Abstract
To resolve the phylogeny of the autochthonous mitochondrial DNA (mtDNA) haplogroups of India and determine the relationship between the Indian and western Eurasian mtDNA pools more precisely, a diverse subset of 75 macrohaplogroup N lineages was chosen for complete sequencing from a collection of >800 control-region sequences sampled across India. We identified five new autochthonous haplogroups (R7, R8, R30, R31, and N5) and fully characterized the autochthonous haplogroups (R5, R6, N1d, U2a, U2b, and U2c) that were previously described only by first hypervariable segment (HVS-I) sequencing and coding-region restriction-fragment–length polymorphism analysis. Our findings demonstrate that the Indian mtDNA pool, even when restricted to macrohaplogroup N, harbors at least as many deepest-branching lineages as the western Eurasian mtDNA pool. Moreover, the distribution of the earliest branches within haplogroups M, N, and R across Eurasia and Oceania provides additional evidence for a three-founder-mtDNA scenario and a single migration route out of Africa.
Introduction
The “population genomics” era has emerged in the research of human mtDNA (Hedges 2000; Richards and Macaulay 2001) by utilization of complete or nearly complete mtDNA sequences to infer the prehistoric dispersal of modern humans and the phylogeny of the major mtDNA lineages in Europe, Africa, America, Oceania (Australia and Papua New Guinea), and East Asia (Ingman et al. 2000; Finnilä et al. 2001; Maca-Meyer et al. 2001, 2003; Torroni et al. 2001; Derbeneva et al. 2002b; Herrnstadt et al. 2002, 2003; Ingman and Gyllensten 2003; Kong et al. 2003; Mishmar et al. 2003; Reidla et al. 2003). However, complete phylogenetic information was hitherto not available for South Asia and for India in particular, an important area that served as a major corridor of modern human dispersal out of Africa (Cann 2001) and that hosts a diverse conglomerate of people with different morphological, genetic, cultural, and linguistic characteristics. Quite a number of mtDNA studies that focus on the first hypervariable segment (HVS-I) of the control region have been applied to various Indian populations and have provided some insights into the genetic structure of the populations in this area (Kaur et al. 2002; Basu et al. 2003; Kivisild et al. 2003a; Roy et al. 2003 and references therein). In addition to India-specific M subhaplogroups (i.e., M2, M3, M4, M5, and M6), some autochthonous haplogroups, including U2a, U2b, U2c, and many unclassified lineages within the nested macrohaplogroups R and N, have been observed in Indian populations (Passarino et al. 1996; Kivisild et al. 1999a, 1999b, 2003a, 2003b; Bamshad et al. 2001; Basu et al. 2003; Quintana-Murci et al. 2004). However, since these studies were based mainly on HVS-I plus a few coding-region RFLPs, none of those haplogroups had yet been fully characterized. Moreover, some western Eurasian haplogroups also occur in India at low frequencies (Passarino et al. 1996; Kivisild et al. 2003b; Quintana-Murci et al. 2004)—or even at high frequencies, in particular regions (Forster et al. 2002). Although rather detailed phylogenies of western Eurasian mtDNA lineages have been obtained by Finnilä et al. (2001), Maca-Meyer et al. (2001), and Herrnstadt et al. (2002) (which were, however, in minor conflict with one another), no Indian counterpart was hitherto available for comparison. Although most of the mtDNA lineages of western Eurasian ancestry must have a rather recent entry date (<10 thousand years ago [kya] [Kivisild et al. 1999a]), it is not clear whether one would also have to consider some early offshoots of the western Eurasian mtDNA phylogeny that are specific to India and neighboring regions. To resolve these problems and prepare the grounds for future extensive phylogeographic screening, it is necessary to contrast the mtDNA phylogeny of Indians with that of western Eurasians, on the basis of complete sequencing executed on a diverse collection of mtDNAs.
Complete sequence information from India is also urgently needed in the forensic field. In the past, forensic studies have predominantly described the general pattern of mtDNA control sequence variation in western Eurasian and East Asian individuals (Allard et al. 2002, 2004). Recently, however, coding-region SNPs in European populations have increasingly been utilized in forensics, especially for discriminating frequent control-region haplotypes that are poorly characterized by HVS-I and -II alone (Brandstätter et al. 2003; Coble et al. 2004; Quintáns et al. 2004). Similar studies of the mtDNA variation in other (sub)continents are still pending.
Complete mtDNA information for South Asia is also highly relevant to medical studies of mitochondrial diseases. To perform systematic studies of the major mitochondrial diseases (Leber hereditary optic neuropathy [LHON]; mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes [MELAS]; etc.) in patients with South Asian matrilineal ancestries, at least a basal outline of the total mtDNA phylogeny in this subcontinent is indispensable—see, for example, articles by Rocha et al. (1999) and Kong et al. (2004) for studies involving African, European, and East Asian haplogroup backgrounds.
Since control-region sequences of the Asian-specific macrohaplogroup M can hardly be confused with western Eurasian sequences (except for subhaplogroup M1 [Quintana-Murci et al. 1999]), we focused on macrohaplogroup N in India, which appeared to be totally missing or at least severely underrepresented in all previous worldwide studies of complete mtDNA variation (Ingman et al. 2000; Maca-Meyer et al. 2001; Cavalli-Sforza and Feldman 2003; Mishmar et al. 2003).
Material and Methods
Sampling
In the present study, 75 mtDNA lineages were selected from >800 samples across India, with the goal that at least one representative was chosen from each western Eurasian haplogroup and each group of previously unclassified haplotypes (authors’ unpublished data). Samples were from the following populations: Reddy (R) and Thogataveera (T) from Andhrapradesh, South India; Bhargava (A), Chaturvedi (B), and other Brahmin (C) from Uttarpradesh, North India; and Rajbhansi from West Bengal (SW) and the Khasi population from Meghalaya (S), both located in Northeast India.
DNA Amplification and Sequencing
DNA was amplified using 15 pairs of primers (Kong et al. 2003) (table 1). After being purified on spin columns (Watson BioTechnologies), the 15 overlapping fragments were sequenced by means of PCR primers and 47 internal primers (Kong et al. 2003) (table 2) and BigDye Terminator chemistry (Applied Biosystems). Sequencing was performed on a 3700 DNA Analyzer (Applied Biosystems), and the resulting sequences were handled with the DNASTAR software (DNASTAR). Mutations were scored relative to the revised Cambridge Reference Sequence (rCRS [Andrews et al. 1999]). The 75 complete sequences have been submitted to GenBank (accession numbers AY713976–AY714050).
Table 1.
Primer Pairs for Amplification and Sequencing[Note]
|
Light mtDNA Strand |
Heavy mtDNA Strand |
|||
| Primer | Location (5′→3′)in rCRS | Primer | Location (5′→3′)in CRS | |
| L394 | 375–394 | H1782 | 1801–1782 | |
| L1466 | 1445–1466 | H3054 | 3074–3054 | |
| L2796 | 2777–2796 | H3674 | 3693–3674 | |
| L3644 | 3625–3644 | H5099 | 5118–5099 | |
| L4887 | 4866–4887 | H5832 | 5851–5832 | |
| L5781 | 5762–5781 | H6899 | 6918–6899 | |
| L6869 | 6850–6869 | H7990 | 8009–7990 | |
| L7882 | 7861–7882 | H9212 | 9231–9212 | |
| L9198 | 9181–9198 | H10660 | 10679–10660 | |
| L10519 | 10498–10519 | H11689 | 11708–11689 | |
| L11338 | 11319–11338 | H12603 | 12623–12603 | |
| L12334 | 12315–12334 | H13666 | 13685–13666 | |
| L13612 | 13593–13612 | H14591 | 14610–14591 | |
| L14575 | 14556–14575 | H16048 | 16067–16048 | |
| L15996 | 15975–15996 | H408 | 429–408 | |
Note.— The annealing temperature for all primer pairs was 53°C.
Table 2.
The Inner Primers for the Complete Sequencing
| Primera | Location (5′→3′) in rCRS |
| L713 | 696–712 |
| H902 | 922–902 |
| L1156 | 1138–1156 |
| H1172 | 1190–1172 |
| L2025 | 2004–2025 |
| H2053 | 2073–2053 |
| L2415 | 2395–2415 |
| H2426 | 2444–2426 |
| L3179 | 3160–3179 |
| H3274 | 3293–3274 |
| L4210 | 4189–4210 |
| H4227 | 4247–4227 |
| L4499 | 4480–4499 |
| H4792 | 4813–4792 |
| L5278 | 5259–5278 |
| H5442 | 5461–5442 |
| L6337 | 6318–6337 |
| H6367 | 6387–6367 |
| L7356 | 7337–7356 |
| H7406 | 7427–7406 |
| L8215 | 8196–8215 |
| H8345 | 8366–8345 |
| L8581 | 8563–8581 |
| H8861 | 8882–8861 |
| L9794 | 9774–9794 |
| H9848 | 9867–9848 |
| L10170 | 10147–10170 |
| H10356 | 10376–10356 |
| L11004 | 10985–11004 |
| H11081 | 11100–11081 |
| L11718 | 11692–11718 |
| H11944 | 11963–11944 |
| L12028 | 12008–12028 |
| H12341 | 12361–12341 |
| L12572 | 12553–12572 |
| H12878 | 12897–12878 |
| L13049 | 13031–13049 |
| H13124 | 13143–13124 |
| L14054 | 14035–14054 |
| H14186 | 14206–14186 |
| L14989 | 14970–14989 |
| H15086 | 15105–15086 |
| L15391 | 15372–15391 |
| H15400 | 15419–15400 |
| L16209 | 16190–16209 |
| H16498 | 16517–16498 |
| L29 | 8–29 |
“L” and “H” refer to light and heavy strands of mtDNA, respectively.
Quality Control
To avoid the kinds of errors and artifacts that have affected some earlier analyses of mtDNA coding-region variation, we utilized a strict quality-control procedure, similar to that of Kong et al. (2003). First, each base was sequenced from at least two independent amplifications. Second, all private mutations (i.e., mutations on a terminal branch of a single sequence) and all indels (insertions and deletions), as well as some seemingly unusual recurrent substitutions (such as transition 16266 in A65 and B53 and transition 15326 in A165 and S4), were confirmed by independent PCR and sequencing.
Database Comparison
To differentiate between the Indian and western Eurasian mtDNA lineages, it is necessary to combine all published sequences sampled from western Eurasia (Ingman et al. 2000; Finnilä et al. 2001; Maca-Meyer et al. 2001; Rose et al. 2001; Taylor et al. 2001; Derbeneva et al. 2002a; Herrnstadt et al. 2002, 2003; Ingman and Gyllensten 2003; Mishmar et al. 2003; Reidla et al. 2003; Coble et al. 2004) and compare them with the Indian sequences. To clearly display and distinguish these lineages from our Indian samples and avoid confusion with haplogroup designation, we refer to particular samples from the articles cited above by use of the initials (first name and surname) of the first author as a prefix—that is, MI, SF, NM, GR, RT, OD, CH, IG, DM, MR, and MC, followed by “#” and the original sample code, albeit in lowercase instead of capital letters.
Haplogroup Nomenclature
We adopt the nomenclature system of Richards et al. (1998). For new basal subhaplogroups of N and R, we would reserve the codes N5–N8, R5–R8, and R30–R39 for South Asian mtDNAs. Although we regard it as admissible to widen the definition of some previously named haplogroups in light of new complete sequence information, the correction of haplogroup names (such as “J2a” for the incorrect “J1a”) is done with the understanding that the obsolete names should not be recycled and are thus blocked henceforth to avoid misunderstanding.
Results
Autochthonous Indian Haplogroups
In our study, a number of novel haplogroups have emerged and then been named according to the rules of the notational system (see the “Material and Methods” section). The new haplogroup N5 is characterized by (at most) six transitions in the coding region (at sites 1719, 5063, 7076, 9545, 11626, and 13434) and two in the control region (at sites 16111 and 16311), as supported by lineage R148 and the authors' unpublished data. The two lineages T1 and C35, which share seven coding-region mutations (at sites 1442, 6248, 9051, 9110, 10289, 13105, and 13830) and four control-region mutations (at sites 16260, 16261, 16319, and 16362), constitute a new subhaplogroup of haplogroup R, designated as “R7.” Another new haplogroup, R8, is recognizable by five specific mutations (at sites 2755, 3384, 7759, 9449, and 13215). We tentatively group five R-derived branches into haplogroup R30 on the basis of the shared 8584 transition and combine three branches with a common 15884 transition into haplogroup R31. Since recurrent mutations have been observed at each of these sites, this classification needs to be corroborated by future screening for related lineages.
With the present mtDNA phylogeny, we can redefine or revise the definitions of several haplogroups that were previously characterized only by control-region sequence and/or coding-region RFLPs. Haplogroup N1d is broadened by requiring only two control-region mutations (16301 and 16356) plus one coding-region mutation, 953 (recognizable by RFLP site −951MboI [Quintana-Murci et al. 2004]). Haplogroup U2b is further characterized by mutations 146, 2706, 5186T, 12106, and 13149 in addition to 15049 (i.e., −15047HaeIII [Quintana-Murci et al. 2004]). Haplogroup U2c is redefined by requiring five mutations: 152, 5790A, 14935, 15061, and 16234, of which mutations 5790A and 15061 were previously recognized by RFLP typing (i.e., as +5789TagI and −15060MboI [Quintana-Murci et al. 2004]). The transition at 8023 (recognized by +8020MboI/+8022TaqI [Quintana-Murci et al. 2004]), shared by our sample R94 and samples from Quintana-Murci et al. (2004), may characterize a major subbranch of U2c. In addition to having mutations at 8594 (corresponding to −8592MboI) and 16304 (Quintana-Murci et al. 2004), haplogroup R5 is further characterized by another three mutations, at 10754, 14544, and 16524. It is not clear at this point whether the 16266 transition defines R5 as a whole or just the main subbranch of R5, since 16266 seems to be prone to back mutation in this haplogroup. A similar caveat holds for hypervariable site 16129 in haplogroup R6. Finally, it has now become evident that U2a is defined only by the rare transversion 16206C (Kivisild et al. 1999b), and no additional motif is found in the coding region.
Haplogroups Shared by Indians and Western Eurasians: Reappraisal of the Western Eurasian mtDNA Phylogeny
Many of the Indian mtDNA lineages belonging to typical western Eurasian haplogroups (such as I, H, V, J1, T2, K, U2e, U7, and U5a) can be integrated into the western Eurasian phylogeny unambiguously (fig. 1). Some haplogroups, however, have to be redefined in order to accommodate deep-rooted lineages, which were newly identified in our Indian samples. For instance, haplogroup I1 is broadened by requiring only two mutations, 6734 and 16311, and embraces one subbranch, designated as “I1a” (the former haplogroup I1 [Herrnstadt et al. 2002]), which is further characterized by the 3447 and 8616T mutations. The definition of haplogroup W1 is also broadened by requiring only the 7864 transition. Then, three subhaplogroups can be identified: W1a and W1b (corresponding to the former haplogroups W1 and W2, respectively [Finnilä et al. 2001]) and W1c (defined by mutation 14148). A new haplogroup, N2, was erected to capture the sister lineage of haplogroup W found in the Mansi (Derbeneva et al. 2002a). It turns out that haplogroup U3 has only four characteristic mutations (150, 14139, 15454, and 16343), and its main subbranch, formerly identified as “U3” by Richards et al. (2000) and Herrnstadt et al. (2002), is designated as “U3a” here. Our study also fully characterized some haplogroups that were previously described by use of control-region data and RFLP analysis, such as N1, (pre-HV)1, HV2, J1b, J1b1, U1, R1, and R2 (Richards et al. 1998, 2000; Macaulay et al. 1999; Quintana-Murci et al. 2004), and identified a number of subhaplogroups: N1b1, V1, V1a, V2, V2a, J1c, J1c1, J2a, J2b, T1a, T1b, U2d, U5a1, K1a, K1a1, K1a2, K1b, K1c, and K2a (fig. 1). In particular, the former haplogroup J1a (Richards et al. 1998), however, is proven to be one subbranch of J2 on the basis of coding-region sequences (Finnilä et al. 2001; Rose et al. 2001; Herrnstadt et al. 2002, 2003) and is therefore renamed “J2a” here (thus retaining the suffix “a”). Since virtually all haplogroup T1b sequences bearing the mutational motif 16126-16189-16243-16294 (Richards et al. 2000) are also mutated at 16163, we assume that 16163 is a basal mutation of T1; therefore, the T1b sequences from Finnilä et al. (2001) seem to have experienced a back mutation at this site. The provisional definition of haplogroup U2d was inferred from the coding-region mutations reported in patient 3 from the study by Crimi et al. (2002) and from the control-region mutations in some other samples (Richards et al. 2000; Bulayeva et al. 2003; Comas et al. 2004; Quintana-Murci et al. 2004). This haplogroup seems to have a predominantly Near Eastern distribution and might share the 16189 mutation with U2e by ancestry.
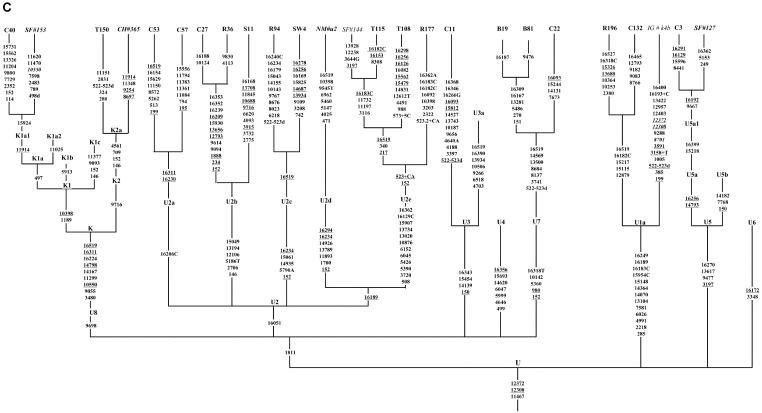
Figure 1.
Phylogenetic tree of 75 Indian complete mtDNA sequences. Parts A, B, C, and D of the figure are the phylogenies of the N, pre-HV and JT, U, and Indian autochthonous R haplogroups, respectively. Mutations are scored relative to the rCRS (Andrews et al. 1999). Indian populations: A = Bhargava, B = Chaturvedi, C = Brahmin, R = Reddy, S = Khasi, SW = Rajbhansi, T = Thogataveera. Twenty-five additional complete sequences were taken from the literature (Finnilä et al. 2001; Maca-Meyer et al. 2001; Taylor et al. 2001; Derbeneva et al. 2002a; Herrnstadt et al. 2002, 2003; Ingman and Gyllensten 2003; Mishmar et al. 2003; Coble et al. 2004), and we referred to particular samples from these articles by SF, NM, RT, OD, CH, IG, DM, and MC, respectively, followed by “#” and the original sample code. Suffixes A, C, G, and T indicate transversions; “d” denotes a deletion, and a plus sign (+) denotes an insertion; recurrent mutations are underlined; “h” indicates heteroplasmy; and italics highlight likely oversights. Mutations in the single reported haplogroup N1a lineage labeled by an asterisk (*) are our reconstruction. The linkage between coding- and control-region mutations in the new haplogroup U2d is tentative. Since the variation at 16519 is extremely hypervariable, only the most parsimonious solution is offered here. For haplogroups H and U5, see articles by Loogväli et al. (2004) and Tambets et al. (2004), respectively.
Discussion
Reconciliation of the Conflicts among Published Western Eurasian Data Sets
With the help of the basal western Eurasian phylogeny constructed here (fig. 1), some obvious conflicts among different data sets could be reconciled. Take the data of Finnilä et al. (2001) as an example. There, transition 12414 had likely been transferred from the haplogroup W sequences to the haplogroup X sequences by mistake. The 10550 transition (characteristic of haplogroup K) had been omitted from all K1 sequences and the 3447 transition from all but one I1 sequence (i.e., SF#104). In other data sets, the typical shortcomings include random oversights of mutations (e.g., at sites 8701, 12308, and 12372 in the haplogroup U1a lineage IG#k4b) or recombination by sample mix-up in singular cases. Particularly problematic are the early data of Maca-Meyer et al. (2001), which were generated through manual sequencing. Many of their western Eurasian lineages lack one to three mutations; the haplogroup I lineage might even have received the 15452A transversion from some haplogroup JT lineage through sample mix-up. We have therefore made only limited use of these data in estimating and representing the basal western Eurasian mtDNA variation. As for the coding-region data published (or corrected) more recently (Herrnstadt et al. 2002, 2003; Mishmar et al. 2003; Coble et al. 2004), the congruence in the basal parts of the phylogeny is assured.
The Phylogeny of mtDNA Haplogroup N in India
The total phylogeny of all mtDNA lineages found in India is partially interwoven with the western Eurasian mtDNA phylogeny but includes numerous basal branches that are absolutely absent in Europe. These findings, first demonstrated by Kivisild et al. (1999a, 1999b) on the basis of HVS-I data, stand the test of complete sequence information. In India, a minority of lineages are of western Eurasian ancestry; the ancestral population probably entered Pakistan and India either from the west (Iran) or the north (via Central Asia) (see Quintana-Murci et al. 2004). In the opposite direction, gene flow was apparently more limited and not very far-reaching: for example, in the data provided by Quintana-Murci et al. (2004), we can find 4 (i.e., 1 haplogroup R5, 1 N5, and 2 M lineages) of 42 mtDNA lineages from (southern) central Iran but only a single lineage (from macrohaplogroup M) of 95 in northern and western Iran that belongs to (potentially) autochthonous South Asian haplogroups. Only one R5 lineage is found in the Iraqi sample of Al-Zahery et al. (2003). Farther to the (north)west, in the Caucasus area and Turkey, such lineages are virtually absent. In the Central Asian data set of Comas et al. (2004), only 6 of 232 lineages belong to South Asian haplogroups (2 from U2a, 1 from U2c, 2 from R5, and possibly 1 from M4). This clear-cut phylogeographic pattern underscores the autochthonous status of the frequent Indian haplogroups U2a, U2b, U2c, and R5–R7. For the less frequent haplogroups (R8, R30, R31, N1d, and N5), comparative HVS-I information would suggest their indigenous status, too, but focused searches of neighboring mtDNA pools by screening characteristic coding-region sites are necessary to confirm this.
Recognition of Indigenous Indian Haplogroup N Lineages
In the study of the mtDNA of patients with mitochondrial diseases whose matrilines are of unknown continental origin, the total worldwide complete mtDNA pool is necessary for comparison. When MITOMAP is consulted for mutation status, mtDNA lineages from South Asia would inevitably go unidentified, and many of their characteristic mutations would be shunted into the category of “novel” mutations. Then, some of these seemingly novel mutations, which might even be regarded as candidates for pathological mutations, are, in fact, specific to new haplogroups. For instance, Pulkes et al. (2003) analyzed 13 mitochondrial genomes from patients with classic mitochondrial phenotypes but reported only those “novel” mutations that were not listed in MITOMAP at the time. Even with respect to this restricted information, five of those mtDNA lineages can then be classified unambiguously here: the mtDNA types of patients 3, 5, and 15 belong to western Eurasian haplogroups (T2a, I1a, and (pre-HV)1, respectively), whereas those of patients 7 and 10 are of South Asian ancestry. Indeed, the mtDNA of patient 7 harbors the three transitions at 8594, 10754, and 14544, which are characteristic of haplogroup R5; moreover, the 8987 and 9708 mutations indicate some closer relationship with haplotype A26/A30 from our Indian sample. Patient 10 carries four specific mutations (3384, 7759, 9449, and 13215) that define haplogroup R8.
When only HVS-I data together with RFLP results (for the coding region) are available, one can nevertheless often predict haplogroup status by near-matching with the haplogroup motifs. Take the haplogroup N data from Kivisild et al. (2003a), for example. We can assign their haplotype 39 to N5 and haplotypes 45 and 46 to R7 without ambiguity. Further, haplotypes 40, 41, and 48 may belong to haplogroup R6, whereas haplotype 42 may belong to R5. Therefore, we are optimistic that our basal phylogeny of haplogroup N in South Asia covers most of the Indian autochthonous lineages. However, we have to concede that there may still be some minor haplogroups that remain to be determined for the coding region.
In the absence of any coding-region information, one can still predict (sub)haplogroup status for the vast majority of Indian HVS-I and HVS-II sequences from haplogroup N via (near-)matching with well-classified haplotypes. For instance, the data of Forster et al. (2002) are conspicuous for their extremely high proportion of haplogroup U1a lineages. This haplogroup status can be confirmed in view of the presence of the full U1a motif (i.e., 16189-16249-73-263-285). We arrive at the same count of 16 sequences belonging to haplogroup U2 as do Forster et al. (2002): namely, five to U2a, seven to U2b, and four to U2c. Then, five lineages are members of R5 (#133, #152, #221, #63b, and #25), two of R6 (#77 and #78), one of R8 (#141), and nine of R31 (#84, #101, #107, #117, #135, #140, #168, #200, and #201). Only ∼10 potential haplogroup R lineages cannot be classified unambiguously (some of which may belong to R8). Haplogroups N5 and N1d do not seem to appear in these data.
Different Layers of mtDNA Haplogroups in India
Since there is thus no evidence that the offshoots R5, R6, R7, R8, R30, and R31 arose west of the Indian subcontinent, one can approximately calculate the time of the arrival of the predicted founder R haplotype in India by stipulating that all of the variation seen in this part of the phylogeny arose in situ. Then, we estimate the age of the root type of R on the basis of these lineages, in the same way as Kong et al. (2003), by assuming the rate of one substitution (other than a deletion or insertion) in the coding region per 5,140 years (Mishmar et al. 2003). This yields an age (mean ± SD) of 64,200±6,300 years, so the actual entry of the R root has a point estimate of ∼65 kya. The only other branch of macrohaplogroup N that could have arisen in India that early is haplogroup N5, but more data on the diversity and geographic distribution of this haplogroup are needed.
Haplogroup U2 as a subhaplogroup of haplogroup U is definitely younger than this early diversification of R in India. Taking the mtDNA lineages from the autochthonous subhaplogroups U2a, U2b, and U2c, we estimate a potential founder age of 49,900±7,900 years for this part of the phylogeny. The age calculated here, however, depends on the relative frequency of U2a versus U2b, since the latter contributes to higher ages in view of the large number of coding-region mutations in the U2b stem of the phylogeny. Control-region estimates for the Indian U2 lineages are slightly higher (∼50–55 kya [Kivisild et al. 1999a]). On the other hand, unless further screening of the Near Eastern mtDNA pool would exhibit early offshoots of U2a, U2b, or U2c, an entry of U2 in India more recent than 40 kya is not plausible. A recent estimate giving a very young age for U (25.6 kya [Baig et al., in press]), however, results from insufficient data, miscalculation (of ρ and σ), and an inadequate merging of U2 lineages with the other U lineages that came to India definitely much later.
Much later migrations, during the Holocene, from the Near East or Central Asia to India are well discernible in the Indian mtDNA pool and are believed to be related to the spread of agriculture and the Aryan invasion, respectively (Kivisild et al. 1999a, 1999b, 2000; McElreavey and Quintana-Murci 2002). Our analysis of the complete sequence variation supports the recent entry of those typical European haplogroups, at least for those haplogroups that have been widely sampled in western Eurasia, such as H, V, K, U5, J, and T. The entry time (calculated from the potentially private mutations) for members of these haplogroups is bounded by <11.5 kya (see Kivisild et al. [1999a] for an estimate based on HVS-I). The actual arrival times will be much more recent, since the Near Eastern mtDNA pool is insufficiently screened for the coding region.
South Asia as the Gate from Africa to Southeast Asia
In regard to the evolution and spread of modern humans, the genetic evidence obtained from high-resolution uniparental (i.e., mtDNA and Y chromosome) markers clearly supports the recent African origin of modern humans. Although this key feature of the Out-of-Africa scenario is widely accepted, the specific question of the routes used by modern humans to leave Africa is still being disputed. The traditional view, born out of the analysis of classic markers and the interpretation of population trees, is that there were two distinct exit routes: a southern route along the Asian coastline and a northern route through the Levant via Central Asia. The modernized variant of this model, however, suggests that both migrations stemmed from a single source in Africa (Cavalli-Sforza and Feldman 2003). Since the Eurasian/Oceanian mtDNA pool consists of two macrohaplogroups, a simplistic interpretation would tag one macrohaplogroup (M) to the southern root and the other (N) to the northern route (Maca-Meyer et al. 2001). This “one haplogroup–one migration” model, however, does not sit easily with the geographic distribution (fig. 2) and with estimated ages of the most basal branches of these macrohaplogroups and of R in particular. Moreover, the mtDNA record from Oceania indicates three autochthonous subhaplogroups of R and N (i.e., P, O, and S; see fig. 2) as well as two autochthonous M subhaplogroups (Ingman et al. 2000). Our analysis of the indigenous haplogroup R lineages in India points to a common first spread of the root haplotypes of M, N, and R along the southern route some 60–70 kya, since haplogroup M was estimated to be essentially of this age in India (Kivisild et al. 2003a) as well as in East Asia (Kong et al. 2003). The analysis further shows that the intrusion of haplogroup U2 (from the Near East ∼50 kya) postdated the protosettlement along the southern route, thus giving further support to the migration scenario proposed by Kivisild et al. (1999a, 1999b). Equating these two events, as Cann (2001) apparently suggested in her figure 1, is then difficult to reconcile with the mtDNA data. At the other extreme, a very early protomigration along the southern Asian coast, before the Toba event (∼74 kya), as proposed by Oppenheimer (2003), does not receive support from the complete sequence data, at least given the employed calibration of the molecular clock. It must be noted, however, that this calibration is inevitably fraught with uncertainty; calibrating mtDNA founder events directly against well documented archeological records may provide more precise estimates in the future.
Figure 2.
Subcontinental ancestry of the most basal Eurasian/Oceanian branches of the mtDNA phylogeny. South Asian and western Eurasian haplogroups are defined as described in the present study; for East Asian haplogroups, see Kong et al. (2003); for haplogroup P, see Forster et al. (2001); O and S are newly defined here on the basis of the data from Ingman and Gyllensten (2003). Potential coalescences based only on a single highly variable site are disregarded.
Acknowledgments
We warmly thank Hui Pan, Guo-Dong Wang, Liang Xie and Yong-Yi Shen for technical assistance. We are also grateful to an anonymous reviewer for drawing our attention to the sister lineage of haplogroup W. The work was supported by National Natural Science Foundation of China grant 30021004, National Key Technologies R&D Program of China and Science and Technology Committee of Yunnan Province grant 2004BA901A07, and Chinese Academy of Sciences grant KSCX2-SW-2010.
Electronic-Database Information
Accession numbers and the URL for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for accession numbers AY713976–AY714050)
References
- Allard MW, Miller K, Wilson MR, Monson KL, Budowle B (2002) Characterization of the Caucasian haplogroups present in the SWGDAM forensic mtDNA data set for 1771 human control region sequences. J Forensic Sci 47:1215–1223 [PubMed] [Google Scholar]
- Allard MW, Wilson MR, Monson KL, Budowle B (2004) Control region sequences for East Asian individuals in the Scientific Working Group on DNA Analysis Methods forensic mtDNA data set. Legal Med 6:11–24 [DOI] [PubMed] [Google Scholar]
- Al-Zahery N, Semino O, Benuzzi G, Magri C, Passarino G, Torroni A, Santachiara-Benerecetti AS (2003) Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol 28:458–472 [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 [DOI] [PubMed] [Google Scholar]
- Baig M, Khan A, Kulkarni K. Mitochondrial DNA diversity in tribal and caste groups of Maharashtra (India) and its implication on their genetic origins. Ann Hum Genet (in press) [DOI] [PubMed] [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, Papiha SS, Villems R, Redd AJ, Hammer MF, Nguyen SV, Carroll ML, Batzer MA, Jorde LB (2001) Genetic evidence on the origins of Indian caste populations. Genome Res 11:994–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, Roychoudhury S, Majumder PP (2003) Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res 13:2277–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstätter A, Parsons TJ, Parson W (2003) Rapid screening of mtDNA coding region SNPs for the identification of west European Caucasian haplogroups. Int J Legal Med 117:291–298 [DOI] [PubMed] [Google Scholar]
- Bulayeva K, Jorde LB, Ostler C, Watkins S, Bulayev O, Harpending H (2003) Genetics and population history of Caucasus populations. Hum Biol 75:837–853 [DOI] [PubMed] [Google Scholar]
- Cann RL (2001) Genetic clues to dispersal in human populations: retracing the past from the present. Science 291:1742–1748 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Feldman MW (2003) The application of molecular genetic approaches to the study of human evolution. Nat Genet 33 Suppl:266–275 [DOI] [PubMed] [Google Scholar]
- Coble MD, Just RS, O’Callaghan JE, Letmanyi IH, Peterson CT, Irwin JA, Parsons T (2004) Single nucleotide polymorphisms over the entire mtDNA genome that increase the power of forensic testing in Caucasians. Int J Legal Med 118:137–146 [DOI] [PubMed] [Google Scholar]
- Comas D, Plaza S, Wells RS, Yuldaseva N, Lao O, Calafell F, Bertranpetit J (2004) Admixture, migrations, and dispersals in Central Asia: evidence from maternal DNA lineages. Eur J Hum Genet 12:495–504 [DOI] [PubMed] [Google Scholar]
- Crimi M, Sciacco M, Galbiati S, Bordoni A, Malferrari G, Del Bo R, Biunno I, Bresolin N, Comi GP (2002) A collection of 33 novel human mtDNA homoplasmic variants. Hum Mutat 20:409 [DOI] [PubMed] [Google Scholar]
- Derbeneva OA, Starikovskaia EB, Volod’ko NV, Wallace DC, Sukernik RI (2002a) Mitochondrial DNA variation in Kets and Nganasans and its implications for the initial peopling of Northern Eurasia. Russian J Genet 38:1316–1321 [PubMed] [Google Scholar]
- Derbeneva OA, Sukernik RI, Volodko NV, Hosseini SH, Lott MT, Wallace DC (2002b) Analysis of mitochondrial DNA diversity in the Aleuts of the Commander Islands and its implications for the genetic history of Beringia. Am J Hum Genet 71:415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä S, Lehtonen MS, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster L, Forster P, Lutz-Bonengel S, Willkomm H, Brinkmann B (2002) Natural radioactivity and human mitochondrial DNA mutations. Proc Natl Acad Sci 99:13950–13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Torroni A, Renfrew C, Röhl A (2001) Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol Biol Evol 18:1864–1881 [DOI] [PubMed] [Google Scholar]
- Hedges SB (2000) A start for population genomics. Nature 408:652–653 [DOI] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 (erratum 71:448–449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Preston G, Howell N (2003) Errors, phantom and otherwise, in human mtDNA sequences. Am J Hum Genet 72:1585–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Gyllensten U (2003) Mitochondrial genome variation and evolutionary history of Australian and New Guinean aborigines. Genome Res 13:1600–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2000) Mitochondrial genome variation and the origin of modern humans. Nature 408:708–713 [DOI] [PubMed] [Google Scholar]
- Kaur I, Roy S, Chakrabarti S, Sarhadi VK, Majumder PP, Bhanwer AJS, Singh JR (2002) Genomic diversities and affinities among four endogamous groups of Punjab (India) based on autosomal and mitochondrial DNA polymorphisms. Hum Biol 74:819–836 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, Papiha SS, Mastana SS, Mir MR, Ferak V, Villems R (1999a) Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol 9:1331–1334 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha SS, Villems R (1999b) The place of the Indian mitochondrial DNA variants in the global network of maternal lineages and the peopling of the Old World. In: Deka R, Papiha SS (eds) Genomic diversity. Kluwer/Academic/Plenum Publishers, New York, pp 135–152 [Google Scholar]
- Kivisild T, Papiha SS, Rootsi S, Parik J, Kaldma K, Reidla M, Laos S, Metspalu M, Pielberg G, Adojaan M, Metspalu E, Mastana SS, Wang Y, Gölge M, Demirtas H, Schnakenberg E, De Stefano GF, Geberhiwot T, Claustres M, Villems R (2000) An Indian ancestry: a key for understanding human diversity in Europe and beyond. In: Renfrew C, Boyle K (eds) Archaeogenetics: DNA and the population prehistory of Europe. McDonald Institute for Archaeological Research, University of Cambridge, Cambridge, UK, pp 267–279 [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, Gölge M, Usanga E, Papiha SS, Cinnioğlu C, King R, Cavalli-Sforza L, Underhill PA, Villems R (2003a) The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72:313–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Metspalu E, Parik J, Katrin K, Usanga E, Mastana S, Papiha SS, Villems R (2003b) Genetics of the language and farming spread in India. In: Renfrew C, Boyle K (eds) Examining the farming/language dispersal hypothesis. McDonald Institute Monographs Series, McDonald Institute for Archaeological Research, Cambridge, UK, pp 215–222 [Google Scholar]
- Kong Q-P, Yao Y-G, Sun C, Bandelt H-J, Zhu C-L, Zhang Y-P (2003) Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet 73:671–676 (erratum 75:157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Q-P, Yao Y-G, Sun C, Zhu C-L, Zhong L, Wang C-Y, Cai W-W, Xu X-M, Xu A-L, Zhang Y-P (2004) Phylogeographic analysis of mitochondrial DNA haplogroup F2 in China reveals T12338C in the initiation codon of the ND5 gene not to be pathogenic. J Hum Genet 49:414–423 [DOI] [PubMed] [Google Scholar]
- Loogväli EL, Roostalu U, Malyarchuk BA, Derenko MV, Kivisild T, Metspalu E, Tambets K, et al (2004) Disuniting uniformity: a pied cladistic canvas of mtDNA haplogroup H in Eurasia. Mol Biol Evol. http://mbe.oupjournals.org/cgi/reprint/msh209v1 (electronically published July 14, 2004; accessed September 28, 2004) [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N, González AM, Larruga JM, Flores C, Cabrera VC (2001) Major genomic mitochondrial lineages delineate early human expansions. BMC Genetics 2:13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maca-Meyer N, González AM, Pestano J, Flores C, Larruga JM, Cabrera VC (2003) Mitochondrial DNA transit between West Asia and North Africa inferred from U6 phylogeography. BMC Genetics 4:15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonne-Tamir B, Sykes B, Torroni A (1999) The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElreavey K, Quintana-Murci L (2002) Understanding inherited disease through human migrations: a south-west Asian perspective. Community Genet 5:153–156 [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC (2003) Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA 100:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer S (2003) Out of Eden: the peopling of the world. Constable, London. Republished as: The real Eve: modern man’s journey out of Africa. Carroll & Graf, New York [Google Scholar]
- Passarino G, Semino O, Bernini LF, Santachiara-Benerecetti AS (1996) Pre-Caucasoid and Caucasoid genetic features of Indian population revealed by mtDNA polymorphisms. Am J Hum Genet 59:927–934 [PMC free article] [PubMed] [Google Scholar]
- Pulkes T, Liolitsa D, Nelson IP, Hanna MG (2003) Classical mitochondrial phenotypes without mtDNA mutations: the possible role of nuclear genes. Neurology 61:1144–1147 [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, Coppa A, Ayub Q, Mohyuddin A, Tyler-Smith C, Mehdi SQ, Torroni A, McElreavey K (2004) Where West meets East: the complex mtDNA landscape of the southwest and central Asian corridor. Am J Hum Genet 74:827–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt H-J, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 [DOI] [PubMed] [Google Scholar]
- Quintáns B, Álvarez-Iglesias V, Salas A, Phillips C, Lareu MV, Carracedo A (2004) Typing of mitochondrial DNA coding region SNPs of forensic and anthropological interest using SNaPshot minisequencing. Forensic Sci Int 140:251–257 [DOI] [PubMed] [Google Scholar]
- Reidla M, Kivisild T, Metspalu E, Kaldma K, Tambets K, Tolk H-V, Parik J, et al (2003) Origin and diffusion of mtDNA haplogroup X. Am J Hum Genet 73:1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M, Macaulay V (2001) The mitochondrial gene tree comes of age. Am J Hum Genet 68:1315–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Bandelt H-J, Sykes B (1998) Phylogeography of mitochondrial DNA in western Europe. Ann Hum Genet 62:241–260 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Rocha H, Flores C, Campos Y, Arenas J, Vilarinho L, Santorelli FM, Torroni A (1999) About the “pathological” role of the mtDNA T3308C mutation… Am J Hum Genet 65:1457–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Passarino G, Carrieri G, Altomare K, Greco V, Bertolini S, Bonafè M, Franceschi C, De Benedictis G (2001) Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians. Eur J Hum Genet 9:701–707 [DOI] [PubMed] [Google Scholar]
- Roy S, Thakur CM, Majumder PP (2003) Mitochondrial DNA variation in ranked caste groups of Maharashtra (India) and its implication on genetic relationship and origins. Ann Hum Biol 30:443–454 [DOI] [PubMed] [Google Scholar]
- Tambets K, Rootsi S, Kivisild T, Help H, Serk P, Loogväli EL, Tolk H-V, et al (2004) The western and eastern roots of the Saami—the story of genetic “outliers” told by mitochondrial DNA and Y chromosomes. Am J Hum Genet 74:661–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Taylor GA, Durham SE, Turnbull DM (2001) The determination of complete human mitochondrial DNA sequences in single cells: implications for the study of somatic mitochondrial DNA point mutations. Nucleic Acids Res 29:E74–E81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon FL, Simionati B, Valle G, Richards M, Macaulay V, Scozzari R (2001) Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet 69:1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]