Abstract
Prostate cancer is one of the most common cancers among men and has long been recognized to occur in familial clusters. Brothers and sons of affected men have a 2–3-fold increased risk of developing prostate cancer. However, identification of genetic susceptibility loci for prostate cancer has been extremely difficult. Although the suggestion of linkage has been reported for many chromosomes, the most promising regions have been difficult to replicate. In this study, we compare genome linkage scans using microsatellites with those using single-nucleotide polymorphisms (SNPs), performed in 467 men with prostate cancer from 167 families. For the microsatellites, the ABI Prism Linkage Mapping Set version 2, with 402 microsatellite markers, was used, and, for the SNPs, the Early Access Affymetrix Mapping 10K array was used. Our results show that the presence of linkage disequilibrium (LD) among SNPs can lead to inflated LOD scores, and this seems to be an artifact due to the assumption of linkage equilibrium that is required by the current genetic-linkage software. After excluding SNPs with high LD, we found a number of new LOD-score peaks with values of at least 2.0 that were not found by the microsatellite markers: chromosome 8, with a maximum model-free LOD score of 2.2; chromosome 2, with a LOD score of 2.1; chromosome 6, with a LOD score of 4.2; and chromosome 12, with a LOD score of 3.9. The LOD scores for chromosomes 6 and 12 are difficult to interpret, because they occurred only at the extreme ends of the chromosomes. The greatest gain provided by the SNP markers was a large increase in the linkage information content, with an average information content of 61% for the SNPs, versus an average of 41% for the microsatellite markers. The strengths and weaknesses of microsatellite versus SNP markers are illustrated by the results of our genome linkage scans.
Introduction
Prostate cancer (MIM 176807) is one of the most common cancers in men in the Western world, as well as a leading cause of mortality, yet little is known about its causes. Old age, African American ancestry, and a family history of prostate cancer have long been recognized as important risk factors, yet we are only at the early stage of unraveling the complex genetic and environmental influences on this disease. Over the past 20 years, the body of evidence that genetics plays a key role has grown immensely, including a range of studies—from familial aggregation and twin studies, to family-based linkage studies, to detection of likely functional genes via mutation screening, to molecular epidemiological studies of both rare and common polymorphisms of candidate genes (Schaid 2004). However, the evidence also points toward a much more complex genetic basis of prostate cancer than was initially anticipated. Early linkage results have provided targeted candidate regions for prostate cancer–susceptibility loci, including HPC1 (MIM 601518) on chromosome 1q23-25 (Smith et al. 1996; Xu and International Consortium for Prostate Cancer Genetics 2000; Carpten et al. 2002), PCAP (MIM 602759) on chromosome 1q42-43 (Berthon et al. 1998), CAPB (MIM 603688) on chromosome 1p36 (Gibbs et al. 1999), chromosome 8p22-23 (Xu et al. 2001), HPC2 (MIM 605367) on chromosome 17p (Tavtigian et al. 2001), HPC20 (MIM 608656) on chromosome 20q13 (Berry et al. 2000), and HPCX (MIM 300146) on chromosome Xq27-28 (Xu et al. 1998). However, further reports and 10 genome linkage screens based on microsatellite markers (for reviews, see Easton et al. [2003] and Schaid [2004]) have demonstrated the difficulty in replicating linkage findings for prostate cancer susceptibility. Some causes of this complexity are likely to be a high rate of phenocopies; a lack of complete genetic information, because parents and older ancestors of men with prostate cancer are not available to genotype; low to moderate penetrance of susceptibility genes; multiple genes; and a variety of heterogeneous environmental risk factors.
One way to enrich genetic linkage information is to increase the marker density. Like many other groups using microsatellite markers, we have used the ABI Prism Linkage Mapping Set (Applied Biosystems) of STRs that have an average spacing of 10 cM. In contrast, the Early Access Affymetrix Mapping 10K array, which contains ∼10,000 SNP markers, is estimated to have an average spacing of 0.34 cM. To avoid confusion between the abbreviations STR and SNP, we will hereafter use the abbreviation “M-STR” for microsatellite marker. An advantage of the M-STRs is that they are highly polymorphic, much more so than the diallelic SNPs. The advantage of the SNPs is that they are much more plentiful, and the hope is that their greater density will compensate for the smaller amount of information per SNP, by creating local haplotypes of SNPs that function as “super” alleles, which, jointly as a haplotype, have greater linkage information content. There has been considerable debate on the advantages and disadvantages of M-STRs versus SNPs, in terms of their relative linkage information content, the ability of software to analyze the large number of SNPs, and the effect of linkage disequilibrium (LD) among the SNPs (current software for analyzing a large number of markers assumes that the markers are in linkage equilibrium). The present study has two objectives. The first aim is to perform a dense linkage analysis of the families with prostate cancer in our study, to extract nearly the maximum linkage information content. The second aim is to summarize our experience with using both M-STRs and SNPs on a common set of pedigrees, to assist in the scientific evaluation of the strengths and weaknesses of both genomic technologies.
Methods
Selection of Families
Each family was selected through a proband who received treatment for prostate cancer at the Mayo Clinic, with the requirement of at least three men with prostate cancer in the family, of whom at least two were alive for recruitment. The details of our large-scale survey, telephone follow-up, and family recruitment can be found elsewhere (Schaid et al. 1998; Cunningham et al. 2003). For the genotyping of M-STRs, we used 160 families, which included 437 men affected with prostate cancer and 157 unaffected men and women. For the genotyping of SNPs, we used 433 of these affected men from 159 families (four men were excluded because of degraded DNA, and one family was excluded, because it was no longer informative, as a result of degraded DNA). Because of cost constraints, we did not genotype the SNPs for the unaffected members of these original families. In addition, nine new affected subjects have been added to these pedigrees, and eight new pedigrees were recruited (25 affected men and 17 unaffected men and women). Hence, for the SNPs, all affected men in the original pedigrees were genotyped, and all members of the new pedigrees were genotyped, resulting in 167 families with 467 affected men. For the comparisons of linkage results from M-STRs versus SNPs, we used only the 159 families with 433 affected men that were genotyped by both technologies.
The research protocol and informed consents were approved by the Mayo Clinic Institutional Review Board. DNA was isolated from peripheral blood lymphocytes by standard methods.
Genetic Markers and Genotyping
For the M-STRs, the ABI Prism Linkage Mapping Set version 2 (Applied Biosystems), with 402 markers, was used as described by Cunningham et al. (2003). For the SNPs, the Early Access Affymetrix Mapping 10K array was used in accordance with the manufacturer’s recommendations. In brief, 250 ng of high-quality genomic DNA was digested with XbaI (20,000 U/ml), and adaptor sequences were ligated using T4 ligase on the digested DNA. After PCR amplification with Xba primers (250 μM dNTPs, 2.5 mM MgCl2, 0.75 μM each primer, and 0.1 U of AmpliTaq Gold), amplicons were processed via MinElute plate with a QIAvac 96 vacuum manifold and were quantified; 20 μg was required for the subsequent steps of fragmentation with DNase I, end labeling with biotinylated ddATP, and hybridization to the 10K array. Hybridization was detected by streptavidin-phycoerythrin conjugates. Arrays were processed through an Affymetrix microfluidics station and then were scanned on an Agilent reader. Quality controls included check gel electrophoresis after the PCR (2% agarose) and fragmentation (4% agarose) steps. Robotic workstations were used whenever possible, to minimize the chance of specimen-handling errors.
Statistical Analyses
Genetic Maps
The chromosome genetic maps for the M-STRs are based on the CEPH data and the Genethon linkage map (Gyapay et al. 1994). The average intermarker distance is 9.4 cM (25th–75th percentiles 7.1–11.3 cM). Because these maps were based on many more informative meioses than were available in our pedigrees, the maps were assumed to be correct. The chromosome genetic maps of the SNPs were created by Affymetrix, by (1) placing the deCODE microsatellites (Kong et al. 2002) and SNPs on the physical sequence map, (2) using the deCODE microsatellite map as a framework, and (3) using linear interpolation to place the SNPs on inferred genetic maps. Because the SNP genetic maps were not based on meioses within families, we used our families with prostate cancer to perform a crude validity check (crude, because our families are not very informative for construction and validation of fine-scale genetic-marker maps). To perform this check, we used the software CRIMAP (Lander and Green 1987) with the “flips2” option. This procedure flips adjacent marker loci along the assumed genetic-map sequence and reports LOD scores for the original sequence versus the flipped sequence. If the LOD score favored the flipped sequence by a LOD score of at least 1.0, then the order of the two markers involved was considered suspicious, and so the marker with the least heterozygosity was excluded from further analyses.
LOD-Score Calculations
The frequencies of all marker alleles, both M-STRs and SNPs, were estimated across the pool of all subjects, ignoring genetic relationships. Because founders of our pedigrees were not available to genotype, allele frequencies of the marker alleles can have a large impact on our linkage results. To avoid potential bias caused by rare alleles, we present analyses based only on SNPs with minor-allele frequencies of at least 5%. Multipoint model-free analyses based on the Kong and Cox (1997) exponential model were conducted by the software Merlin (Abecasis et al. 2002). Although we attempted to analyze the SNP data by use of Genehunter Plus, a modified version of Genehunter version 1.3 (Kruglyak et al. 1996), this software was limited by the large number of SNPs on some chromosomes. The information content of the genotypes was estimated by Merlin, by use of the entropy information described by Kruglyak et al. (1996).
Results
Families and Affected Men
The 167 families used for our genome linkage screen are described in table 1. There was one family with Hispanic heritage, one with African American heritage, and the remaining with white heritage. The average age at diagnosis per family ranged from 47 years to 75 years, with 21 families having an average age at diagnosis <60 years. The number of affected men per family ranged from 3 to 11, and the number of affected men with DNA available ranged from 2 to 7. To classify pedigrees as having hereditary prostate cancer (HPC) or not, we used the Carter criteria (Carter et al. 1993), which require a pedigree to have at least one of the following three criteria to be classified as a pedigree with HPC: (1) three consecutive generations of prostate cancer along a line of descent, (2) at least three first-degree relatives with a diagnosis of prostate cancer, or (3) two or more relatives with a prostate cancer diagnosis at an age ⩽55 years. Although 77% of our families were classified as having HPC, only 40% had five or more men with prostate cancer, 48% had both a father and a son with prostate cancer (“paternal transmission”), and 50% had an average age at diagnosis <66 years. Seventy-five families had two affected men whose DNA was available, 62 families had three, 22 families had four; and 8 families had five or more.
Table 1.
Characteristics of the 167 Families Used in Linkage Analysis
|
Affected Individuals with DNA Available |
|||||
| Families | No. of Families | Average No. ofAffected Individualsper Family (range) | Average Age at Diagnosisin Families (SD)(years) | No. ofIndividuals | Average perFamily (Range) |
| All | 167 | 4.5 (3–11) | 65.5 (5.0) | 467 | 2.8 (2–7) |
| With average age at diagnosis: | |||||
| <66 years | 84 | 4.6 (3–10) | 61.7 (3.7) | 230 | 2.7 (2–5) |
| ⩾66 years | 83 | 4.5 (3–11) | 69.5 (2.2) | 237 | 2.9 (2–7) |
| With no. of affected men: | |||||
| <5 | 100 | 3.4 (3–4) | 65.4 (5.3) | 241 | 2.4 (2–4) |
| ⩾5 | 67 | 6.2 (5–11) | 65.7 (4.4) | 226 | 3.4 (2–7) |
| With paternal transmission: | |||||
| Yes | 80 | 4.9 (3–10) | 65.1 (5.3) | 217 | 2.7 (2–5) |
| No | 87 | 4.2 (3–11) | 66.0 (4.6) | 250 | 2.9 (2–7) |
| With HPC: | |||||
| Yes | 129 | 4.7 (3–11) | 65.8 (5.2) | 365 | 2.8 (2–7) |
| No | 38 | 3.9 (3–8) | 64.6 (4.0) | 102 | 2.7 (2–5) |
Among the 467 affected men with available DNA, prostate cancer was confirmed by review of medical records for all but two men. A majority of men (90%) received diagnoses through a clinically indicated biopsy, and a majority (68%) received diagnoses after 1990, when prostate-specific antigen (PSA) screening became more widely used. Although most men had a PSA level ⩾4 at the time of diagnosis, PSA at diagnosis was missing for 24% of the men, and 10% had PSA levels <4. Gleason grade tended to be low for the group of men, with 58% having Gleason grade <7, and 18% with a missing grade. Most men did not have nodal involvement or metastatic prostate cancer at the time of diagnosis (82%). In addition, most men (66%) had a BMI <28 (BMI computed as the height [cm] squared, divided by weight [kg]). The most common type of treatment was radical prostatectomy alone (69%), followed by external-beam radiation therapy alone (16%).
Genotype Quality
The quality of the M-STR genotype data was checked in numerous ways, including Mendelian inheritance checks, checks for departures from Hardy-Weinberg genotype proportions, range checks on allele sizes, and verification of extremely rare alleles (for a summary of the quality of microsatellite genotype data, see Cunningham et al. 2003). Questionable genotypes were set to missing. After cleaning the M-STR data, we had 96% of the genotypes expected if there were no missing data. Reported relationships were evaluated by the software Relpair (Boehnke and Cox 1997), and subjects with questionable relationships were excluded from analyses. Note that the M-STR analyses were completed before initiation of SNP genotyping, and subjects with questionable relationships were never genotyped for the SNPs. This results in potentially fewer erroneous relationships than those detected by SNPs alone.
For the Early Access Affymetrix Mapping 10K array, there were 10,043 SNPs available for analysis. Because some of the SNPs on the early-access array were replaced in the Affymetrix final production array for various reasons (see reasons stated in the table 2 footnotes), we took a conservative approach by analyzing only those SNPs that were included in the final production chip (1,096 SNPs were excluded). For each SNP, the call rate (percentage of successful genotype calls among subjects) was used as a measure of quality, and we excluded SNPs with call rates <90% (849 excluded). Mendelian inheritance was evaluated, although, of the 167 pedigrees in our study, only 6 were capable of showing Mendelian-inheritance errors for diallelic SNPs, if errors existed. Three SNPs were excluded as a result of Mendelian inconsistencies in multiple pedigrees. To test for Hardy-Weinberg equilibrium (HWE), one subject from each pedigree was randomly sampled, an exact test for HWE was performed, and this process was repeated 100 times, to compute an average P value per SNP. This simulation process avoids using related subjects in the test for HWE but uses an average over a large number of random samples to be sure that conclusions are not based on an unusual random sample. Seven SNPs were excluded as a result of departure from HWE. After the above-mentioned data cleaning, additional genotypes that were likely erroneous, as determined by the default error-detection option of Merlin (Abecasis et al. 2002), were removed. A summary of the reasons for excluding SNPs from analyses are listed in sequential order in table 2.
Table 2.
Reduction from 10,043 SNPs in the Affymetrix Early-Access Chip to 5,656 SNPs Used in Analyses
| Reason Excluded (in Sequential Order) | No. Excluded |
| Excluded from production chipa | 1,096 |
| Missing genetic-map positionb | 484 |
| Call rate per SNP <90% | 849 |
| Conflicting map positionsc | 4 |
| Low informationd | 778 |
| Questionable chromosome ordere | 8 |
| Failed HWE, P<.001 | 7 |
| Mendelian errors in multiple pedigrees | 3 |
| High LD with neighboring SNPsf | 1,158 |
Reasons for exclusion include unacceptable call rates, poor cluster scores, reproduction problems, Mendelian errors, visually unacceptable SNPs, SNPs in the same physical position, Hardy-Weinberg disequilibrium, chromosome-X heterozygotes, cross-hybridization, and discordance between SNP calls and single base extension.
Map position was determined by linear interpolation of physical sequence onto deCODE genetic map and was supplied by Affymetrix, but genetic-map position or chromosome number was missing at the time of analysis.
Genetic-map position from Affymetrix for early-access (EA) chip and production (P) chip differed: SNP 52738 (EA on chromosome 3, P on chromosome 14); SNP 55401 (EA on chromosome 5, P on chromosome 12); SNP 1511280 (EA on chromosome 3, P on chromosome 16); and SNP 1525228 (EA on chromosome 7, P on chromosome 11). The chromosome location of the production chip of two of these four SNPs were questionable (SNPs 52738 and 1511280 were far outside the map lengths of chromosomes reported by deCODE), so all four SNPs were excluded.
119 SNPs were not polymorphic; 130 SNPs had only 1–4 total copies of the rare allele (allele frequency <0.5%); 529 SNPs had a minor allele frequency 0.5%–4.9%.
Chromosome order of SNPs evaluated by CRIMAP multipoint with the “flips2” option gave a LOD score of 1.0 that favored the flipped order, compared with the order provided by Affymetrix.
For each cluster of SNPs with allele frequencies at least 5%, pairwise |D′| >0.7 for any two SNPs in a cluster was used to indicate excessive LD; only the most informative (highest heterozygosity) SNP from each cluster was used in linkage analyses, and all other SNPs with high LD were excluded.
For the SNP analysis, a total of 510 Affymetrix Mapping 10K arrays were processed (467 affected males, 9 unaffected males, 9 females, 11 internal controls, and 14 repeats), resulting in >5 million genotypes. Throughout the project, quality was assessed by monitoring a number of parameters. These included monitoring the overall call rate per array (i.e., over all SNPs for a subject), the overall call rate per SNP (i.e., over all subjects for a SNP), the heterozygosity frequency per array, and, for SNPs on the X chromosome, checking for heterozygotes. The mean call rate per array was 0.95, with a range of 0.81–0.97. All arrays that had a call rate <0.90 were retested (n=14), and all retests resulted in call rates >0.89. The mean call rate per SNP across all subjects was 0.95 (range 0.006–1.0), with 2,829 SNPs having a call rate of 1.00. For the 5,656 SNPs used in the final analyses (see table 2), the mean call rate per SNP was 0.99 (range 0.90–1.0), with 1,912 SNPs having a call rate of 1.00. For all men in this study, it was possible to examine markers on the X chromosome for errors due to miscalls or PCR contamination (i.e., heterozygotes should not be observed). No SNPs were heterozygous for males. Finally, to estimate the genotyping error rate, five samples were tested multiple times. For the pairwise comparisons of replicate samples, 157 genotype discrepancies were found, giving an estimated error rate of 0.08%. Overall, the quality of results was excellent, with a very low error rate.
Impact of LD on Linkage Results
Current software available to compute multipoint linkage analyses for a large number of genetic markers assumes that the markers are in linkage equilibrium. Although this may be reasonable for the widely spaced M-STR markers, LD is likely to exist among some of the closely spaced SNPs. To evaluate the impact of LD on our linkage results, we performed analyses both with and without the SNPs in high LD, restricted to SNPs with minor-allele frequencies of at least 5%. To identify high-LD SNPs, we computed the pairwise LD measure |D′| between each SNP and all other SNPs within 5-cM distance from it. Because linkage phase of the SNPs is not directly observed, we simplified our approach by ignoring relationships among family members and by using the expectation-maximization algorithm (Excoffier and Slatkin 1995) to estimate 2-locus haplotype frequencies, from which |D′| could be calculated. Although ignoring relationships may result in a loss of efficiency, it should not introduce bias into our estimates. Figure 1 (top panel) illustrates the distribution of |D′| according to the pairwise distance of SNPs within 5 cM. We used a threshold of |D′|>0.7 to define the high-LD SNPs, and their distribution (fig. 1, lower left panel) illustrates that the majority of the high-LD SNPs had |D′|=1.0, and the distribution of pairwise distances between the high-LD SNPs (fig. 1, lower right panel) illustrates that the majority of high-LD SNPs were within 0.5 cM of each other (median distance 0.16 cM; 25th–75th percentiles 0.01–0.55 cM). Among the clusters of SNPs that had |D′|>0.7, only one SNP (the most informative) from each cluster was used in the linkage analyses. A total of 1,158 SNPs were excluded, because they were in strong LD with remaining SNPs.
Figure 1.
Distribution of pairwise |D′| values according to the pairwise distance of SNPs within 5 cM (top panel). The solid horizontal line in the top panel is the threshold for high LD at |D′|>0.7. The distribution of the high-LD SNPs is given in the lower left panel, and the distribution of the pairwise distances between the high-LD SNPs is given in the lower right panel.
The LOD scores for the analyses with and without the high-LD SNPs are illustrated in figure 2. The panels of this figure illustrate that the high-LD SNPs can lead to inflated LOD scores. In some cases, the inflated LOD scores were extreme, such as for chromosomes 4, 6, and 14. The tick marks along the bottom panels of figure 2 illustrate where the SNPs of high LD were excluded. The information contents for the analyses in figure 2 were nearly identical, with a mean of 0.65 when the high-LD SNPs were included versus a mean of 0.63 when the high-LD SNPs were excluded. Plots of information content per chromosome (not shown) illustrated that exclusion of the high-LD SNPs had little impact on the information content. We interpret these findings to mean that the presence of LD among SNPs artificially increases the LOD scores. All results in figure 2 excluded SNPs with minor-allele frequencies <5%. Because we were curious about the impact of rare alleles, we reanalyzed our genome scan, including SNPs with rare alleles, for the analyses that included high-LD SNPs. The resulting LOD scores were indistinguishable from those presented in figure 2, for which the high-LD SNPs were included (data not shown). Hence, it appears that the high-LD SNPs had the greatest impact on the LOD scores, rather than the allele frequencies. All these analyses are based on removal of likely genotype errors, determined by the error-detection option of Merlin. A total of 1,391 genotypes were declared likely to be erroneous by Merlin, resulting in a genotype error rate of 0.05%, which is close to the rate of 0.08% estimated in our replicate samples. To evaluate the impact of this genotype elimination, we ran the analyses, after removing the high-LD SNPs, both before and after removing the likely genotype errors. The difference in maximum LOD scores (after vs. before) ranged from 0.42 to −0.08 across the chromosomes. Because genotyping errors were difficult to detect with SNPs and because their presence can falsely deflate LOD scores (Douglas et al. 2000; Sobel et al. 2002), all subsequent results are based on removal of likely errors by Merlin’s algorithm.
Figure 2.
LOD scores for SNPs, shown with versus without the high-LD SNPs. In each panel, the solid line excludes the high-LD SNPs, and the broken line includes them. At the bottom of each panel are tick marks that indicate the positions of the high-LD SNPs.
Because some of the high-LD SNPs had a large impact on the linkage findings, we excluded the high-LD SNPs from further analyses. For the remaining 5,656 SNPs used in the linkage analyses, the average inter-SNP distance was 0.63 cM (25th–75th percentile 0.06–0.77 cM). The chromosomes that had LOD scores of at least 2 were chromosomes 2, 8, 12, 20, and X. The solid lines in figure 2 show where these maximum values occurred, and table 3 gives numeric LOD scores and flanking M-STR markers. The results for chromosome 20 are consistent with the linkage results we reported elsewhere for M-STR markers (Berry et al. 2000), which are further evaluated below. The results for chromosome 8 are consistent with the linkage findings of Xu et al. (2001), and the results for chromosome X are consistent with our prior analyses with M-STRs (Xu et al. 1998). The linkage findings for chromosomes 2, 6, and 12 are a bit odd, given that the maximum LOD scores occurred at the extreme ends of the chromosomes. Further genotyping at the ends of these chromosomes may help validate these findings.
Table 3.
Location of Maximum LOD Scores >2.0 and Flanking M-STR Markers
| M-STR |
||||
| Chromosome | Maximum LODScore | SNP Position(cM) | Upstream | Downstream |
| 2 | 2.1 | 242.48 | D2S2205 | D2S2973 |
| 6 | 4.2 | 183.26 | D6S297 | D6S1697 |
| 8 | 2.2 | 54.91 | D8S1750 | D8S571 |
| 12 | 3.9 | 169.62 | D12S1723 | D12S357 |
| 20 | 2.4 | 75.81 | D20S436 | D20S897 |
| X | 2.2 | 161.05 | DXS8073 | DXS8045 |
SNP versus M-STR LOD Scores
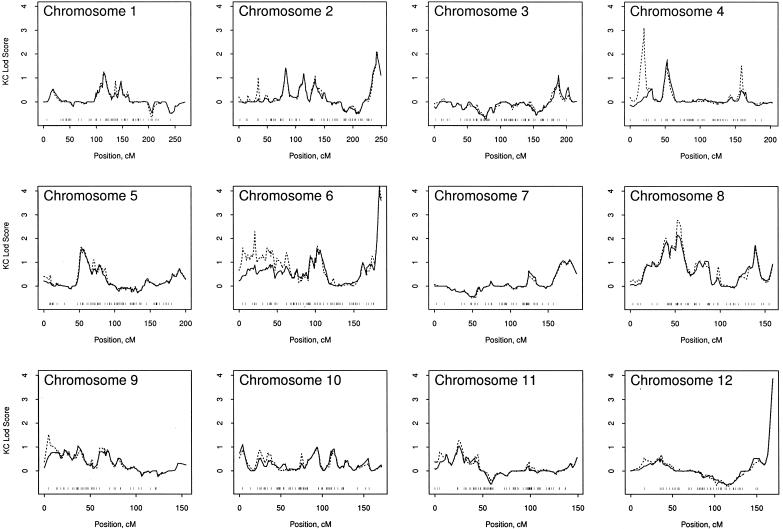
After excluding the high-LD SNPs, we compared the linkage findings from three types of genetic marker data: (1) M-STRs alone, (2) SNPs alone, and (3) the combination of M-STRs and SNPs. For these comparisons, we used only those subjects who had genotypes available for both genotyping technologies. The LOD scores from these three types of genetic marker data are presented in figure 3, along with their information content. For many regions of the chromosomes, the LOD-score curves for SNPs, M-STRs, and the combination of both were fairly similar, although it was not unusual to find a number of slightly higher peaks for the SNPs, compared with the M-STRs (e.g., see fig. 3, chromosome 1). For a number of chromosomes, the LOD-score peak was dramatically higher for the SNP markers (either alone or combined with M-STRs) than for the M-STRs alone (e.g., see fig. 3, chromosomes 2–8, 12, 14, and 16). The unusual aspect of some of these differences is the narrow width of the LOD-score peaks for the SNPs (see fig. 3, chromosome 4). We speculate that these narrow peaks could be caused by several factors, including the fact that the high density of the SNPs could provide more narrow peaks than the M-STR markers and that unaccounted LD could bias the LOD scores for SNPs to be spuriously high. The only place where the SNPs had a much lower LOD score than the M-STR markers was on the X chromosome, at <50 cM, where the SNPs were less informative than the M-STRs.
Figure 3.
LOD scores for SNP markers (dotted line), M-STR markers (dashed line), and the combination of both SNPs and M-STRs (solid line). The bottom of each panel shows the information content of the three types of analyses.
On the basis of only M-STR markers, we have reported positive linkage to chromosome 20 (Cunningham et al. 2003). It is interesting to note that the SNP markers tend to refine the region of the maximum LOD score (fig. 3). For the M-STR markers, the maximum LOD score was 2.96 at 66.1 cM, and the width of the 1-LOD region of support was 29.3 cM. For the SNPs, there were two peaks separated by 53 cM: the maximum LOD score at 22.5 cM was 2.16 (width of 1-LOD support 15.8 cM), and the maximum LOD score at 75.7 cM was 2.85 (width of 1-LOD support 19.8 cM). These results suggest that there may be two susceptibility loci on chromosome 20. Hence, the SNPs provided greater linkage resolution for chromosome 20 than M-STRs alone. It is also interesting that the M-STR markers do not contribute additional information to that already provided by the SNPs.
The contrast of information content in figure 3 illustrates that the SNPs provided much more linkage information than the M-STRs, despite the lower information content per SNP, compared with that per M-STR. Over all chromosome positions, the average information for the M-STRs was 41% (SD 7.5%); for the SNPs, 61% (SD 6.1%); and for the combination of both M-STRs and SNPs, 64% (SD 4.7%). To provide a more complete global view of the information content of the three types of genetic marker data, we computed the percentage of the genome that had information content above a range of thresholds. The genome percentage was computed by determining the length of the genome that exceeded an information threshold, divided by the total length of the genome. The contrast of these genome information percentages is illustrated in figure 4. To understand this figure, it is instructive to examine the X-axis at a threshold of 50% information content. Approximately 10% of the genome had at least 50% information content for the M-STRs, ∼80% of the genome had at least 50% information content for the SNPs, and ∼90% of the genome had at least 50% information content for the combination of both M-STRs and SNPs. This figure illustrates the dramatic increase in the information content provided by the high-density SNPs, in contrast to the little gain provided by combining the M-STRs with the SNPs.
Figure 4.
Percentage of genome exceeding different levels of genetic information for SNP markers (dotted line), M-STR markers (dashed line), and the combination of SNPs and M-STRs (solid line).
Subgroup Analyses
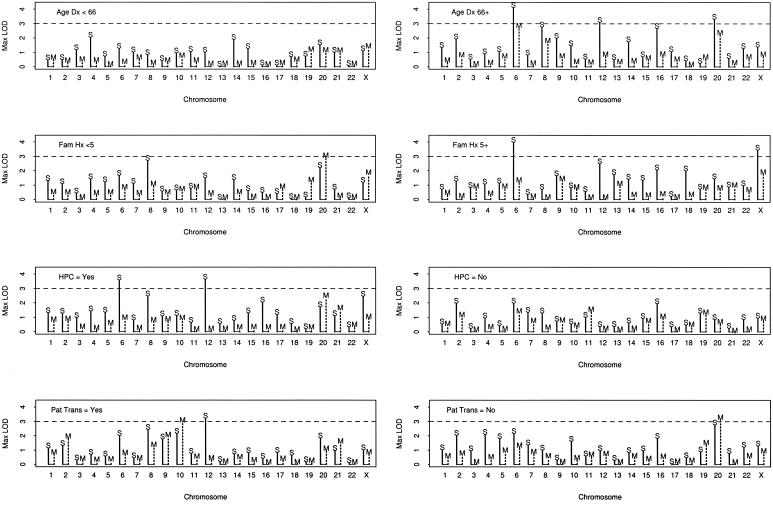
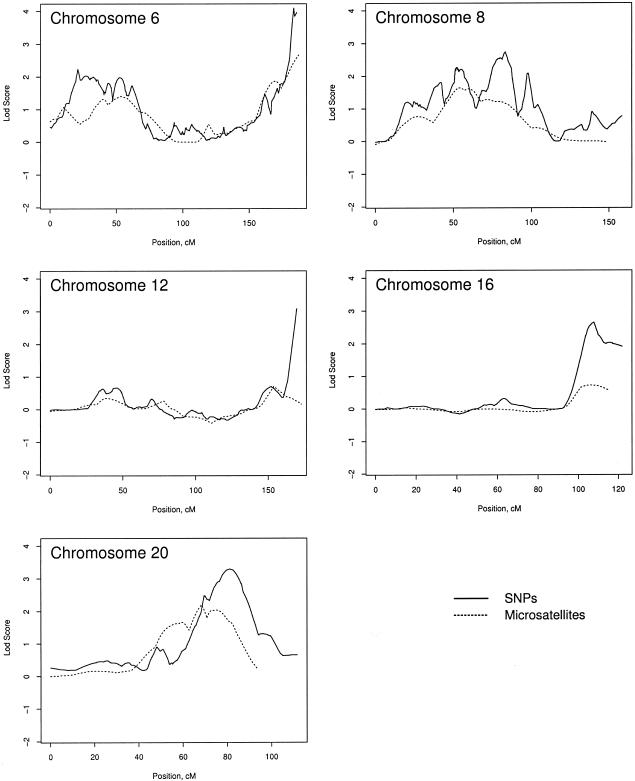
Pedigrees were stratified on the basis of pedigree average age at diagnosis (<66 years vs. ⩾66 years), number of men affected (<5 vs. ⩾5), HPC (yes vs. no), and paternal transmission of disease in a pedigree, in which both a father and son have prostate cancer (yes vs. no), as a surrogate for X linkage. Figure 5 presents the maximum LOD score for each subset, by chromosome and by analysis using SNPs versus M-STRs. This figure shows that, on average, the LOD scores for the SNPs are higher than those for the M-STRs, and LOD scores >3 occurred on chromosomes 6, 12, 20, and X for the SNPs but not for the M-STRs. However, this figure does not show where on the chromosomes the differences occurred. To illustrate some of the differences, we present the LOD-score curves for the subset with an age at diagnosis of ⩾66 years, for chromosomes 6, 8, 12, 16, and 20 (fig. 6). This figure illustrates that, for chromosomes 6, 12, and 16, the differences in LOD scores occurred only at the extreme ends of the chromosomes, whereas, for chromosomes 8 and 20, the larger LOD scores for the SNPs appeared more consistent throughout longer stretches of the chromosomes. These patterns were similar in other subsets (data not shown). On the other hand, for some subsets, there were some chromosomes for which the LOD scores were greater for the M-STRs than for the SNPs—in particular, chromosomes 10 and 20.
Figure 5.
Maximum LOD scores by subset, chromosome, and SNPs versus M-STRs. Age Dx = the mean age at diagnosis per pedigree; Fam Hx = the number of men with prostate cancer in a pedigree; HPC = hereditary prostate cancer by Carter criteria; Pat Trans = paternal transmission. The solid vertical line with the “S” indicates the SNP maximum LOD score, and the broken vertical line with the “M” indicates the M-STR maximum LOD score.
Figure 6.
Example LOD-score plots for the subset of pedigrees with an average age at diagnosis of >66 years, to compare the LOD scores for SNPs versus M-STRs.
Discussion
Until recently, M-STRs have been the primary type of markers used for linkage analyses. They are abundant and equally dispersed throughout the genome, and, because they are highly polymorphic, they are highly informative. Because of these marker characteristics, their use over the years has been extremely valuable. The increased availability of SNPs now provides an opportunity for alternative methodologic approaches. SNPs are more abundant than microsatellites and are also dispersed equally throughout the genome, but they are less informative than microsatellites, because they are only diallelic. Thus, a considerably larger number of SNP markers are required to achieve an information content similar to that of microsatellites. The advent of several high through-put genotyping platforms, including that by Affymetrix, make it now feasible to use SNPs for linkage analysis. Although there is a great deal of experience in the use of microsatellite markers—in terms of both laboratory and statistical methods for linkage analysis—there is very little experience with large-scale SNP mapping projects. In this study, we compare these two analytical approaches to linkage.
Overall, the quality of the SNP genotypes was excellent. We achieved a mean call rate per SNP of 95%, with an estimated error rate of 0.05%–0.08%. Although technology has advanced considerably over the years, we were able to complete the analytical phase of the project in a matter of months for the SNP-based genotyping, compared with years for the M-STR–based genotyping. Although the Affymetrix array contains 10,043 SNPs, only 5,656 were utilized in the final analysis (table 2). The number of usable markers will certainly increase over time, because we used an early-access array and because our conservative approach preferred exclusion of SNPs. In addition to the laboratory analytical issues that need to be addressed, software challenges remain, including the need for programs that can handle large data sets for parametric linkage analyses, as well as the presence of LD.
Our results suggest that the presence of LD among SNPs can lead to inflated LOD scores, and this seems to be an artifact due to the assumption of linkage equilibrium that is required by the current genetic-linkage software. We excluded SNPs with high LD, defined as |D′|>0.7. Although this criterion likely captures the most extreme impact of LD on the linkage results, it is possible for any remaining LD, either pairwise values with |D′|⩽0.7 or higher-order disequilibria, to still influence our linkage results. With this caveat, we found, using SNPs, a number of new LOD-score peaks with values of at least 2.0 that were not found using the M-STR data. Chromosome 8, with a maximum LOD score of 2.2, supports recent linkage evidence for chromosome 8 (Xu et al. 2001), along with the candidate gene MSR, which maps to this region (Xu et al. 2002). Chromosome 2, with a maximum LOD score of 2.1, is consistent with three other reports of suggestive linkage (LOD scores between 1 and 3) to this chromosome (Suarez et al. 2000; Edwards et al. 2003; Xu et al. 2003). Chromosome 6, with a maximum LOD score of 4.2, is consistent with the suggestive linkage (LOD scores between 1 and 3) reported by three different groups (Edwards et al. 2003; Janer et al. 2003; Xu et al. 2003). Chromosome 12, with a maximum LOD score of 3.9, is consistent with two reports of LOD scores between 1 and 2 for this chromosome (Suarez et al. 2000; Hsieh et al. 2001). For chromosomes 2, 6, and 12, the meaning of the maximum LOD scores occurring at the extreme ends of these chromosomes is not entirely clear. Although it is possible for susceptibility genes to exist at the extreme ends, there may be unknown errors in the genetic map. For chromosome 6, there is a series of LOD-score peaks >1.0 at ∼80 cM. It may be that genetic map errors have “pushed” a high LOD score to the extreme end of chromosome 6, because the multipoint analyses are not robust to map errors (Risch and Giuffra 1992). Chromosomes 2 and 12 may be subject to this same problem, although the strength of evidence is weaker for these chromosomes.
Our finding of new linkage peaks by use of SNPs is consistent with two recent studies that compared SNP versus M-STR linkage results, for bipolar disorder (Middleton et al. 2004) and for rheumatoid arthritis (John et al. 2004). The study by Middleton et al. (2004) was based on 148 genotyped subjects from 25 families, with an average of 5.9 genotyped subjects per family. In contrast, we genotyped 467 affected men from 167 families—an average of 2.8 genotyped subjects per family. Hence, our families each had a lower genetic-linkage information content, so our evaluation of the differences between SNP and M-STR linkage results is based on different types of families than those used by Middleton et al. (2004). This was reflected in our average information content of 61% for the SNPs, versus an average of 84% reported by Middleton et al. (2004). A strength of our study was the larger number of families, allowing more critical evaluation of the impact of LD on the resulting LOD scores. The study by John et al. (2004) was based on 157 families, of which 37% had DNA available from one or both parents, with an average information content of 75%. Furthermore, John et al. (2004) reported that some SNPs in high LD slightly increased NPL scores, whereas other high-LD SNPs led to a modest reduction in NPL scores, and they concluded that the cause of change in NPL scores because of exclusion of high-LD SNPs could be a result of either the software not accounting for LD or a loss of genetic information. Their definition of high LD differed from ours; we used |D′|>0.7 to define high LD, in contrast to r2⩾0.4, used by John et al. (2004). With our definition of high LD, our results suggest that it is likely that high LD influences LOD scores, because only minor differences in the information content occurred when we excluded the high-LD SNPs. Another difference between our study and that of John et al. (2004) is that we used the Kong and Cox (1997) LOD score, in contrast to the NPL score used by John et al. It is not clear whether this difference in choice of statistic can lead to different conclusions regarding the impact of LD on linkage statistics.
In conclusion, for our pedigrees, the SNP markers provided a higher information content than the M-STR markers, with averages of 61% versus 41%, respectively. These averages are remarkably consistent with recent simulations that confirm that a dense map of SNPs provides substantially greater information content than the traditional map of M-STRs spaced at 1 marker/∼10 cM when parents are not genotyped (Evans and Cardon 2004). These simulation studies also showed that more-sparse marker maps are just as informative if parental genotypes are available. Furthermore, the SNPs identified more linkage peaks with more-narrow widths than did M-STR markers; some linkage signals would have gone undetected by our 10-cM genome scan with M-STRs. Further follow-up of the linkage signals in our families, with selection of additional SNPs and evaluation of candidate genes, is warranted. Although the presence of LD among the SNPs complicated the linkage analyses, we were able to at least partially address this by removing those SNPs that were in high LD with other SNPs. Another limitation of using SNPs in our families was the inability to detect Mendelian errors, although we circumvented this by using the Merlin software that statistically identifies and eliminates likely genotyping errors. Perhaps one of the most difficult issues, at least in the study of our small families, was the inability to accurately validate genetic maps. As more information becomes available from other linkage studies using SNPs, this concern may diminish. One can envision pooling a large number of studies in order to validate SNP genetic maps.
Acknowledgments
This study was supported by the Mayo Clinic Genomic Center and Comprehensive Cancer Center and by the U.S. Public Health Service, National Institutes of Health (grants CA72818, CA15083, CA89600, CA91956, and CA94919).
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for prostate cancer, HPC1, PCAP, CAPB, HPC2, HPC20, and HPCX)
References
- Abecasis G, Cherny S, Cookson W, Cardon L (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flowtrees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Berry R, Schroeder JJ, French AJ, McDonnell SK, Peterson BJ, Cunningham JM, Thibodeau SN, Schaid DJ (2000) Evidence for a prostate cancer–susceptibility locus on chromosome 20. Am J Hum Genet 67:82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon P, Valeri A, Cohen-Akenine A, Drelon E, Paiss T, Wöhr G, Latil A, et al (1998) Predisposing gene for early-onset prostate cancer, localized on chromosome 1q42.2-43. Am J Hum Genet 62:1416–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ (1997) Accurate inference of relationships in sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, Faruque M, et al (2002) Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet 30:181–184 10.1038/ng823 [DOI] [PubMed] [Google Scholar]
- Carter B, Bova G, Beaty T, Steinberg G, Childs B, Isaacs W, Walsh P (1993) Hereditary prostate cancer: epidemiologic and clinical features. J Urol 150:797–802 [DOI] [PubMed] [Google Scholar]
- Cunningham JM, McDonnell SK, Marks A, Hebbring S, Anderson SA, Peterson BJ, Slager S, French A, Blute ML, Schaid DJ, Thibodeau SN (2003) Genome linkage screen for prostate cancer susceptibility loci: results from the Mayo Clinic Familial Prostate Cancer Study. Prostate 57:335–346 10.1002/pros.10308 [DOI] [PubMed] [Google Scholar]
- Douglas JA, Boehnke M, Lange K (2000) A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data. Am J Hum Genet 66:1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Schaid DJ, Whittemore AS, Isaacs WJ (2003) Where are the prostate cancer genes? a summary of eight genome wide searches. Prostate 57:261–269 10.1002/pros.10300 [DOI] [PubMed] [Google Scholar]
- Edwards S, Meitz J, Eles R, Evans C, Easton D, Hopper J, Giles G, Foulkes WD, Narod S, Simard J, Badzioch M, Mahle L (2003) Results of a genome-wide linkage analysis in prostate cancer families ascertained through the ACTANE consortium. Prostate 57:270–279 10.1002/pros.10301 [DOI] [PubMed] [Google Scholar]
- Evans DM, Cardon LR (2004) Guidelines for genotyping in genomewide linkage studies: single-nucleotide-polymorphism maps versus microsatellite maps. Am J Hum Genet 75:687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927 [DOI] [PubMed] [Google Scholar]
- Gibbs M, Stanford JL, McIndoe RA, Jarvik GP, Kolb S, Goode EL, Chakrabarti L, Schuster EF, Buckley VA, Miller EL, Brandzel S, Li S, Hood L, Ostrander EA (1999) Evidence for a rare prostate cancer–susceptibility locus at chromosome 1p36. Am J Hum Genet 64:776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J (1994) The 1993–94 Genethon human genetic linkage map. Nat Genet 7:246–338 10.1038/ng0694supp-246 [DOI] [PubMed] [Google Scholar]
- Hsieh C-L, Oakley-Girvan I, Balise RR, Halpern J, Gallagher RP, Wu AH, Kolonel LN, O’Brien LE, Lin IG, Van Den Berg DJ, Teh CZ, West DW, Whittemore AS (2001) A genome screen of families with multiple cases of prostate cancer: evidence of genetic heterogeneity. Am J Hum Genet 69:148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janer M, Friedrichsen DM, Stanford JL, Badzioch MD, Kolb S, Deutsch K, Peters MA, Goode EL, Welti R, DeFrance HB, Iwasaki L, Li S, Hood L, Ostrander EA, Jarvik GP (2003) Genomic scan of 254 hereditary prostate cancer families. Prostate 57:309–319 10.1002/pros.10305 [DOI] [PubMed] [Google Scholar]
- John S, Shephard N, Liu G, Zeggini E, Cao M, Chen W, Vasavda N, Mills T, Barton A, Hinks A, Eyre S, Jones KW, Ollier W, Silman A, Gibson N, Worthington J, Kennedy GC (2004) Whole-genome scan, in a complex disease, using 11,245 single-nucleotide polymorphisms: comparison with microsatellites. Am J Hum Genet 75:54–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson D, Sainz J, Jonsdottir G, Gudjonsson S, Richardsson B, Sigurdardottir S, Barnard B, Hallbeck B, Masson M, Shlien A, Palsson S, Frigge M, Thorgeirsson T, Gulcher J, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Pato MT, Gentile KL, Morley CP, Zhao X, Eisener AF, Brown A, Petryshen TL, Kirby AN, Medeiros H, Carvalho C, Macedo A, Dourado A, Coelho I, Valente J, Soares MJ, Ferreira CP, Lei M, Azevedo MH, Kennedy JL, Daly MJ, Sklar P, Pato CN (2004) Genomewide linkage analysis of bipolar disorder by use of a high-density single-nucleotide–polymorphism (SNP) genotyping assay: a comparison with microsatellite marker assays and finding of significant linkage to chromosome 6q22. Am J Hum Genet 74:886–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Giuffra L (1992) Model misspecification and multipoint linkage analysis. Hum Hered 42:77–92 [DOI] [PubMed] [Google Scholar]
- Schaid D (2004) The complex genetic epidemiology of prostate cancer. Hum Mol Genet 13:R103–R121 10.1093/hmg/ddh072 [DOI] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Blute ML, Thibodeau SN (1998) Evidence for autosomal dominant inheritance of prostate cancer. Am J Hum Genet 62:1425–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Freije D, Carpten JD, Grönberg H, Xu J, Isaacs SD, Brownstein MJ, Bova GS, Guo H, Bujinovszky P, Nusskern DR, Damber JE, Bergh A, Emanuelsson M, Kallioniemi OP, Walker-Daniels J, Bailey-Wilson JE, Beaty TH, Meyers DA, Walsh PC, Collins FS, Trent JM, Isaacs WB (1996) Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science 274:1371–1374 10.1126/science.274.5291.1371 [DOI] [PubMed] [Google Scholar]
- Sobel E, Papp JC, Lange K (2002) Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet 70:496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez BK, Lin J, Burmester JK, Broman KW, Weber JL, Banerjee TK, Goddard KAB, Witte JS, Elston RC, Catalona WJ (2000) A genome screen of multiplex sibships with prostate cancer. Am J Hum Genet 66:933–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian S, Simard J, Teng D, Abtin V, Baumgard M, Beck A, Camp N, et al (2001) A strong candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet 27:172–180 10.1038/84808 [DOI] [PubMed] [Google Scholar]
- Xu J, Gillanders EM, Isaacs SD, Chang BL, Wiley KE, Zheng SL, Jones M, Gildea D, Riedesel E, Albertus J, Freas-Lutz D, Markey C, Meyers DA, Walsh PC, Trent JM, Isaacs WB (2003) Genome-wide scan for prostate cancer susceptibility genes in the Johns Hopkins hereditary prostate cancer families. Prostate 57:320–325 10.1002/pros.10306 [DOI] [PubMed] [Google Scholar]
- Xu J, International Consortium for Prostate Cancer Genetics (2000) Combined analysis of hereditary prostate cancer linkage to 1q24-25: results from 772 hereditary prostate cancer families from the International Consortium for Prostate Cancer Genetics. Am J Hum Genet 66:945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Meyers D, Freije D, Isaacs S, Wiley K, Nusskern D, Ewing C, et al (1998) Evidence for a prostate cancer susceptibility locus on the X chromosome. Nat Genet 20:175–179 10.1038/2477 [DOI] [PubMed] [Google Scholar]
- Xu J, Zheng SL, Hawkins GA, Faith DA, Kelly B, Isaacs SD, Wiley KE, Chang B, Ewing CM, Bujinovszky P, Carpten JD, Bleecker ER, Walsh PC, Trent JM, Meyers DA, Isaacs WB (2001) Linkage and association studies of prostate cancer susceptibility: evidence for linkage at 8p22-23. Am J Hum Genet 69:341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zheng SL, Komiya A, Mychaleckyj JC, Isaacs SD, Hu JJ, Sterling D, et al (2002) Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat Genet 32:321–325 10.1038/ng994 [DOI] [PubMed] [Google Scholar]