, is necessary to cause the ALS phenotype, suggesting a dominant negative mechanism rather than one of haploinsufficiency. Even after >10 years of investigation, the exact mechanism of SOD1-mediated pathogenesis remains uncertain.
There is considerable phenotypic variation in SOD1-mediated ALS, including age at onset and severity and rate of decline; however, this can only partly be explained by the spectrum of mutations. Because clinical variation occurs among patients of the same SOD1 genotype and members of the same family (Andersen et al. 1997), it is apparent that the phenotype is modified by other genetic and/or environmental factors. One example is the D90A SOD1 mutation, which is recessive in some genetic backgrounds but dominant in others (Al-Chalabi et al. 1998). The recessive D90A SOD1 mutations share a common founder haplotype, suggesting that there is a linked cis-acting protective factor that makes this mutation recessive in this specific genetic background (Parton et al. 2002). Genetic background also affects other forms of SOD1-mediated ALS. Although it causes one of the most severe forms of the disease, with death typically occurring <18 mo after diagnosis, the penetrance of the A4V SOD1 mutation is only 91% (Cudkowicz et al. 1997). Similarly, the A89V SOD1 mutation shows incomplete penetrance and variable age at onset (Rezania et al. 2003). The variable penetrance and age at onset caused by SOD1 mutations can be mimicked in transgenic mouse models of ALS, by varying the mouse strain on which a mutation is carried (Kunst et al. 2000).
Transgenic mice that develop ALS-like phenotypes have been constructed with at least nine forms of mutant human SOD1: A4V, G93A, G85R, G37R, D90A, L126Z, H46R/H48Q, H46R/H48Q/H63G/H120G, and G127insTGGG (Dal Canto and Gurney 1995; Wong et al. 1995; Bruijn et al. 1997b; Brannstrom et al. 1998; Deng et al. 1999; Wang et al. 2002, 2003; Jonsson et al. 2004). Another mouse model of ALS was generated using the mouse gene with a G86R mutation that corresponds to the human G85R mutation (Ripps et al. 1995). Transgenic rats, carrying G93A or H46R SOD1, also develop ALS-like phenotypes (Nagai et al. 2001; Howland et al. 2002). Although each model of ALS is phenotypically consistent for a given mutation, they vary in their age at onset, disease progression, and certain histopathological features, mimicking the diversity of phenotypes observed in human ALS. In the mouse and rat models of ALS, the mutant SOD1 allele is expressed in the presence of the two endogenous copies of the wild-type SOD1 gene; thus, the mice and rats have either normal or increased SOD1 activity levels, depending on the activity of the SOD1 mutant expressed. However, SOD1 overexpression is not the cause of ALS, since mice overexpressing wild-type human SOD1 do not develop an ALS-like phenotype (Dal Canto and Gurney 1995; Wong et al. 1995). In G93A SOD1 mice (Jaarsma et al. 2000) but not in G85R SOD1 mice (Bruijn et al. 1998), overexpression of wild-type SOD1 accelerates disease onset and progression. Lack of SOD1 is also not sufficient to cause ALS in mice. Mice with SOD1 null alleles have a number of interesting phenotypes but do not develop symptoms of ALS (Reaume et al. 1996). Together, these results suggest that mutations in SOD1 cause a novel toxic gain of function that is lethal to motor neurons.
Although ALS is predominantly a disease of motor neuron loss, neuronal expression of mutant SOD1 is not sufficient to cause ALS. Overexpression of mutant SOD1 in neurons or astrocytes alone does not cause ALS or motor neuron death in transgenic mice (Gong et al. 2000; Pramatarova et al. 2001; Lino et al. 2002). Studies in chimeric mice, created from mixtures of normal and mutant SOD1-expressing cells, reveal that toxicity to motor neurons requires damage from mutant SOD1 acting from within nonneuronal cells (Clement et al. 2003). In the chimeras, motor neurons expressing only wild-type SOD1 develop ALS pathology when adjacent nonneuronal cells express mutant SOD1. Furthermore, nonneuronal cells expressing only wild-type SOD1 delay degeneration and extend survival of nearby motor neurons expressing mutant SOD1 in these chimeric mice (Clement et al. 2003). Because ALS pathology is seen in the chimeras but not in the mice with mutant SOD1 overexpression in the neurons or astrocytes alone, it appears that expression of mutant SOD1 in both cell types is necessary to initiate the disease process. From there, SOD1 expression from nonneuronal cells modulates disease progression, with the effect being dependent on the form of SOD1 expressed.
Although the toxic gain of function of mutant SOD1 has not yet been elucidated, there are numerous hypotheses of SOD1-mediated toxicity. These have been recently and expertly reviewed by others (e.g., Cleveland and Rothstein 2001; Hand and Rouleau 2002; Heath and Shaw 2002; McGeer and McGeer 2002; Ischiropoulos and Beckman 2003; Bruijn et al. 2004); however, several major hypotheses, including oxidative stress, mitochondrial dysfunction, excitotoxicity, inflammation, and aggregation, will be summarized here. These mechanisms are not mutually exclusive, and the complicated pathogenic process of ALS may include features of all of them.
Because of its role of preventing cellular damage from superoxide, one of the first hypotheses of SOD1-mediated toxicity was copper-mediated oxidative and peroxidative damage by a promiscuous mutant SOD1 enzyme. In support of this, there is evidence of lipid peroxidation and nitrotyrosine formation in both transgenic models and human patients with ALS (Dal Canto and Gurney 1995; Beal et al. 1997). Some mutations in SOD1 make the protein more susceptible to forming a zinc-deficient variant (Crow et al. 1997; Estévez et al. 1999). The copper in the zinc-deficient SOD1 becomes more accessible, allowing SOD1 to participate in a number of deleterious reactions, including oxidizing endogenous antioxidants such as ascorbate, transferring electrons to oxygen to produce superoxide, and creating peroxynitrite from nitric oxide (Estévez et al. 1998, 1999). In vitro experiments by Estévez et al. (1999) have shown that zinc-deficient SOD1 kills motor neurons through a peroxynitrite dependent mechanism. In support of this mechanism acting in vivo, a specific inhibitor of neuronal nitric oxide synthase (AR-R 17,477) was able to delay onset in the G93A SOD1 mice (Facchinetti et al. 1999). However, G93A SOD1 mice that also lack the neuronal nitric oxide synthase gene develop ALS without a delay in disease onset (Facchinetti et al. 1999), making the role of neuronal nitric oxide and its by-products uncertain.
Additional experiments raise questions about the copper-dependent hypothesis of SOD1-mediated toxicity. SOD1 that has been engineered not to bind copper by mutating the histidine residues required for copper binding (H46R/H48Q/H63G/H120G) causes ALS in transgenic mice (Wang et al. 2003). Furthermore, knocking out the gene for the copper chaperone (CCS [MIM 603864]) that inserts copper into SOD1 has no effect on the development of ALS in transgenic mice (Subramaniam et al. 2002). However, even in the CCS null mice, there is residual SOD1 activity; therefore, this mechanism cannot be ruled out entirely.
Substantial evidence links mutations in SOD1 to mitochondrial dysfunction. Previously, SOD1 was considered an exclusively cytoplasmic protein; however, recent studies show that ∼1%–2% of SOD1 is located in the intermembrane space of mitochondria (Mattiazzi et al. 2002). Some researchers postulate that it is this mitochondrial pool of mutant SOD1 that triggers disease. One of the first pathological changes in G93A and G37R SOD1 transgenic mice is the development of large membrane-bound vacuoles derived from degenerating mitochondria in motor neurons (Dal Canto and Gurney 1995; Wong et al. 1995). G93A SOD1 mice also develop metabolic defects in mitochondrial energy generation, in both spinal cord and motor regions of the brain (Browne et al. 1998; Jung et al. 2002; Mattiazzi et al. 2002). Kong and Xu (1998) found evidence of a burst of degenerating mitochondria within motor neurons immediately prior to symptom onset in G93A SOD1 mice. Recently, Liu et al. (2004) found that mutant SOD1—but not wild-type SOD1—is selectively and aberrantly recruited to the cytoplasmic face of mitochondria only in tissues affected by ALS. This recruitment was independent of enzymatic activity and the copper chaperone for SOD1. Although some mutant SOD1 was correctly imported into the intermembrane space, covalently damaged adducts of mutant SOD1 accumulated on the cytoplasmic face of mitochondria in spinal cord. This tissue-specific recruitment suggests that mitochondrial abnormalities may be involved in disease initiation.
One observation in both sporadic and familial ALS is the selective loss of the glial glutamate transporter EAAT2 (MIM 600300) (Rothstein et al. 1995; Howland et al. 2002) in some but not all patients and animal models. EAAT2 (GLT1 in rodents) is responsible for clearing 90% of the glutamate near motor neurons (Cleveland and Rothstein 2001). Glutamate-mediated excitotoxicity is thought to occur from the repetitive firing or elevation of intracellular calcium by calcium-permeable glutamate receptors. A role of glutamate-mediated excitotoxicity in both sporadic and familial disease is bolstered by the efficacy of riluzole, a compound that antagonizes glutamate excitotoxicity, which is effective in slowing disease in both mice and humans (Gurney et al. 1998; Miller et al. 2003).
There is growing evidence that inflammation and microglial activation play a role in the pathogenesis of ALS (McGeer and McGeer 2002). Reactive microglia and astrocytes accumulate in the areas surrounding degenerating motor neurons (reviewed by McGeer and McGeer 2002). Obal et al. (2001) demonstrated that intraperitoneal injections of immunoglobulin G from human patients with ALS caused the recruitment of activated microglia to the ventral horn of the spinal cord of mice. Numerous biochemical markers of inflammation are observed in ALS spinal cord tissue. Both caspase 1 and cyclooxygenase 2 are increased in spinal cord of mutant SOD1 transgenic mice (McGeer and McGeer 2002; Bruijn et al. 2004). These enzymes generate mature interleukin-1β and prostaglandin E2, respectively. Both of these diffusible compounds are proinflammatory and can activate cell death. Supporting their role in ALS, inhibitors of caspases and cyclooxygenase 2 have been shown to prolong survival in G93A SOD1 transgenic mice (Li et al. 2000; Klivenyi et al. 2004). Other markers of inflammation are also increased in ALS. For example, tumor necrosis factor α, which can activate apoptosis, is up-regulated in the spinal cord of mutant G93A SOD1 mice (Elliott 2001). Taken together, these data suggest that mutations in SOD1 directly or indirectly induce a variety of inflammatory responses that may play a role in the selective death of motor neurons.
A final, and perhaps favored, potential mechanism of SOD1-mediated toxicity is aggregation of mutant SOD1. Aggregates of misfolded mutant SOD1 protein in affected motor regions are a common pathologic feature of mutant SOD1 mouse models of ALS (Dal Canto and Gurney 1995; Bruijn et al. 1997a, 1998; Wang et al. 2002; Jonsson et al. 2004). The effect of SOD1 aggregation may be analogous to the effects of aggregates of mutant proteins in other neurodegenerative diseases, such as Alzheimer disease, Huntington disease, and Parkinson disease, in which aggregation of mutated proteins causes oxidative stress, depletes important cellular proteins, and disrupts proteasome and chaperone function. In vitro experiments have linked SOD1 aggregation to apoptotic cell death (Durham et al. 1997; Roy et al. 1998). Watanabe et al. (2001) found additional proteins in SOD1 aggregates, including CCS; ubiquitin; neurofilaments; glial fibrillary acidic protein (GFAP [MIM 137780]); two neuronal glutamate transporters, GLAST (MIM 600111) and EAAC1 (MIM 133550); and proteins involved in chaperone and proteasome functions. Overexpression of chaperones can suppress mutant SOD1 aggregation, protect neuronal function, and enhance survival of motor neurons in culture (Takeuchi et al. 2002). Arimoclomol, an inducer of heat shock proteins, increased life span by 22% in the G93A SOD1 mice (Kieran et al. 2004), further supporting the hypothesis that aggregated SOD1 is toxic to motor neurons.
Although SOD1 is ubiquitously expressed, aggregates of mutant SOD1 are found only within the nervous system of mutant SOD1 transgenic mice, despite the very high concentrations of mutant SOD1 in other organs, higher in liver or kidney than in spinal cord (Wang et al. 2002; Puttaparthi et al. 2003). Puttaparthi et al. (2003) used an organotypic spinal cord slice culture system from G93A SOD1 mice to show that proteasome-mediated protein degradation represents the major clearance mechanism for SOD1 aggregates in spinal cord. It is interesting that proteasome activity decreases most prominently in spinal cord during aging, and this decrease correlates with accumulation and aggregation of mutant SOD1 in vivo (Puttaparthi et al. 2003). These observations may help to explain the selective vulnerability of motor neurons to mutant SOD1 with increased age.
Juvenile-Onset ALS Genes
There are three loci for juvenile onset ALS (table 1): one is autosomal dominant, on chromosome 9q34 (ALS4 [MIM 602433]) (Chen et al. 2004); and two are autosomal recessive, on chromosomes 2q33 (ALS2 [MIM 205100]) (Hadano et al. 2001; Yang et al. 2001) and 15q15.1-q21.1 (ALS5 [MIM 602099]) (Hentati et al. 1998). In general, survival time from diagnosis is longer and disease progression slower in the juvenile-onset cases. The chromosome 2 and chromosome 9 genes have been identified, whereas the chromosome 15 locus remains to be identified.
ALS2: Alsin
Two groups (Hadano et al. 2001; Yang et al. 2001) identified the chromosome 2 recessive ALS gene known as “alsin” or ALS2 (MIM 606352). Alsin/ALS2 is alternatively spliced to produce a long and a short transcript. It was originally hypothesized that deletions affecting both transcripts result in ALS2, whereas homozygous deletions affecting just the long transcript cause a related disease, juvenile primary lateral sclerosis (Hadano et al. 2001; Yang et al. 2001). However, Eymard-Pierre et al. (2002) found that mutations in alsin/ALS2 could also cause infantile-onset ascending hereditary spastic paralysis (IAHSP), with no overt genotype-phenotype correlation.
Alsin/ALS2 is an 184-kDa protein with three putative guanine-nucleotide-exchange factor (GEF) domains. The function of alsin/ALS2 is not yet well understood. Alsin/ALS2, which has Rab5 activity (Otomo et al. 2003), can act as a guanine nucleotide exchange factor for Rac1 (Topp et al. 2004) and appears to be important for endosomal dynamics (Kunita et al. 2004). At least two groups have created alsin/ALS2 knockout mice (Kriz et al. 2003; Cai et al. 2003); however, no major phenotypes consistent with ALS or other motor neuron disease have yet been described. It is interesting that Kanekura et al. (2004) recently discovered that the long isoform of alsin/ALS2 specifically binds to mutant—but not to wild-type—SOD1, via its RhoGEF domain. Expression of the long isoform of alsin/ALS2 protected motor neurons in vitro from mutant SOD1-mediated toxicity. The physical interactions between mutant SOD1 and alsin/ALS2 may link the motor neuron-specific pathways of pathogenesis in these two forms of familial ALS.
ALS4: Senataxin
The ALS4 locus, mapped to chromosome 9q34, was originally identified in a single large family with autosomal dominant juvenile ALS. This family was unusual, because life expectancy was normal, although the clinical criteria were sufficient to diagnose ALS. Other motor neuron disorders, including distal spinal muscular atrophy or spinal Charcot-Marie-Tooth syndrome, have also been linked to this locus (De Jonghe et al. 2002), but later clinical re-examination of these families resulted in ALS diagnoses (Chen et al. 2004). Chen et al. (2004) identified missense mutations in the senataxin (SETX [MIM 608465]) gene in three families with autosomal dominant juvenile ALS. Each family had a distinct mutation—L389S, R2136H, and T3I—in the SETX gene. Senataxin is a large protein with a superfamily I DNA/RNA helicase domain (Chen et al. 2004). The majority of the protein appears novel, with no domain conservation or homology to other proteins for much of its length (Chen et al. 2004). The exact function of senataxin is not known, but DNA/RNA helicases are involved in DNA repair, replication, recombination, transcription, RNA processing, transcript stability, and the initiation of translation. Recessive loss-of-function mutations in SETX are associated with ataxia-oculomotor apraxia type 2 (Moreira et al. 2004). Ataxia-oculomotor apraxia is a heterogeneous disorder characterized by cerebellar ataxia/atrophy, oculomotor apraxia, loss of reflexes, late peripheral neuropathy, and immunodeficiency. The phenotypic differences between these disorders and their distinct patterns of inheritance suggest that the dominant ALS4 mutations cause a toxic gain of function, resulting in a motor neuron–specific phenotype, whereas the recessive loss-of-function mutations cause a pleiotropic phenotype.
Atypical ALS
The symptoms of ALS can occasionally occur together with Parkinson disease and dementia or frontotemporal dementia (FTD [MIM 600274]) alone. FTD is a neurodegenerative disorder involving degeneration of the frontal and temporal cortices, accompanied by dementia. Hosler et al. (2000) identified five families with autosomal dominant ALS and FTD (ALS-FTD [MIM 105550]). The causative gene was mapped to a 17-cM interval between D9S301 and D9S167 on chromosome 9q21 (Hosler et al. 2000). The ALS-FTD locus is associated with a range of phenotypes, and not all patients develop all symptoms of ALS and FTD, suggesting that the phenotype can be modulated by other genetic or environmental influences. Although Hosler et al. (2000) did not find any evidence for ALS-FTD genes on other chromosomes, Ostojic et al. (2003) recently identified a Swedish family with ALS-FTD without linkage to this locus, suggesting that additional ALS-FTD loci remain to be identified. Prudlo et al. (2004) identified a single patient with ALS-FTD and a chromosomal translocation, t(18;21)(q23;q22), that may be associated with the disease. Because this is an isolated case, it is difficult to predict whether this balanced translocation is incidental or associated with disease. In similar studies, Meyer et al. (2003) demonstrated that patients with sporadic ALS have, in general, a higher rate of constitutional chromosomal rearrangement than does the general population, but these studies have not been verified with a larger patient population. If causally related to disease, these unique translocations would provide a valuable resource for the identification of additional ALS causative genes.
As described above, the symptoms of ALS can occasionally occur together with Parkinson disease and dementia. Mutations in the microtubule-associated protein tau (MAPT [MIM 157140]) gene are associated with FTD with Parkinson disease, with ALS symptoms sometimes associated with the phenotype (Clark et al. 1998; Hutton et al. 1998). The mutant tau, which is encoded by the MAPT gene, forms insoluble aggregates and filamentous inclusions that are associated with neurodegeneration. Not all individuals with symptoms of familial ALS with FTD and Parkinson disease have MAPT mutations (Kowalska et al. 2003; Wilhelmsen et al. 2004), suggesting that, as with ALS and ALS-FTD, additional genes causing this constellation of symptoms remain to be identified.
Puls et al. (2003) characterized a family with a disorder related to ALS that had a progressive, autosomal dominant form of lower motor neuron disease without sensory symptoms (MIM 607641). A single base-pair change in the dynactin (DCTN1 [MIM 601143]) gene, causing a G59S mutation in a single North American family, was identified. The G59S mutation is located in a highly conserved domain that binds to microtubules. The interaction of dynactin with dynein is thought to be required for the retrograde axonal transport of vesicles and organelles. Impaired axonal transport in motor neurons has been proposed as a mechanism for neuronal degeneration in motor neuron disease.
Genetics of Sporadic ALS
Glutamate Transporters and Receptors
As described above, suppressed expression of the glial glutamate transporter EAAT2 occurs in ∼60% of patients with sporadic ALS (Rothstein et al. 1995), implicating glutamate excitotoxicity in the pathogenesis of sporadic ALS. The mechanism of the selective loss of EAAT2 in affected spinal cord regions may be the aberrant processing of the EAAT2 transcript (Lin et al. 1998), but ALS-specific aberrant splicing events have not been observed by all investigators (e.g., Flowers et al. 2001). Additional evidence for a role of glutamate excitotoxicity in ALS comes from recent work by Kawahara et al. (2004). They investigated the RNA editing of the GluR2 (MIM 138247) subunit of the glutamate AMPA receptor in patients with sporadic ALS and in controls. Under normal conditions, RNA editing changes a glutamine to arginine in virtually 100% of transcripts, rendering the channel impermeable to calcium. In RNA-specific adenosine deaminase (ADAR2 [MIM 601218]) null mice, defects in the GluR2 editing process lead to premature neuronal death that can be rescued by restoring normal RNA editing function (Higuchi et al. 2000). GluR2 editing was observed to be defective in spinal motor neurons from patients with sporadic ALS but not in Purkinje cells, which were isolated by laser capture microdissection. Control samples showed 100% editing in all analyzed cells, suggesting that there is an ALS-specific motor neuron defect in GluR2 RNA editing that may be involved in the process of motor neuron death (Kawahara et al. 2004).
Neurofilaments
The abnormal accumulation of neurofilaments in the cell bodies and proximal axons of motor neurons is a hallmark of the pathogenesis of ALS (Rouleau et al. 1996). There is evidence that mutations in the neurofilament heavy (NF-H, or NEFH [MIM 162230]) gene are associated with a small fraction of ALS in a subset of cases and may predispose to disease development (Al-Chalabi et al. 1999). These mutations do not segregate with disease in familial ALS (Cleveland 1999) and, thus, are either not directly causative or act at low penetrance. However, genetic manipulations of neurofilament subunit expression in transgenic mice have confirmed the importance of neurofilaments in motor neuron integrity. First, the overexpression of NF-H, NF-L, and peripherin, as well as the disrupted activity of the microtubule-motor dynein, all cause development of paralytic phenotypes associated with motor neuron degeneration and muscle denervation (reviewed by Lariviere and Julien [2004]). Other alterations in neurofilament gene expression are beneficial in the mutant SOD1 mouse models of ALS. For example, disease development in the G85R SOD1 mice is delayed on an NF-L null background (Williamson et al. 1998). NF-L is the major neurofilament subunit required for filament assembly; thus, in NF-L null mice, NF-M and NF-H are not assembled and transported correctly, resulting in reduced levels in axons but increased levels in motor neuron cell bodies. This may explain why the overexpression of human NF-H increased the mean life span of the G37R SOD1 mice by 65% (Couillard-Després et al. 1998).
Vascular Endothelial Growth Factor
Lambrechts et al. (2003) have shown that vascular endothelial growth factor (VEGF [MIM 192240]) is a modifier of ALS in both mice and humans. When the hypoxia-responsive element was deleted from the VEGF promoter, mice developed a late-onset motor neuron disease reminiscent of ALS (Oosthuyse et al. 2001). When these VEGF mice were bred to G93A SOD1 mice, ALS onset was accelerated, reducing the mean age at death from 124 to 107 d (P=.001 [Lambrechts et al. 2003]). Further studies revealed that certain SNPs in the human VEGF gene were associated with both reduced VEGF expression and increased ALS risk (Lambrechts et al. 2003), suggesting a link between VEGF expression levels and ALS susceptibility.
Survival of Motor Neuron
Homozygous deletions of the survival of motor neuron gene (SMN1 [MIM 600354]) on chromosome 5 cause spinal muscular atrophy (SMA [MIM 253300]), a fatal childhood-onset neuromuscular disease characterized by the death of spinal motor neurons and subsequent muscle paralysis. A second highly conserved gene, SMN2 (MIM 601627), has five nucleotide differences between intron 6 and exon 8 that distinguish it from SMN1. One of these polymorphisms causes frequent skipping of exon 7 and very low levels of intact SMN2 protein as a result. One study of 110 patients with ALS and 100 controls found that SMN2 gene deletions were overrepresented in patients with sporadic ALS (16%) when compared with controls (4%) and may be risk factors for motor neuron disease development (Veldink et al. 2001). In a similar study, Corcia et al. (2002) investigated 167 patients with ALS and their unaffected spouses for SMN1 and SMN2 copy number. Surprisingly, 16% of patients with ALS had an abnormal copy number of SMN1 (one or three copies), versus 4% of controls. In contrast to the results of Veldink et al. (2001), no differences in SMN2 copy number were observed between the groups. Although the SMN gene involved differs between the studies, both implicate a role for SMN copy number in the risk of developing ALS. Further studies are needed to clarify the role of the SMN1 and SMN2 genes in sporadic ALS.
Ciliary Neurotrophic Factor
Although there is considerable phenotypic heterogeneity within families with mutant SOD1–mediated ALS, Giess et al. (2002) searched for modifier loci in an unusual family with ALS. A 25-year-old man carrying a V148G SOD1 mutation died from ALS 11 mo after disease onset. His mother and three other family members developed ALS between the ages of 43 and 62 years. His 35-year-old carrier sister remained asymptomatic. Because of the early onset and rapid progression of disease in this individual, several candidate modifier loci were analyzed. A homozygous null mutation in his ciliary neurotrophic factor (CNTF [MIM 118945]) gene, a potent survival factor for motor neurons, was identified. The other patients with ALS and the unaffected sister were either wild-type or heterozygous at the CNTF locus. Because of this result, Giess et al. (2002) bred CNTF null mice to G93A SOD1 transgenic mice to create CNTF−/−/G93A SOD1 mice. In these mice, ALS developed significantly earlier (P<.001), although disease duration was unaffected. Similarly, ALS onset occurred ∼10 years earlier (48.6±15 versus 58.4±9 years) in 8 people with the CNTF−/− genotype compared with 30 CNTF+/+ controls. As with the mouse model, disease duration was not affected, suggesting that CNTF genotype affects susceptibility to disease initiation but not disease progression.
Apolipoprotein E
The apolipoprotein E (ApoE [MIM 107741]) ɛ4 genotype is known to be associated with a lowered age at Alzheimer disease onset; therefore, Drory et al. (2001) genotyped 100 consecutive patients with ALS and 133 controls for the ApoE ɛ4 allele. Although the frequency of the ApoE ɛ4 allele was slightly higher in patients with ALS (15.1%) versus controls (10.9%), there was no association between ApoE genotype and age at ALS onset. However, Kaplan Meier survival analysis demonstrated that the ApoE ɛ4 genotype correlated with a shortened survival (32 mo; P=.03) after diagnosis compared with other ApoE genotypes, suggesting that ApoE ɛ4 can effect disease progression but not onset. This result is somewhat controversial, since it has not been observed by all groups (e.g., Siddique et al. 1998b).
Gene-Environment Interactions
Many genes that play a role in the pathogenesis of ALS have been identified or mapped; however, because ALS is predominantly sporadic in origin, environmental triggers are clearly involved in disease initiation. Very few ALS environmental risks have been identified, perhaps because the triggers act only in a genetically susceptible individual. There is an unusual ALS variant in Guam and other regions of the Western Pacific called “ALS-PDC” (parkinsonism-dementia complex) that appears to result from eating toxins from the cycad nut that have been concentrated in flying foxes (Banack and Cox 2003). This diet-induced disease can be recapitulated in mice by feeding them washed cycad flour (Wilson et al. 2002). The mouse model shares many of the features of traditional ALS, including reduced expression of the glial glutamate transporter EAAT2/GLT-1 (Wilson et al. 2003). The identification of other environmental risk factors for ALS has been difficult, but a number of potential disease triggers have been identified, including smoking, BMI, a glutamate-rich diet, heavy-metal exposure, and military service including the first Gulf War (Kamel et al. 1999, 2003; Nelson et al. 2000a, 2000b; Scarmeas et al. 2002; Armon 2003; Haley 2003; Weisskopf et al. 2004).
ALS Therapy Development
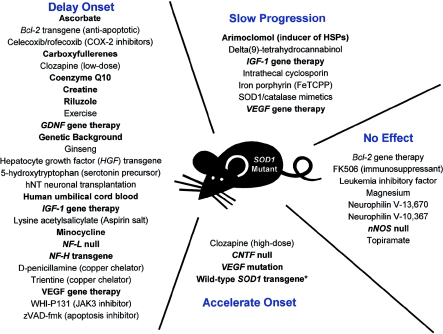
Riluzole, the only FDA-approved treatment for ALS, works as well in the SOD1 mouse model as it does in people with sporadic ALS (e.g., Gurney et al. 1998). Therefore, the mutant SOD1 models are used extensively for drug screening. However, these model organisms have several significant limitations. Because of the rapid disease progression in these mice, treatment typically begins presymptomatically, something that is currently not possible in human sporadic ALS. Furthermore, therapies that work presymptomatically may target disease initiation from mutant SOD1 rather than disease propagation, which is ultimately the necessary target for human ALS treatment and which may be a process distinct from disease initiation. In spite of these weaknesses, the SOD1 mutant mice and rats remain the best animal models for ALS. Drugs tested for efficacy in ALS have targeted many of the pathways implicated in disease pathogenesis, including protein aggregation, apoptosis, cell cycle regulation, excitotoxicity, immune system regulation, inflammation, mitochondrial function, and oxidative stress (fig. 1). Treatment regimens including vitamins, antibiotics, pain medication, carboxyfullerenes, bone marrow transplantation, and stem cells have been tried as therapeutics in the mice (fig. 1) (see, e.g., Bruijn 2002). While many trials are carried out in academic laboratories, the ALS Therapy Development Foundation initiates four to six drug-screening studies each month in the G93A SOD1 mouse model, with ∼12 drug studies ongoing at any given time (ALS Therapy Development Foundation Treatment Targets Web site). A number of compounds, such as creatine and coenzymeQ10, extend life span, delay the onset of motor impairment, protect against motor neuron loss, and decrease the evidence of oxidative stress in mouse models of disease (Matthews et al. 1998; Klivenyi et al. 1999). However, not all compounds that are efficacious in mice are effective in patients with ALS. For example, Groeneveld et al. (2003) demonstrated that, although creatine is effective in the G93A SOD1 mouse model of ALS, it did not effect survival or disease progression in patients with ALS in a recent clinical trial.
Figure 1.
Therapeutic approaches utilized in the mutant SOD1 trangenic mice. A variety of therapeutic genes and agents have been tested in the mutant SOD1 transgenic mice. Treatments discussed in the text are in boldface type. The asterisk (*) reflects a result that was observed in G93A but not G85R SOD1 mice.
Because of their striking neuroprotective effects in vitro, a variety of neurotrophic factors, such as CNTF, glial cell–derived neurotrophic factor (GDNF [MIM 600837]), brain-derived neurotrophic factor (BDNF [MIM 113505]), and insulin growth factor 1 (IGF-1 [MIM 113505]), have been largely unsuccessful in human clinical trials for the treatment of ALS (see, e.g., Bruijn 2002). Only IGF-1 has had marginal success in one of two clinical trials (Mitchell et al. 2002). Although these agents are strongly neuroprotective in vitro, their limited efficacy in vivo may be due to the limited ability of these compounds to cross the blood-brain barrier. Two recent studies exploited the retrograde transport ability of some recombinant viral vectors to deliver IGF-1, GDNF, and VEGF to motor neurons and surrounding cells in ALS mouse models (Kaspar et al. 2003; Azzouz et al. 2004). Retrograde transport from motor neurons that innervate muscles requires the virus to bind to receptors on the axon terminal, with subsequent transport to the motor neuron nucleus, allowing sustained gene expression. These studies are extraordinarily promising, since the treatment regimens not only delayed disease onset but could slow disease progression when initiated after onset of symptoms.
Kaspar et al. (2003) used the retrograde transport ability of adeno-associated virus (AAV) to directly target affected motor neurons, to test the efficacy of IGF-1 and GDNF in the G93A SOD1 mouse model of ALS. In injections into the quadriceps muscle, as much as 1.1% of the virus injected at a dose of 1×1010 viral particles was transported to the lumbar region of the spinal cord, as assessed by quantitative PCR. The AAV vectors expressing GDNF or IGF-1 were bilaterally injected into the hindlimb quadriceps and intercostal muscles of G93A SOD1 animals before disease onset at 60 d of age, with a dosage of 1×1010 particles per injection (Kaspar et al. 2003). IGF-1 and GDNF treatment delayed ALS onset by 31 and 16 d and increased median survival by 37 and 16 d, respectively, compared with GFP-treated controls. Injections of IGF-1 not only delayed the onset but also slowed the rate of disease progression in some mice. In contrast, GDNF delayed the onset of symptoms but did not alter disease progression.
To test the ability of the IGF-1 and GDNF treatments to affect disease after it had begun, Kaspar et al. (2003) used the same treatment protocol on mice that were 90 d old, the age at which symptoms begin. In this treatment regimen, GDNF treatment caused a 7-d extension in survival (P<.0001). In contrast, IGF-1 treatment extended the median life span by 22 d and slowed progression of the disease, as assessed by body mass loss, rotarod performance, and grip strength. These combined results suggest that treatment with IGF-1 after the onset of overt motor dysfunction results not only in an extension of life but also in a delay in the functional decline associated with the disease.
In similar studies, Azzouz et al. (2004) used a recombinant lentiviral vector (rabies-G pseudotyped equine infectious anemia virus), which also exhibits retrograde transport, to test the effect of human VEGF expression on ALS onset in the G93A SOD1 mice. This experiment was based on the previous observation that low levels of VEGF expression correlate with ALS susceptibility (Lambrechts et al. 2003). Bilateral injections of VEGF viral vectors into hindlimb gastrocnemius, diaphragm, intercostal, facial, and tongue muscles at 3 wk of age significantly (P<.0001) delayed ALS onset (95–123 d) and increased the average life span (125–163 d) (Azzouz et al. 2004). Like Kaspar et al. (2003), Azzouz et al. (2004) also treated the symptomatic 90-d-old G93A SOD1 mice, increasing survival from 127 d to 146 d (P<.0001). Like IGF-1, VEGF is an effective treatment after disease onset and appears to both delay initiation and slow propagation of disease in the G93A SOD1 mice.
Future Directions
It is difficult to predict the future, but the identification of both additional ALS genes and ALS modifier genes will allow the creation of new models for study. The utility of such models for understanding the pathogenesis of ALS has been demonstrated in the mutant SOD1 mice. Mechanistic insights gleaned from new ALS genes and mouse models will uncover new targets for therapy development. The testing of synergistic combinations of therapeutics targeting multiple pathogenic mechanisms has already begun, and such studies will likely be expanded. If safety issues with gene therapy vectors can be ameliorated, clinical trials with IGF-1 or VEGF would be worth pursuing, since they are effective in model organisms after symptomatic onset of disease. One potential approach to ALS therapy, not discussed in detail above, is treatment with stem cells. Human safety studies for intraspinal cord implantation of autologous mesenchymal stem cells have already begun in patients with ALS (Mazzini et al. 2003), and clinical trials with stem cells, perhaps as delivery vehicles for neurotrophic factors, may be conducted in the future. Therapy development has unfortunately lagged behind the elucidation of the genetic and pathogenic mechanisms involved in ALS. The future, however, is bright. Because it is possible to slow disease progression in mouse models of familial ALS after symptomatic onset of disease, the creation of effective therapies for ALS is likely an achievable task.
Note added in proof.—The ALS8 locus (table 1) was recently identified. A missense mutation in the vesicle-associated membrane protein/synaptobrevin-associated membrane protein B (VAPB [MIM 605704]) was discovered in several families with ALS and related motor neuron diseases (Nishimura et al. 2004b).
Acknowledgments
This work was supported by National Institutes of Health grant NS041646. I would like to thank Dr. David Patterson, Dr. Patrick Bosque, Dr. Miles Brennan, and Sharon Trilk, for helpful comments and corrections.
Electronic-Database Information
The URLs for data presented herein are as follows:
- alsod.org: The ALS Online Database, http://www.alsod.org/
- ALS Therapy Development Foundation, http://www.als.net/
- ALS Therapy Development Foundation Treatment Targets, http://www.als.net/research/treatments/targetClasses.asp
- Motor Syndromes, http://www.neuro.wustl.edu/neuromuscular/motor.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ALS1, SOD1, CCS, EAAT2, GFAP, GLAST, EAAC1, ALS4, ALS2, ALS5, alsin, SETX, ALS-FTD, MAPT, FTD, progressive motor neuron disease without sensory symptoms, DCTN1, GluR2, ADAR2, NF-H, VEGF, SMN1, SMA, SMN2, CNTF, ApoE, GDNF, BDNF, IGF-1, ALS3, ALS6, ALS7, ALS8, VAPB, and ALS X)
- Revised Criteria for the Diagnosis of Amyotrophic Lateral Sclerosis (El Escorial Revisited), http://www.wfnals.org/guidelines/1998elescorial/elescorial1998.htm [DOI] [PubMed]
References
- Abalkhail H, Mitchell J, Habgood J, Orrell R, de Belleroche J (2003) A new familial amyotrophic lateral sclerosis locus on chromosome 16q12.1-16q12.2. Am J Hum Genet 73:383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, Andersen PM, Chioza B, Shaw C, Sham PC, Robberecht W, Matthijs G, Camu W, Marklund SL, Forsgren L, Rouleau G, Laing NG, Hurse PV, Siddique T, Leigh PN, Powell JF (1998) Recessive amyotrophic lateral sclerosis families with the D90A SOD1 mutation share a common founder: evidence for a linked protective factor. Hum Mol Genet 7:2045–2050 10.1093/hmg/7.13.2045 [DOI] [PubMed] [Google Scholar]
- Al-Chalabi A, Andersen PM, Nilsson P, Chioza B, Andersson JL, Russ C, Shaw CE, Powell JF, Leigh PN (1999) Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum Mol Genet 8:157–164 10.1093/hmg/8.2.157 [DOI] [PubMed] [Google Scholar]
- Andersen PM, Nilsson P, Keranen M-L, Forsgren L, Hagglund J, Karlsborg M, Ronnevi L-O, Gredal O, Marklunk SL (1997) Phenotypic heterogeneity in motor neuron disease patients with CuZn-superoxide dismutase mutations in Scandinavia. Brain 120:1723–1737 10.1093/brain/120.10.1723 [DOI] [PubMed] [Google Scholar]
- Andersen PM, Sims KB, Xin WW, Kiely R, O’Neill G, Ravits J, Pioro E, Harati Y, Brower RD, Levine JS, Heinicke HU, Seltzer W, Boss M, Brown RH Jr (2003) Sixteen novel mutations in the Cu/Zn superoxide dismutase gene in amyotrophic lateral sclerosis: a decade of discoveries, defects and disputes. Amyotroph Lateral Scler Other Motor Neuron Disord 4:62–73 [DOI] [PubMed] [Google Scholar]
- Armon C (2003) Epidemiology of amyotrophic lateral sclerosis/motor neuron disease. In: Shaw PJ, Strong MJ (eds) Motor neuron disease (Blue Book). Butterworth-Heinemann, Philadelphia, pp 167–205 [Google Scholar]
- Azzouz M, Ralph GS, Storkebaum E, Walmsley LE, Mitrophanous KA, Kingsman S M, Carmeliet P, Mazarakis ND (2004) VEGF delivery with retrogradely transported lentivector prolongs survival in a mouse ALS model. Nature 429:413–417 10.1038/nature02544 [DOI] [PubMed] [Google Scholar]
- Banack SA, Cox PA (2003) Biomagnification of cycad neurotoxins in flying foxes: implications for ALS-PDC in Guam. Neurology 61:387–389 [DOI] [PubMed] [Google Scholar]
- Beal MF, Ferrante RJ, Browne SE, Matthews RT, Kowall NW, Brown RH Jr (1997) Increased 3-nitrotyrosine in both sporadic and familial amyotrophic lateral sclerosis. Ann Neurol 42:644–654 [DOI] [PubMed] [Google Scholar]
- Brannstrom T, Ernhill K, Marklund S, Nilsson P (1998) Transgenic mice homozygous for the Asp90Ala human SOD1 mutation develop ALS clinically and histologically. Paper presented at the Annual Meeting of the Society for Neuroscience, Los Angeles, November 7–12 [Google Scholar]
- Browne SE, Bowling AC, Baik MJ, Gurney M, Brown RH Jr, Beal MF (1998) Metabolic dysfunction in familial, but not sporadic, amyotrophic lateral sclerosis. J Neurochem 71:281–287 [DOI] [PubMed] [Google Scholar]
- Bruijn LI (2002) Amyotrophic lateral sclerosis: from disease mechanisms to therapies. BioTechniques 32:1112–1121 [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Beal MF, Becher MW, Schulz JB, Wong PC, Price DL, Cleveland DW (1997a) Elevated free nitrotyrosine levels, but not protein-bound nitrotyrosine or hydroxyl radicals, throughout amyotrophic lateral sclerosis (ALS)-like disease implicate tyrosine nitration as an aberrant in vivo property of one familial ALS-linked superoxide dismutase 1 mutant. Proc Natl Acad Sci USA 94:7606–7611 10.1073/pnas.94.14.7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruijn LI, Becher MW, Lee MK, Anderson KL, Jenkins NA, Copeland NG, Sisodia SS, Rothstein JD, Borchelt DR, Price DL, Cleveland DW (1997b) ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron 18:327–338 10.1016/S0896-6273(00)80272-X [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, Ohama E, Reaume AG, Scott RW, Cleveland DW (1998) Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 281:1851–1854 10.1126/science.281.5384.1851 [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Miller TM, Cleveland DW (2004) Unraveling the mechanisms involved in motor neuron degeneration in ALS. Ann Rev Neurosci 27:723–749 10.1146/annurev.neuro.27.070203.144244 [DOI] [PubMed] [Google Scholar]
- Cai H, Wen H, Chaing HC, Price DL, Wong PC (2003) Physiological role of ALS2: selective vulnerability and generation of ALS2 knockout mice. Amyotroph Lateral Scler Other Motor Neuron Disord 4 Suppl 1:11–12 [Google Scholar]
- Chen Y-Z, Bennet CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, Nicholson GA, Auer-Grumbach M, Wagner K, De Jonghe P, Griffin JW, Fischbeck KH, Timmerman V, Cornblath DR, Chance PF (2004) DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am J Hum Genet 74:1128–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LN, Poorkaj P, Wszolek Z, Geschwind DH, Nasreddine ZS, Miller B, Li D, Payami H, Awert F, Markopoulou K, Andreadis A, D’Souza I, Lee VM, Reed L, Trojanowski JQ, Zhukareva V, Bird T, Schellenberg G, Wilhelmsen KC (1998) Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proc Natl Acad Sci USA 95:13103–13107 10.1073/pnas.95.22.13103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement AM, Nguyen MD, Roberts EA, Garcia ML, Boillee S, Rule M, McMahon AP, Doucette W, Siwek D, Ferrante RJ, Brown RH Jr, Julien JP, Goldstein LS, Cleveland DW (2003) Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science 302:113–117 10.1126/science.1086071 [DOI] [PubMed] [Google Scholar]
- Cleveland DW (1999) From Charcot to SOD1: mechanisms of selective motor neuron death in ALS. Neuron 24:515–520 10.1016/S0896-6273(00)81108-3 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD (2001) From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci 2:806–819 10.1038/35097565 [DOI] [PubMed] [Google Scholar]
- Corcia P, Mayeux-Portas V, Khoris J, de Toffol B, Autret A, Muh JP, Camu W, Andres C, French ALS Research Group (2002) Abnormal SMN1 gene copy number is a susceptibility factor for amyotrophic lateral sclerosis. Ann Neurol 51:243–246 10.1002/ana.10104 [DOI] [PubMed] [Google Scholar]
- Couillard-Després S, Zhu Q, Wong PC, Price DL, Cleveland DW, Julien JP (1998) Protective effect of neurofilament heavy gene overexpression in motor neuron disease induced by mutant superoxide dismutase. Proc Natl Acad Sci USA 95:9626–9630 10.1073/pnas.95.16.9626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JP, Sampson JB, Zhuang Y, Thompson JA, Beckman JS (1997) Decreased zinc affinity of amyotrophic lateral sclerosis-associated superoxide dismutase mutants leads to enhanced catalysis of tyrosine nitration by peroxynitrite. J Neurochem 69:1936–1944 [DOI] [PubMed] [Google Scholar]
- Cudkowicz ME, McKenna-Yasek D, Sapp PE, Chin W, Geller B, Hayden DL, Schoenfeld DA, Hosler BA, Horvitz HR, Brown RH (1997) Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann Neurol 41:210–221 [DOI] [PubMed] [Google Scholar]
- Dal Canto MC, Gurney ME (1995) Neuropathological changes in two lines of mice carrying a transgene for mutant human Cu,Zn SOD, and in mice overexpressing wild type human SOD: a model of familial amyotrophic lateral sclerosis (FALS). Brain Res 676:25–40 10.1016/0006-8993(95)00063-V [DOI] [PubMed] [Google Scholar]
- De Jonghe P, Auer-Grumbach M, Irobi J, Wagner K, Plecko B, Kennerson M, Zhu D, De Vriendt E, Van Gerwen V, Nicholson G, Hartung H-P, Timmerman V (2002) Autosomal dominant juvenile amyotrophic lateral sclerosis and distal hereditary motor neuronopathy with pyramidal tract signs: synonyms for the same disorder? Brain 125:1320–1325 10.1093/brain/awf127 [DOI] [PubMed] [Google Scholar]
- Deng H-X, Fu R, Zhai H, Siddique T (1999) A truncation mutation (L126Z) of SOD1 gene leads to ALS-like phenotype in transgenic mice. Am J Hum Genet Suppl 65:A292 [Google Scholar]
- Drory VE, Birnbaum M, Korczyn AD, Chapman (2001) Association of APOE ɛ4 allele with survival in amyotrophic lateral sclerosis. J Neurol Sci 190:17–20 10.1016/S0022-510X(01)00569-X [DOI] [PubMed] [Google Scholar]
- Durham HD, Roy J, Dong L, Figlewicz DA (1997) Aggregation of mutant Cu/Zn superoxide dismutase proteins in a culture model of ALS. J Neuropathol Exp Neurol 56:523–530 [DOI] [PubMed] [Google Scholar]
- Elliott JL (2001) Cytokine upregulation in a murine model of familial amyotrophic lateral sclerosis. Brain Res Mol Brain Res 95:172–178 [DOI] [PubMed] [Google Scholar]
- Estévez AG, Crow JP, Sampson JB, Reiter C, Zhuang Y, Richardson GJ, Tarpey MM, Barbeito L, Beckman JS (1999) Induction of nitric oxide-dependent apoptosis in motor neurons by zinc-deficient superoxide dismutase. Science 286:2498–2500 10.1126/science.286.5449.2498 [DOI] [PubMed] [Google Scholar]
- Estévez AG, Spear N, Manuel SM, Radi R, Henderson CE, Barbeito L, Beckman JS (1998) Nitric oxide and superoxide contribute to motor neuron apoptosis induced by trophic factor deprivation. J Neurosci 18:923–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eymard-Pierre E, Lesca G, Dollet S, Santorelli FM, di Capua M, Bertini E, Boespflug-Tanguy O (2002) Infantile-onset ascending hereditary spastic paralysis is associated with mutations in the alsin gene. Am J Hum Genet 71:518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti F, Sasaki M, Cutting FB, Zhai P, MacDonald JE, Reif D, Beal MF, Huang PL, Dawson TM, Gurney ME, Dawson VL (1999) Lack of involvement of neuronal nitric oxide synthase in the pathogenesis of a transgenic mouse model of familial amyotrophic lateral sclerosis. Neuroscience 90:1483–1492 10.1016/S0306-4522(98)00492-8 [DOI] [PubMed] [Google Scholar]
- Flowers JM, Powell JF, Leigh PN, Andersen P, Shaw CE (2001) Intron 7 retention and exon 9 skipping EAAT2 mRNA variants are not associated with amyotrophic lateral sclerosis. Ann Neurol 49:643–649 10.1002/ana.1029.abs [DOI] [PubMed] [Google Scholar]
- Giess R, Holtmann B, Braga M, Grimm T, Muller-Myhsok B, Toyka KV, Sendtner M (2002) Early onset of severe familial amyotrophic lateral sclerosis with a SOD-1 mutation: potential impact of CNTF as a candidate modifier gene. Am J Hum Genet 70:1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong YH, Parsadanian AS, Andreeva A, Snider WD, Elliott JL (2000) Restricted expression of G86R Cu/Zn superoxide dismutase in astrocytes results in astrocytosis but does not cause motoneuron degeneration. J Neurosci 20:660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeneveld GJ, Veldink JH, van der Tweel I, Kalmijn S, Beijer C, de Visser M, Wokke JH, Franssen H, van den Berg LH (2003) A randomized sequential trial of creatine in amyotrophic lateral sclerosis. Ann Neurol 53:437–445 10.1002/ana.10554 [DOI] [PubMed] [Google Scholar]
- Gurney ME, Fleck TJ, Himes CS, Hall ED (1998) Riluzole preserves motor function in a transgenic model of familial amyotrophic lateral sclerosis. Neurology 50:62–66 [DOI] [PubMed] [Google Scholar]
- Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH Jr, Scherer SW, Rouleau GA, Hayden MR, Ikeda JE (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29:166–173 10.1038/ng1001-166 [DOI] [PubMed] [Google Scholar]
- Haley RW (2003) Excess incidence of ALS in young Gulf War veterans. Neurology 61:750–756 [DOI] [PubMed] [Google Scholar]
- Hand CK, Khoris J, Salachas F, Gros-Louis F, Lopes AAS, Mayeux-Portas V, Brown RH Jr, Meininger V, Camu W, Rouleau GA (2002) A novel locus for familial amyotrophic lateral sclerosis on chromosome 18q. Am J Hum Genet 70:251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand CK, Rouleau GA (2002) Familial amyotrophic lateral sclerosis. Muscle Nerve 25:135–159 10.1002/mus.10001.abs [DOI] [PubMed] [Google Scholar]
- Heath PR, Shaw PJ (2002) Update on the glutamatergic neurotransmitter system and the role of excitotoxicity in amyotrophic lateral sclerosis. Muscle Nerve 26:438–458 10.1002/mus.10186 [DOI] [PubMed] [Google Scholar]
- Hentati A, Ouahchi K, Pericak-Vance MA, Nijhawan D, Ahmad A, Yang Y, Rimmler J, Hung W, Schlotter B, Ahmed A, Ben Hamida M, Hentati F, Siddique T (1998) Linkage of a commoner form of recessive amyotrophic lateral sclerosis to chromosome 15q15-q22 markers. Neurogenetics 2:55–60 10.1007/s100480050052 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406:78–81 10.1038/35017558 [DOI] [PubMed] [Google Scholar]
- Hosler BA, Siddique T, Sapp PC, Sailor W, Huang MC, Hossain A, Daube JR, Nance M, Fan C, Kaplan J, Hung W-Y, McKenna-Yasek D, Haines JL, Pericak-Vance MA, Horvitz HR, Brown RH Jr (2000) Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. JAMA 284:1664–1669 [DOI] [PubMed] [Google Scholar]
- Howland DS, Liu J, She Y, Goad B, Maragakis NJ, Kim B, Erickson J, Kulik J, DeVito L, Psaltis G, DeGennaro LJ, Cleveland DW, Rothstein JD (2002) Focal loss of glutamate transporter EAAT2 in transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 99:1604–1609 10.1073/pnas.032539299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, et al (1998) Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393:702–705 10.1038/31508 [DOI] [PubMed] [Google Scholar]
- Ince PG (2000) Neuropathology. In: Brown RH Jr, Meininger K, Swash M (eds) Amyotrophic lateral sclerosis. Martin Dunitz, London, pp 83–112 [Google Scholar]
- Ischiropoulos H, Beckman JS (2003) Oxidative stress and nitration in neurodegeneration: cause, effect, or association? J Clin Invest 111:163–169 10.1172/JCI200317638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaarsma D, Haasdijk ED, Grashorn JA, Hawkins R, van Duijn W, Verspaget HW, London J, Holstege JC (2000) Human Cu/Zn superoxide dismutase (SOD1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant SOD1. Neurobiol Dis 7:623–643 10.1006/nbdi.2000.0299 [DOI] [PubMed] [Google Scholar]
- Jonsson PA, Ernhill K, Andersen PM, Bergemalm D, Brannstrom T, Gredal O, Nilsson P, Marklund SL (2004) Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain 127:73–88 10.1093/brain/awh005 [DOI] [PubMed] [Google Scholar]
- Jung C, Higgins CM, Xu Z (2002) A quantitative histochemical assay for activities of mitochondrial electron transport chain complexes in mouse spinal cord sections. J Neurosci Methods 114:165–172 10.1016/S0165-0270(01)00524-6 [DOI] [PubMed] [Google Scholar]
- Kamel F, Umbach DM, Lehman TA, Park LP, Munsat TL, Shefner JM, Sandler DP, Hu H, Taylor JA (2003) Amyotrophic lateral sclerosis, lead, and genetic susceptibility: polymorphisms in the delta-aminolevulinic acid dehydratase and vitamin D receptor genes. Environ Health Perspect 111:1335–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel F, Umbach DM, Munsat TL, Shefner JM, Sandler DP (1999) Association of cigarette smoking with amyotrophic lateral sclerosis. Neuroepidemiology 18:194–202 10.1159/000026211 [DOI] [PubMed] [Google Scholar]
- Kanekura K, Hashimoto Y, Niikura T, Aiso S, Matsuoka M, Nishimoto I (2004) Alsin, the product of the ALS2 gene, suppresses SOD1 mutant neurotoxicity through RhoGEF domain by interacting with SOD1 mutants. J Biol Chem 279:19247–19256 10.1074/jbc.M313236200 [DOI] [PubMed] [Google Scholar]
- Kaspar BK, Lladó J, Sherkat N, Rothstein JD, Gage FH (2003) Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science 301:839–842 10.1126/science.1086137 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kawk S (2004) RNA editing and the death of motor neurons: there is a glutamate-receptor defect in patients with amyotrophic lateral sclerosis. Nature 427:801 10.1038/427801a [DOI] [PubMed] [Google Scholar]
- Kieran D, Kalmar B, Dick JR, Riddoch-Contreras J, Burnstock G, Greensmith L (2004) Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nat Med 10:345–347 10.1038/nm0404-345 [DOI] [PubMed] [Google Scholar]
- Klivenyi P, Ferrante RJ, Matthews RT, Bogdanov MB, Klein AM, Andreassen OA, Mueller G, Wermer M, Kaddurah-Daouk R, Beal MF (1999) Neuroprotective effects of creatine in a transgenic animal model of amyotrophic lateral sclerosis. Nature Med 5:347–350 10.1038/6568 [DOI] [PubMed] [Google Scholar]
- Klivenyi P, Kiaei M, Gardian G, Calingasan NY, Beal MF (2004) Additive neuroprotective effects of creatine and cyclooxygenase 2 inhibitors in a transgenic mouse model of amyotrophic lateral sclerosis. J Neurochem 88:576–582 [DOI] [PubMed] [Google Scholar]
- Kong J, Xu Z (1998) Massive mitochondrial degeneration in motor neurons triggers the onset of amyotrophic lateral sclerosis in mice expressing a mutant SOD1. J Neurosci 18:3241–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalska A, Konagaya M, Sakai M, Hashizume Y, Tabira T (2003) Familial amyotrophic lateral sclerosis and parkinsonism-dementia complex—tauopathy without mutations in the tau gene? Folia Neuropathol 41:59–64 [PubMed] [Google Scholar]
- Kriz J, Millecamps S, Zhu Q, Julien J-P (2003) Creation of a mouse model for juvenile amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 4 Suppl 1:11 [Google Scholar]
- Kunita R, Otomo A, Mizumura H, Suzuki K, Showguchi-Miyata J, Yanagisawa Y, Hadano S, Ikeda JE (2004) Homo-oligomerization of ALS2 through its unique carboxy-terminal regions is essential for the ALS2-associated Rab5 guanine nucleotide exchange activity and its regulatory function on endosome trafficking. J Biol Chem 279:38626–38635 10.1074/jbc.M406120200 [DOI] [PubMed] [Google Scholar]
- Kunst C, Messer L, Gordon J, Haines J, Patterson D (2000) Genetic mapping of a mouse modifier gene that can prevent ALS onset. Genomics 70:181–189 10.1006/geno.2000.6379 [DOI] [PubMed] [Google Scholar]
- Lambrechts D, Storkebaum E, Morimoto M, Del-Favero J, Desmet F, Marklund SL, Wyns S, et al (2003) VEGF is a modifier of amyotrophic lateral sclerosis in mice and humans and protects motoneurons against ischemic death. Nat Genet 34:383–394 10.1038/ng1211 [DOI] [PubMed] [Google Scholar]
- Lariviere RC, Julien JP (2004) Functions of intermediate filaments in neuronal development and disease. J Neurobiol 58:131–148 10.1002/neu.10270 [DOI] [PubMed] [Google Scholar]
- Li M, Ona VO, Guegan C, Chen M, Jackson-Lewis V, Andrews LJ, Olszewski AJ, Stieg PE, Lee JP, Przedborski S, Friedlander RM (2000) Functional role of caspase-1 and caspase-3 in an ALS transgenic mouse model. Science 288:335–339 10.1126/science.288.5464.335 [DOI] [PubMed] [Google Scholar]
- Lin C-LG, Bristol LA, Jin L, Dykes-Hoberg M, Crawford T, Clawson L, Rothstein JD (1998) Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron 20:589–602 10.1016/S0896-6273(00)80997-6 [DOI] [PubMed] [Google Scholar]
- Lino MM, Schneider C, Caroni P (2002) Accumulation of SOD1 mutants in postnatal motoneurons does not cause motoneuron pathology or motoneuron disease. J Neurosci 22:4825–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lillo C, Jonsson PA, Velde CV, Ward CM, Miller TM, Subramaniam JR, Rothstein JD, Marklund S, Andersen PM, Brannstrom T, Gredal O, Wong PC, Williams DS, Cleveland DW (2004) Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron 43:5–17 10.1016/j.neuron.2004.06.016 [DOI] [PubMed] [Google Scholar]
- Matthews RT, Yang L, Browne S, Baik M, Beal MF (1998) Coenzyme Q10 administration increases brain mitochondrial concentrations and exerts neuroprotective effects. Proc Natl Acad Sci USA 95:8892–8897 10.1073/pnas.95.15.8892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiazzi M, D’Aurelio M, Gajewski CD, Martushova K, Kiaei M, Beal MF, Manfredi G (2002) Mutated human SOD1 causes dysfunction of oxidative phosphorylation in mitochondria of transgenic mice. J Biol Chem 277:29626–29633 10.1074/jbc.M203065200 [DOI] [PubMed] [Google Scholar]
- Mazzini L, Fagioli F, Boccaletti R, Mareschi K, Oliveri G, Olivieri C, Pastore I, Marasso R, Madon E (2003) Stem cell therapy in amyotrophic lateral sclerosis: a methodological approach in humans. Amyotroph Lateral Scler Other Motor Neuron Disord 4:158–161 10.1080/14660820310014653 [DOI] [PubMed] [Google Scholar]
- McGeer PL, McGeer EG (2002) Inflammatory processes in amyotrophic lateral sclerosis. Muscle Nerve 26:459–470 10.1002/mus.10191 [DOI] [PubMed] [Google Scholar]
- Meyer T, Alber B, Roemer K, Martin T, Kalscheuer VM, Gottert E, Zang KD, Ludolph AC, Ropers H-H, Prudlo J (2003) High rate of constitutional chromosomal rearrangements in apparently sporadic ALS. Neurology 60:1348–1350 [DOI] [PubMed] [Google Scholar]
- Miller RG, Mitchell JD, Lyon M, Moore DH (2003) Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Amyotroph Lateral Scler Other Motor Neuron Disord 4:191–206 10.1080/14660820310002601 [DOI] [PubMed] [Google Scholar]
- Mitchell JD, Wokke JH, Borasio GD (2002) Recombinant human insulin-like growth factor I (rhIGF-I) for amyotrophic lateral sclerosis/motor neuron disease. Cochrane Database Syst Rev 3:CD002064 [DOI] [PubMed] [Google Scholar]
- Moreira MC, Klur S, Watanabe M, Nemeth AH, Ber IL, Moniz JC, Tranchant C, et al (2004) Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat Genet 36:225–227 10.1038/ng1303 [DOI] [PubMed] [Google Scholar]
- Nagai M, Aoki M, Miyoshi I, Kato M, Pasinelli P, Kasai N, Brown RH Jr, Itoyama Y (2001) Rats expressing human cytosolic copper-zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: associated mutations develop motor neuron disease. J Neurosci 21:9246–9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LM (1995) Epidemiology of ALS. Clin Neurosci 3:327–331 [PubMed] [Google Scholar]
- Nelson LM, Matkin C, Longstreth WT Jr, McGuire V (2000a) Population-based case-control study of amyotrophic lateral sclerosis in western Washington State. II. Diet. Am J Epidemiol 151:164–173 [DOI] [PubMed] [Google Scholar]
- Nelson LM, McGuire V, Longstreth WT Jr, Matkin C (2000b) Population-based case-control study of amyotrophic lateral sclerosis in western Washington State. I. Cigarette smoking and alcohol consumption. Am J Epidemiol 151:156–163 [DOI] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Oliveira JR, Vainzof M, Zatz M (2004a) A novel locus for late onset amyotrophic lateral sclerosis/motor neurone disease variant at 20q13. J Med Genet 41:315–320 10.1136/jmg.2003.013029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P, Zatz M (2004b) A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet 75:822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obal I, Jakab JS, Siklos L, Engelhardt JI (2001) Recruitment of activated microglia cells in the spinal cord of mice by ALS IgG. Neuroreport 12:2449–2452 [DOI] [PubMed] [Google Scholar]
- Oosthuyse B, Moons L, Storkebaum E, Beck H, Nuyens D, Brusselmans K, Van Dorpe J, et al (2001) Deletion of the hypoxia-response element in the vascular endothelial growth factor promoter causes motor neuron degeneration. Nat Genet 28:131–138 10.1038/88842 [DOI] [PubMed] [Google Scholar]
- Ostojic J, Axelman, Lannfelt L, Froelich-Fabre S (2003) No evidence of linkage to chromosome 9q21-q22 in a Swedish family with frontotemporal dementia and amyotrophic lateral sclerosis. Neurosci Lett 340:245–247 10.1016/S0304-3940(03)00126-5 [DOI] [PubMed] [Google Scholar]
- Otomo A, Hadano S, Okada T, Mizumura H, Kunita R, Nishijima H, Showguchi-Miyata J, Yanagisawa Y, Kohiki E, Suga E, Yasuda M, Osuga H, Nishimoto T, Narumiya S, Ikeda JE. (2003) ALS2, a novel guanine nucleotide exchange factor for the small GTPase Rab5, is implicated in endosomal dynamics. Hum Mol Genet 12:1671–1687 10.1093/hmg/ddg184 [DOI] [PubMed] [Google Scholar]
- Parton MJ, Broom W, Andersen PM, Al-Chalabi A, Nigel Leigh P, Powell JF, Shaw CE; D90A SOD1 ALS Consortium (2002) D90A-SOD1 mediated amyotrophic lateral sclerosis: a single founder for all cases with evidence for a cis-acting disease modifier in the recessive haplotype. Hum Mutat 20:473 10.1002/humu.9081 [DOI] [PubMed] [Google Scholar]
- Pramatarova A, Laganiere J, Roussel J, Brisebois K, Rouleau GA (2001) Neuron-specific expression of mutant superoxide dismutase 1 in transgenic mice does not lead to motor impairment. J Neurosci 21:3369–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudlo J, Alber B, Kalscheuer VM, Roemer K, Martin T, Dullinger J, Sittinger H, Niemann S, Heutink P, Ludolph AC, Ropers HH, Zang K, Meyer T (2004) Chromosomal translocation t(18;21)(a23;q22.1) indicates novel susceptibility loci for frontotemporal dementia with ALS. Ann Neurol 55:134–138 10.1002/ana.10822 [DOI] [PubMed] [Google Scholar]
- Przedborski S, Vila M, Jackson-Lewis V (2003) Series introduction: neurodegeneration: what is it and where are we? J Clin Invest 111:3–10 10.1172/JCI200317522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH Jr, Ludlow CL, Fischbeck KH (2003) Mutant dynactin in motor neuron disease. Nat Genet 33:455–456 10.1038/ng1123 [DOI] [PubMed] [Google Scholar]
- Puttaparthi K, Wojcik C, Rajendran B, DeMartino GN, Elliott JL (2003) Aggregate formation in the spinal cord of mutant SOD1 transgenic mice is reversible and mediated by proteasomes. J Neurochem 87:851–860 10.1046/j.1471-4159.2003.02028.x [DOI] [PubMed] [Google Scholar]
- Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, Siwek DF, Wilcox HM, Flood DG, Beal MF, Brown RH Jr, Scott RW, Snider WD (1996) Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet 13:43–47 10.1038/ng0596-43 [DOI] [PubMed] [Google Scholar]
- Rezania K, Yan J, Dellefave L, Deng H-X, Siddique N, Pascuzzi RT, Siddique T, Roos RP (2003) A rare Cu/Zn superoxide dismutase mutation causing familial amyotrophic lateral sclerosis with variable age of onset, incomplete penetrance and a sensory neuropathy. Amyotroph Lateral Scler Other Motor Neuron Disord 4:162–166 [DOI] [PubMed] [Google Scholar]
- Riggs JE, Schochet SS Jr (1992) Rising mortality due to Parkinson’s disease and amyotrophic lateral sclerosis: a manifestation of the competitive nature of human mortality. J Clin Epidemiol 45:1007–1012 10.1016/0895-4356(92)90116-5 [DOI] [PubMed] [Google Scholar]
- Ripps ME, Huntley GW, Hof PR, Morrison JH, Gordon JW (1995) Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA 92:689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, et al (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362:59–62 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Van Kammen M, Levey AI, Martin LJ, Kuncl RW (1995) Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. Ann Neurol 38:73–84 [DOI] [PubMed] [Google Scholar]
- Rouleau GA, Clark AW, Rooke K, Pramatarova A, Krizus A, Suchowersky O, Julien JP, Figlewicz D (1996) SOD1 mutation is associated with accumulation of neurofilaments in amyotrophic lateral sclerosis. Ann Neurol 39:128–131 [DOI] [PubMed] [Google Scholar]
- Roy J, Minotti S, Dong L, Figlewicz DA, Durham HD (1998) Glutamate potentiates the toxicity of mutant Cu/Zn-superoxide dismutase in motor neurons by postsynaptic calcium-dependent mechanisms. J Neurosci 18:9673–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddy DM, Parton MJ, Al-Chalabi A, Lewis CM, Vance C, Smith BN, Leigh N, Powell JF, Siddique T, Meyjes EP, Baas F, De Jong V, Shaw CE (2003) Two families with familial amyotrophic lateral sclerosis are linked to a novel locus on chromosome 16q. Am J Hum Genet 73:390–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp PC, Hosler BA, McKenna-Yasek D, Chin W, Gann A, Genise H, Gorenstein J, Huang M, Sailer W, Scheffler M, Valesky M, Haines JL, Pericak-Vance M, Siddique T, Horvitz HR, Brown RH Jr (2003) Identification of two novel loci for dominantly inherited familial amyotrophic lateral sclerosis. Am J Hum Genet 73:397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarmeas N, Shih T, Stern Y, Ottman R, Rowland LP (2002) Premorbid weight, body mass, and varsity athletics in ALS. Neurology 59:773–775 [DOI] [PubMed] [Google Scholar]
- Siddique T, Hong ST, Brooks BR, Hung WY, Siddique NA, Rimmler J, Kaplan JP, Haines JL, Brown RH Jr, Pericak-Vance MA (1998a) X-linked dominant locus for late-onset familial amyotrophic lateral sclerosis. Am J Hum Genet Suppl 63:A308 [Google Scholar]
- Siddique T, Pericak-Vance MA, Caliendo J, Hong ST, Hung WY, Kaplan J, McKenna-Yasek D, Rimmler JB, Sapp P, Saunders AM, Scott WK, Siddique N, Haines JL, Brown RH (1998b) Lack of association between apolipoprotein E genotype and sporadic amyotrophic lateral sclerosis. Neurogenetics 1:213–216 10.1007/s100480050031 [DOI] [PubMed] [Google Scholar]
- Subramaniam JR, Lyons WE, Liu J, Bartnikas TB, Rothstein J, Price DL, Cleveland DW, Gitlin JD, Wong PC (2002) Mutant SOD1 causes motor neuron disease independent of copper chaperone-mediated copper loading. Nat Neurosci 5:301–307 10.1038/nn823 [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Kobayashi Y, Yoshihara T, Niwa J, Doyu M, Ohtsuka K, Sobue G (2002) Hsp70 and Hsp40 improve neurite outgrowth and suppress intracytoplasmic aggregate formation in cultured neuronal cells expressing mutant SOD1. Brain Res 949:11–22 10.1016/S0006-8993(02)02568-4 [DOI] [PubMed] [Google Scholar]
- Topp JD, Gray NW, Gerard RD, Horazdovsky BF (2004) Alsin is a Rab5 and Rac1 guanine nucleotide exchange factor. J Biol Chem 279:24612–24623 10.1074/jbc.M313504200 [DOI] [PubMed] [Google Scholar]
- Veldink JH, van den Berg LH, Cobben JM, Stulp RP, De Jong JM, Vogels OJ, Baas F, Wokke JH, Scheffer H (2001) Homozygous deletion of the survival motor neuron 2 gene is a prognostic factor in sporadic ALS. Neurology 56:749–752 [DOI] [PubMed] [Google Scholar]
- Wang J, Slunt H, Gonzales V, Fromholt D, Coonfield M, Copeland NG, Jenkins NA, Borchelt DR (2003) Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum Mol Genet 12:2753–2764 10.1093/hmg/ddg312 [DOI] [PubMed] [Google Scholar]
- Wang J, Xu G, Gonzales V, Coonfield M, Fromholt D, Copeland NG, Jenkins NA, Borchelt DR (2002) Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol Dis 10:128–138 10.1006/nbdi.2002.0498 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Dykes-Hoberg M, Culotta VC, Price DL, Wong PC, Rothstein JD (2001) Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis 8:933–941 10.1006/nbdi.2001.0443 [DOI] [PubMed] [Google Scholar]
- Weisskopf MG, O’Reilly E, McCullough ML, Calle EE, Thun MJ, Cudkowicz M, Ascherio A (2004) Prospective study of military service and risk of amyotrophic lateral sclerosis. Paper presented at the 56th Annual Meeting of the American Academy of Neurology, San Francisco, April 24–May 1 [Google Scholar]
- Wilhelmsen KC, Forman MS, Rosen HJ, Alving LI, Goldman J, Feiger J, Lee JV, Segall SK, Kramer JH, Lomen-Hoerth C, Rankin KP, Johnson J, Feiler HS, Weiner MW, Lee VM, Trojanowski JQ, Miller BL (2004) 17q-linked frontotemporal dementia-amyotrophic lateral sclerosis without tau mutations with tau and alpha-synuclein inclusions. Arch Neurol 61:398–406 10.1001/archneur.61.3.398 [DOI] [PubMed] [Google Scholar]
- Williamson TL, Bruijn LI, Zhu Q, Anderson KL, Anderson SD, Julien JP, Cleveland DW (1998) Absence of neurofilaments reduces the selective vulnerability of motor neurons and slows disease caused by a familial amyotrophic lateral sclerosis-linked superoxide dismutase 1 mutant. Proc Natl Acad Sci USA 95:9631–9636 10.1073/pnas.95.16.9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JM, Khabazian I, Pow DV, Craig UK, Shaw CA (2003) Decrease in glial glutamate transporter variants and excitatory amino acid receptor down-regulation in a murine model of ALS-PDC. Neuromolecular Med 3:105–118 10.1385/NMM:3:2:105 [DOI] [PubMed] [Google Scholar]
- Wilson JM, Khabazian I, Wong MC, Seyedalikhani A, Bains JS, Pasqualotto BA, Williams DE, Andersen RJ, Simpson RJ, Smith R, Craig UK, Kurland LT, Shaw CA (2002) Behavioral and neurological correlates of ALS-parkinsonism dementia complex in adult mice fed washed cycad flour. Neuromolecular Med 1:207–221 10.1385/NMM:1:3:207 [DOI] [PubMed] [Google Scholar]
- Wong PC, Pardo CA, Borchelt DR, Lee MK, Copeland NG, Jenkins NA, Sisodia SS, Cleveland DW, Price DL (1995) An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron 14:1105–1116 10.1016/0896-6273(95)90259-7 [DOI] [PubMed] [Google Scholar]
- Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T (2001) The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 29:160–165 10.1038/ng1001-160 [DOI] [PubMed] [Google Scholar]