Abstract
Attention-deficit/hyperactivity disorder (ADHD [MIM 143465]) is the most common behavioral disorder of childhood. Twin, adoption, segregation, association, and linkage studies have confirmed that genetics plays a major role in conferring susceptibility to ADHD. We applied model-based and model-free linkage analyses, as well as the pedigree disequilibrium test, to the results of a genomewide scan of extended and multigenerational families with ADHD from a genetic isolate. In these families, ADHD is highly comorbid with conduct and oppositional defiant disorders, as well as with alcohol and tobacco dependence. We found evidence of linkage to markers at chromosomes 4q13.2, 5q33.3, 8q11.23, 11q22, and 17p11 in individual families. Fine mapping applied to these regions resulted in significant linkage in the combined families at chromosomes 4q13.2 (two-point allele-sharing LOD score from LODPAL = 4.44 at D4S3248), 5q33.3 (two-point allele-sharing LOD score from LODPAL = 8.22 at D5S490), 11q22 (two-point allele-sharing LOD score from LODPAL = 5.77 at D11S1998; multipoint nonparametric linkage [NPL] −log [P value] = 5.49 at ∼128 cM), and 17p11 (multipoint NPL −log [P value] >12 at ∼12 cM; multipoint maximum location score 2.48 [α = 0.10] at ∼12 cM; two-point allele-sharing LOD score from LODPAL = 3.73 at D17S1159). Additionally, suggestive linkage was found at chromosome 8q11.23 (combined two-point NPL−log [P value] >3.0 at D8S2332). Several of these regions are novel (4q13.2, 5q33.3, and 8q11.23), whereas others replicate already-published loci (11q22 and 17p11). The concordance between results from different analytical methods of linkage and the replication of data between two independent studies suggest that these loci truly harbor ADHD susceptibility genes.
Introduction
Over the past decade, twin, adoption, family, and association studies have shown that genetic factors substantially contribute to the etiology of attention-deficit/hyperactivity disorder (ADHD [MIM 143465]), a persistent syndrome characterized by difficulty in paying attention, excessive motor activity, and impulsivity (American Psychiatric Association 1994). ADHD is the most common behavioral disorder of childhood, with prevalence figures of 5%–10% and up to 17% when diagnostic criteria are relaxed (Pineda et al. 1999; Castellanos and Tannock 2002). Children with ADHD are at heightened risk for poor educational attainment, low income, and underemployment, as well as difficulties in social relationships (Greenfield et al. 1988; Faraone et al. 1996; Foley et al. 1996). Between 10% and 20% of children with ADHD have mood disorders, and 20% have conduct disorders (Biederman et al. 1991). Between 30% and 45% of patients with ADHD also have oppositional defiant disorder (ODD), and between 61% and 67% of patients with ODD have ADHD (Harada et al. 2002a, 2002b). Furthermore, bipolar disorder is increasingly being recognized in individuals with ADHD (Chang et al. 2000; Giedd 2000; Faraone et al. 2001; Geller et al. 2002).
Genetic studies in twins indicate a substantially high genetic (additive) contribution to phenotypic variation, reaching 0.91 (Gillis et al. 1992; Levy et al. 1997). Adoption studies have also confirmed that genetics, rather than shared environment, causes familial clustering of ADHD (Morrison and Stewart 1973; van den Oord et al. 1994). Family studies have confirmed the observation of increased recurrence risk by comparing the ratio of the prevalence of ADHD in various kinds of relatives with the population prevalence by use of the λ statistic (Risch and Merikangas 1996, 1997; Faraone et al. 2000). Three independent complex segregation analyses consistently demonstrated that the model that best fit the data was that of a major autosomal dominant/codominant gene (Faraone et al. 1992; Lopera et al. 1999; Maher et al. 1999). Additionally, candidate genes were selected because their theoretical and empirical involvement in the physiopathogenesis of ADHD has shown significant association/linkage to the disorder, even though they disclose very small effect sizes (reviewed by Acosta et al. [2004]).
Two independent groups have conducted genomewide screens aimed at uncovering major risk genes for ADHD (Fisher et al. 2002; Bakker et al. 2003; Ogdie et al. 2003). One study, composed of 270 affected sib pairs (ASPs), found suggestive evidence of linkage at 16p13 and 17p11 in a mostly white population from the United States (Fisher et al. 2002; Ogdie et al. 2003). The second genomewide search was performed in 164 Dutch ASPs. Although the most suggestive linkage region was at 15q, additional regions at chromosomes 7p and 9q were also observed. Assuming that these regions really do harbor ADHD susceptibility genes, the nonoverlapping findings from these two studies indicate that either genetic heterogeneity or other causes, such as differences in the phenotype, may represent a substantial obstacle to replication across samples and to definitive gene mapping and identification.
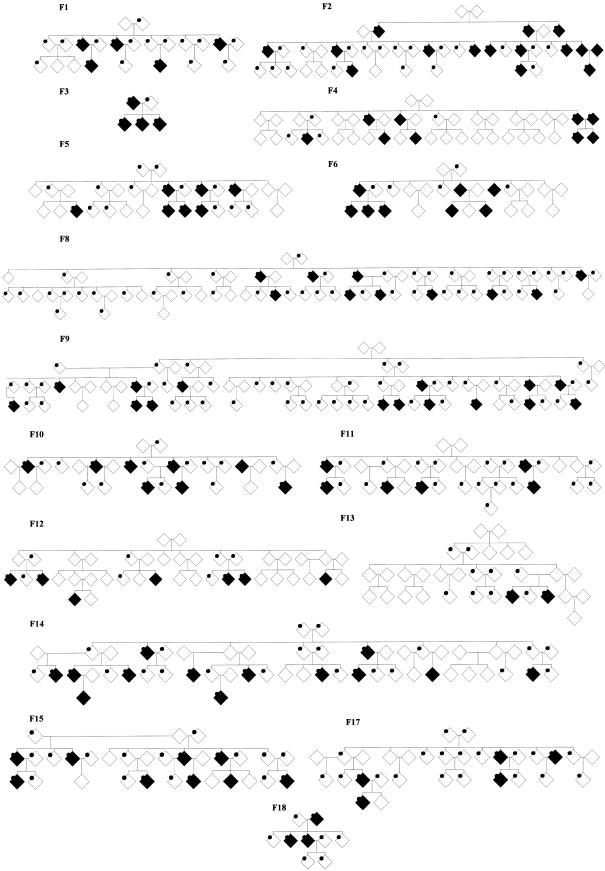
To reduce one source of such heterogeneity, we have assembled a sample from a genetic isolate of the self-designated “Paisa” community in the state of Antioquia, in the region of Medellín, Colombia (Bravo et al. 1996; Jimenez et al. 1996; Arcos-Burgos and Muenke 2002; Arcos-Burgos et al. 2002). We took advantage of the particularly large Paisa families and recruited individuals from 16 multigenerational and extended pedigrees that densely segregated ADHD highly comorbid with conduct and ODDs, as well as with alcohol and tobacco dependence (fig. 1).
Figure 1.
Pedigree structure of 16 multigenerational Paisa families. ADHD affection status is shown in black. DNA samples that were subjected to genotyping are indicated by a dot. Pedigrees have been modified to protect confidentiality.
Methods
Family Ascertainment and Clinical Phenotyping
Ascertainment methods, power simulations for both linkage and association, population genetics, demographics, inclusion and exclusion criteria, clinical methods of assessment, and clinical features of the sample have been described elsewhere (Lopera et al. 2001; Arcos-Burgos et al. 2002, 2004). The sample was selected from Paisa families from the Medellín metropolitan area in the state of Antioquia, Colombia. Families had to be of Paisa descent, to comprise more than two generations, and to have more than two members affected with ADHD. Individuals were considered to be of Paisa descent if all four grandparents originated from the Paisa region of Colombia (i.e., from the former state of Viejo Caldas). Initial coded pedigrees—obtained through a fixed sampling scheme from a parent or grandparent of an index proband (after the provision of written informed consent, as approved by the ethics committee of the University of Antioquia)—were individually reviewed by a genetic statistician (J.B.W.) to minimize the confounding effects of bilineal transmission of ADHD. Bilineality was defined by the presumptive diagnosis of ADHD in both parents on the basis of an informant’s reports of childhood symptoms and/or of academic, occupational, or legal impairment, including alcoholism and related consequences. Full pedigrees that were identified as bilineal were excluded from further study. During the selection phase, pedigrees that contained branches that were bilineal were “pruned” to preserve the presumptively unilineal branches. Individuals in the selected families were then invited to participate in the present study.
As noted above, the first phase of the study, which consisted of the obtainment of pedigrees with provisional diagnoses, was conducted under the auspices and oversight of the ethics committee of the University of Antioquia, which also approved a subsequent collaboration with investigators from the U.S. National Institutes of Health. The proposal to conduct the present study (protocol 00-HG-0058) was jointly approved by the National Human Genome Research Institute institutional review board and the ethics committee of the University of Antioquia. Informed-consent documents were translated into Spanish for use in Colombia and reverse-translated into English for institutional review board review. Both approvals remain active. All adult participants provided written informed consent. Parents of participating minors provided written informed consent; minors ⩾6 years of age who could write also provided signed assent.
Structured diagnostic interviews were conducted in the Neurosciences Clinic or during home visits by bachelor's degree–level psychologists who were blind to participants’ presumptive diagnoses. They were trained by a child-and-adolescent psychiatrist (J.D.P.) who reviewed all interviews and conducted confirmatory clinical interviews with nearly all participants. Parents underwent a full psychiatric structured interview regarding their offspring (Diagnostic Interview for Children and Adolescents, Revised: Parents' Version [DICA-IV-P] [Reich 2000]). Parents and teachers of school-aged children also provided behavior-rating scales. The Composite International Diagnostic Interview was administered to all adult participants (Tacchini et al. 1994), as was the Disruptive Behavior Disorders module from the DICA-IV-P, modified for retrospective use.
Detailed pedigree information was obtained from all surviving and consenting grandparents. In nearly all cases, collateral information was obtained from at least one additional knowledgeable relative. Final diagnoses for each pedigree member were reached by consensus of a committee of four bilingual clinicians (J.D.P., D.P., F.L., and F.X.C.), all of whom have extensive experience in diagnosing ADHD. The diagnoses were made—using the best-estimate procedure (Leckman et al. 1982)—on the basis of the results of structured interviews, collateral historical information, and clinical interviews. The three Colombian physicians (J.D.P., D.P., and F.L.) were personally acquainted with all the unaffected and affected individuals. J.D.P. supervised the psychiatric interviews, D.P. supervised neuropsychological assessments and conducted clinical neurological evaluations, and F.L. was the supervising physician (neurologist and neuropsychologist) for the team and, in many cases, the primary clinician. J.D.P. presented the cases in diagnostic conferences chaired by F.X.C.
Participants were classified in one of four mutually exclusive categories: definitely affected with ADHD, unaffected, possibly affected with ADHD, and unknown. “Definitely affected” subjects generally met full DSM-IV criteria for ADHD during childhood, with onset before the age of 7 years and with persistence of clearly impairing symptoms in more than one setting. In cases of discordance between an individual’s self-report of symptoms and collateral reports, the supervising psychiatrist (J.D.P.) obtained further collateral information and probed more deeply for evidence of early impairment. Individuals were classified as “possibly affected” if they failed to meet DSM-IV criteria for ADHD, particularly with respect to unequivocal impairment (DSM-IV criterion D), or if they met only five of the six DSM-IV “A” criteria in childhood. Individuals who were reported by relatives to meet the criteria for ADHD but from whom interviews were unavailable were also classified as “possibly affected.” Individuals who did not meet DSM-IV criteria for ADHD were classified as “unaffected.” The “unknown” category applied to those subjects for whom complete evaluations could not be performed.
Genomewide Search
Blood samples for DNA extraction (n=375) and, in some cases, for lymphoblastoid cell lines (n=36) were obtained from all participating family members. A genome scan using automated fluorescent microsatellite analysis was performed at the Center for Inherited Disease Research (CIDR). The current CIDR marker set consists of ∼400 primer pairs with an average spacing of 9 cM throughout the genome and an average marker heterozygosity of 0.76. Additional details about genotyping, quality control, marker information, and laboratory protocols can be accessed at the Center for Inherited Disease Research Web site.
After a genomewide search was completed at CIDR and the data were analyzed, we genotyped microsatellite loci spanning five regions that exhibited suggestive linkage, with an average resolution of ∼1.5 cM. Genotyping was performed by deCODE. Additional details about genotyping, quality control, marker information, and laboratory protocols can be accessed at the deCODE Web site.
Genetic Analyses
Parametric analysis of linkage was based on the best-fitting genetic model from a previous complex segregation analysis of Paisa families with ADHD (Lopera et al. 1999). In this analysis, the parsimonious model was the one of a major dominant gene with incomplete penetrance and an ADHD susceptibility-allele frequency of 3%. Because the age of 6 years is the DSM-IV landmark for the diagnosis of ADHD (American Psychiatric Association 1994), we used a penetrance model that assumed that penetrance is 0 at the age of 0 years and increases to 55.3% in male gene carriers, 44.7% in female gene carriers, 2.9% in male noncarriers, and 0.2% in female noncarriers by the age of 10 years (Lopera et al. 2001; Arcos-Burgos et al. 2002). Possibly affected individuals were coded as “unknown” in the analyses. To avoid the effect of inflating the posterior false-positive rate as a consequence of performing multiple comparisons, those models that considered possibly affected individuals as affected or unaffected were not used in the analysis. In the same vein, we did not use models that tested for different models of inheritance for a major gene (e.g., recessive and additive models). Two-point LOD score analyses were estimated using FASTLINK (Cottingham et al. 1993). SIMWALK2 was used to perform the parametric multipoint analysis and the heterogeneity analysis and to obtain two-point nonparametric (model-free) linkage (NPL) A–E statistics (Sobel and Lange 1996). Because of the large size of the pedigrees, we could not use conventional software for families of moderate size to estimate multipoint NPL, and analyses of X-linked markers were constrained to the two-point parametric linkage results.
To address the pedigree-size problem so that model-free multipoint estimates could be obtained, we used sparse inheritance trees for pedigree analysis, as implemented in MERLIN (Abecasis et al. 2002), by trimming affected individuals—mostly single affected cases in a nuclear family—belonging to the central branches of the more extended pedigrees (i.e., F9 and F14 [see fig. 1]).
Additionally, two-point identity-by-descent (IBD)–sharing estimates for the marker loci were obtained with the GENIBD program from the SAGE (Statistical Analysis for Genetic Epidemiology) software package (version 4.5). In accordance with the algorithms implemented in SAGE, if the quantity of 2 × [(number of nonfounders) − (number of founders)] in the pedigree was ⩽18, we used the exact algorithm (Lander and Green 1987). If the pedigree did not meet the size restriction, IBD-sharing estimates were calculated with the Markov chain–Monte Carlo simulation algorithm (Sobel and Lange 1996). With these IBD-sharing estimates, we performed linkage analysis using the conditional logistic model (Olson 1999), a reparameterized version of the LOD score model for ASPs (Risch 1990), as implemented in the module LODPAL in SAGE, version 4.5. Two-point analyses were performed for all relative pairs by use of the one-parameter and two-parameter models for the conditional logistic model (Goddard et al. 2001).
We also used the pedigree disequilibrium test (PDT) (Martin et al. 2000, 2003) to search for evidence of linkage disequilibrium (LD) between ADHD and the marker loci. The PDT program performs both allele-specific and genotype-specific LD analysis of individual markers (Martin et al. 2003). This genotype-based approach allows the detection of additive effects among alleles at the target locus (Martin et al. 2003). Power analysis and design considerations for applying family-based association tests (FBATs), described in several studies by Lange and colleagues (Lange and Laird 2002a, 2002b; Lange et al. 2002), were used to determine the specific power exhibited by this set of families, as implemented in the Power-Based Association Test software (Lange and Laird 2002a). Considering the constraints of our sample size (79 nuclear families embedded in the extended pedigrees; sibship average size 2.4; range 1–12 sibs), we obtained a power of 1.0 at a significance level of α=0.05 and 0.01 for a gene of major effect. The PDT and genoPDT analyses were applied using PDT, version 4.0, and PDT, version 5.1 (Martin et al. 2003), implemented within a Linux environment on a PC provided by CYGWIN.
To test for the presence of linkage heterogeneity (two or more disease loci) with respect to single-marker loci, we used the overall LOD score based on a mixture likelihood, referred to as the “heterogeneity LOD” score, which is maximized over (α, θ), where α represents the proportion of linked families and θ is the fraction of recombination (Ott 1983; Ott and Bhat 1999). For this purpose, we used the HOMOG suite of programs and performed two types of heterogeneity testing: (1) the presence of two family types, one with linkage between a trait to a marker, the other without linkage, and (2) an extension of the first test of homogeneity to any number (m) of trait loci (Bhat et al. 1999), using HOMOGM. Two-point LOD values at the markers with significant linkage or suggestive linkage were used as input for determination of the presence of linkage heterogeneity. The statistical hypotheses contrasted by these tests are defined as follows: H0 is the very basic hypothesis of both homogeneity and absence of linkage, H1 is the usual null hypothesis of homogeneity (i.e., all families belong to a single family type, with linkage between the main locus and the marker locus), and H2 is the hypothesis of heterogeneity, with two family types—one type linked to the trait locus and the other type without linkage (Ott 1983; Ott and Bhat 1999).
To perform family-based association studies involving haplotypes, we used the HAPLOTYPE module of SIMWALK2 to reconstruct the more probable haplotype arrangements for these two loci. This haplotype analysis estimates the most likely arrangement of fully typed maternal and paternal haplotypes of the marker loci for each individual belonging to a particular pedigree (Sobel and Lange 1996). This study used the high-performance computational capabilities of the SGI Origin 2000 system at the Center for Information Technology at the National Institutes of Health in Bethesda, Maryland.
Results
Genomewide Search
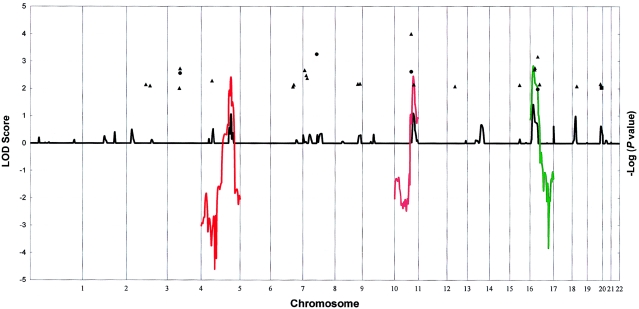
The combined parametric multipoint linkage analysis of all families together, assuming heterogeneity, showed peaks of maximum location scores (MLSs [obtained with SIMWALK2]) >1.0 at chromosomes 5q33.3 (MLS 1.07; 159 cM; α=0.10), 11q22 (MLS 1.09; 121 cM; α=0.10), and 17p11 (MLS 1.42; 12.17 cM; α=0.10). In family-specific parametric (multipoint and two-point) and two-point NPL analyses, we found evidence of significant linkage at one chromosomal region and suggestive linkage in several other regions. In particular, family F9 showed a significant two-point parametric LOD score (obtained with FASTLINK) of 3.22 (θ=0) at 8q11.23 (D8S1110). In this same family, the two-point NPL score obtained with SIMWALK2 at this location had a −log (P value) of 1.44, with similarly significant P values at the adjacent markers, D8S1477 and D8S1771. In addition, family F9 also exhibited some evidence in favor of linkage in two other regions: 4q13.2 (two-point LOD score D4S2367 2.56; two-point NPL −log [P value] =2.74, with NPL significant P values at the adjacent marker D4S3248) and 5q33.3 (multipoint MLS 2.41; 159 cM; two-point NPL −log [P value] = 1.43 at D5S820; adjacent marker D5S1480, NPL −log [P value] = 1.60).
Family F8 showed some evidence of linkage of ADHD to markers at 11q22 (two-point parametric LOD score 2.62 [θ=0] at D11S1998; multipoint MLS 2.45; two-point NPL −log [P value] = 4.0 at D11S1998). Family F14 gave some positive evidence of linkage to 17p11.1 (two-point LOD score 1.98 [θ=0] at D17S799; multipoint MLS 2.83; two-point NPL −log [P value] = 3.06 at D17S799) (see summary of results in table 1 and fig. 2). Other nominal regions exhibiting concordance among different analyses were detected at chromosomes 5p13.3, 8p23.1, 9q33.3, 19p13.2, and 20q13.33. It is intriguing that the PDT showed LD of ADHD with alleles that were strongly clustered at contiguous markers, recapitulating our findings of the linkage analyses. (For extensive and detailed information about the genomewide search on pedigree structure, parametric [two-point LOD scores and multipoint MLS>1.0], two-point NPL scores [P<.05], and PDT [P<.05] results over the whole set of chromosomes, see the ADHD Genetic Research Study Web site.)
Table 1.
Summary of Linkage Results, Assuming Homogeneity, of a Genomewide Scan for 16 Families with ADHD[Note]
| Family IDand Chromosome | Closest Marker | Location(cM) | Parametric Two-Point LODa | Parametric Multipoint MLSb(Location in cM) | Two-Point NPLb,c |
| F9: | |||||
| 4q13.2 | D4S2367 | 78 | 2.56 | .62 (78) | 2.7 |
| 5q33.3 | D5S1480 | 159 | 1.45 | 2.41 (159) | 1.6 |
| 8q11.23 | D8S1110 | 67 | 3.22 | .96 (74) | 1.9 |
| F8: | |||||
| 11q22 | D11S1998 | 113 | 2.62 | 2.45 (121) | 4.0 |
| F14: | |||||
| 17p11 | D17S799 | 32 | 1.98 | 2.829 (12) | 3.0 |
Note.— The appropriate adjustment of significance values has to be taken into account because of multiple comparisons. For a LOD score >3, a simulation study with 10,000 replicates for an unlinked marker in these 16 families results in a posterior rate of false positives of 0.000. Therefore, critical values, as described by Lander and Kruglyak (1995), might be used to consider the significance of linkage. However, because of the heterogeneity pattern exhibited in the 16 families, an adjustment that uses the Bonferroni correction should be taken into account. Two-point LOD scores >2.0, NPL E-STAT −log (P value) >2.0, and MLSs >2.0 are shown, illustrating regions with suggestive linkage and concordance between different methods. Results after combining the whole set of families, assuming heterogeneity, are presented in the “Results” section. No value >2.0 was found in the combined families. (See also fig. 2.)
Estimated with FASTLINK; θ=0.0.
Estimated with SIMWALK2.
−log (P value) E-STAT.
Figure 2.
Results of a genomewide scan of 16 families with ADHD. Two-point LOD scores that are >2.0 per chromosome (as estimated by FASTLINK), for a recombination value of θ=0, are shown as blackened circles; −log (P value) of the two-point NPL E-STAT >2.0 per chromosome for each marker (as estimated by SIMWALK2) are depicted as blackened triangles, and −log (P value) of the PDT Z statistic values >2.0 per chromosome at each marker for the whole set of families together are shown as blackened squares. Two different symbols at the same marker indicate either two positive families or, in the case of NPL statistics, a combined positive result for the whole set of families. Multipoint location scores, for those families exhibiting scores >2.0 (as estimated by SIMWALK2) in any region of the chromosome, are presented as colored lines. Black lines represent the parametric multipoint location scores per chromosome in the combined families. Linkage analysis of the X chromosome does not report any value >2.0
Fine Mapping
We defined minimal critical regions that contained putative ADHD loci on the basis of both a comprehensive assessment of the linkage-analysis results and by direct observation of shared regions in affected individuals, as defined by recombination events over the multigenerational structure of these pedigrees. In fact, establishing a minimal critical region of linkage is one of the advantages of extended and multigenerational pedigrees, compared with the information available from sib pairs. Recombination events that occurred in unaffected individuals were not considered when minimal critical regions were established. Thus far, minimal critical regions for 4q13.2, 5q33.3, 8q11.23, 11q22, and 17p11 were located between the following sets of microsatellite markers: D4S3248 (at 59 Mb) and D4S1647 (at 99 Mb), D5S1505 (at 119 Mb) and D5S816 (at 135 Mb), D8S1477 (at 31 Mb) and D8S1136 (at 65 Mb), D11S1391 (at 110 Mb) and D11S912 (at 128 Mb), and D17S1298 (at 3 Mb) and D17S2196 (at 17 Mb), respectively.
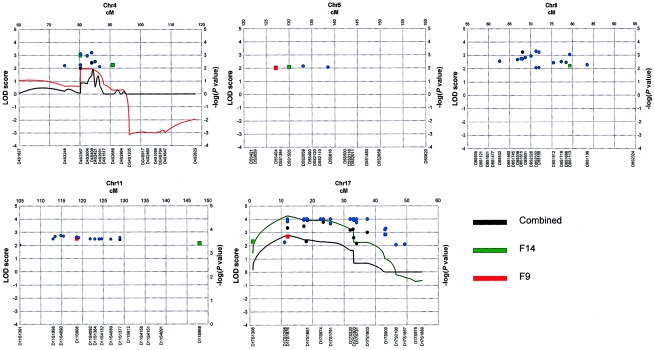
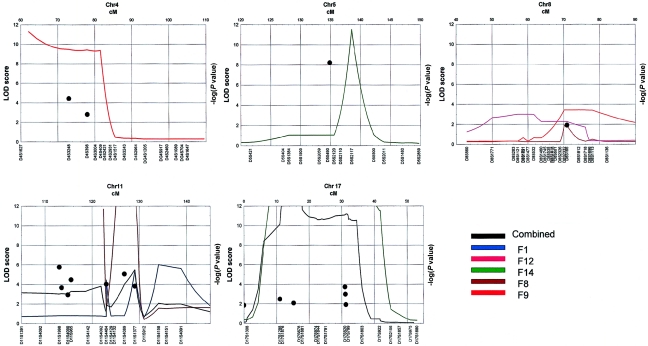
On the basis of the analysis of all families together, we found significant linkage at chromosomes 4q13.2 (two-point NPL allele-sharing LOD score, as implemented in LODPAL = 4.44 at D4S3248), 5q33.3 (two-point NPL LOD score 8.22 at D5S490), 11q22 (two-point NPL LOD score 5.77 at D11S1998; multipoint NPL−log [P value] = 5.49, as implemented in MERLIN, at ∼128 cM), and 17p11 (multipoint MLS 2.48 [proportion of linked families, α=0.10], as implemented in SIMWALK2, at ∼12 cM; multipoint NPL −log [P value] >12, as implemented in MERLIN, at ∼12 cM; two-point NPL LOD score 3.73 at D17S1159). We found significant linkage results in individual families: family F9, at chromosome 4q13.2 (multipoint NPL −log [P value] >10) and chromosome 5q33.3 (multipoint NPL −log [P value] >10); family F14, at chromosome 17p11 (two-point LOD score 3.91; multipoint MLS 4.24; multipoint NPL −log [P value] >12); and family F8, at 11q22 (two-point LOD score 2.43; multipoint NPL −log [P value] >12). Additionally, we found suggestive linkage at chromosome 8q11.23 (two-point NPL −log [P value] >3.0) at D8S2332 in all families combined (see summary of results in figs. 3 and 4 and tables 2 and 3).
Figure 3.
Linkage results of the fine mapping of five regions (4q13.2, 5q33.3, 8q11.23, 11q22, and 17p11) in 16 families with ADHD. Two-point LOD scores >2.0 per chromosome (as estimated by FASTLINK), for a recombination value of θ=0, are shown as blackened circles; −log (P value) of the two-point NPL E-STAT >2.0 per chromosome for each marker (as estimated by SIMWALK2) are shown as blue circles and triangles (significant combined results), and −log (P value) of the PDT Z statistic values >2.0 per chromosome at each marker for the whole set of families are indicated as green (conferring susceptibility) and red (conferring protection) squares. Two different symbols at the same marker indicate either two positive families or, in the case of NPL statistics, a combined positive result for the whole set of families. Two squares at the same marker indicate either that two alleles are in LD or that there was concordance between the two kinds of PDT (i.e., sum and average). Multipoint location scores, as estimated by SIMWALK2, for those families exhibiting scores >2.0 in any region of the chromosome, are presented as colored lines. Black lines represent the parametric multipoint location scores per chromosome in the combined families.
Figure 4.
Model-free linkage results of the fine mapping of five regions (4q13.2, 5q33.3, 8q11.23, 11q22, and 17p11) in 16 families with ADHD. Two-point LOD scores >2.0, estimated by the conditional logistic model (Olson 1999) by use of LODPAL in SAGE, version 4.5, are shown as blackened circles. Continuous lines represent the −log (P value) of the multipoint NPL E-STAT per chromosome, as estimated by MERLIN (colored lines represent specific families, and black lines denote the combined set of all families).
Table 2.
Summary of Family-Specific Linkage Results of the Fine Mapping of Five Regions (4q13.2, 5q33.3, 8q11.23, 11q22, and 17p11)
| Chromosomeand Closest Marker | Location(cM) | Two-Point Parametric LODa (Family ID) | Maximum Multipoint NPLall Zb (Family ID) |
| 4q13.2: | |||
| D4S409 | 81.6 | 2.4 (F9) | >9.0 (F9) |
| 5q33.3: | |||
| D5S2117 | 138.5 | 1.5 (F9) | 11.5 (F14) |
| 8q11.23: | |||
| D8S1110 | 67.0 | 3.2 (F9) | 2.8 (F9) |
| 11q22: | |||
| D11S1377 | 129.0 | 2.4 (F8) | >12.0 (F8) |
| 17p11: | |||
| D17S1876 | 11.9 | 3.4 (F14) | >12.0 (F14) |
| D17S678 | 16.9 | 3.5 (F14) | >12.0 (F14) |
| D17S1881 | 17.9 | 3.9 (F14) | >12.0 (F14) |
| D17S1844 | 22.8 | 3.8 (F14) | >12.0 (F14) |
| D17S1791 | 25.1 | 3.7 (F14) | >12.0 (F14) |
Estimated with FASTLINK.
−log (P value); calculated using MERLIN.
Table 3.
Summary of Linkage Results of the Fine Mapping of Five Regions (4q13.2, 5q33.3, 8q11.23, 11q22, and 17p11) in the Combined Set of Families, Assuming Heterogeneity
| Chromosomeand Closest Marker | Location(cM) | Two-Point Combined Conditional Logistic Analysis of LODa | Multipoint Combined NPLall Zb |
| 4q13.2: | |||
| D4S3248 | 73.0 | 4.4 | <2.0 |
| 5q33.3: | |||
| D5S490 | 134.5 | 8.2 | <2.0 |
| 11q22: | |||
| D11S1998 | 113.0 | 5.8 | <2.0 |
| D11S4089 | 126.8 | 5.1 | 4.0 |
| 17p11: | |||
| D17S1876 | 11.9 | 2.5 | >12.0 |
For affected relative pair; calculated using LODPAL.
−log (P value); calculated using MERLIN.
Additionally, by use of both PDT and genoPDT analyses, we obtained evidence of LD between ADHD and the set of regions submitted to fine mapping. We found significant LD at chromosome 4q13.2 (−log [P value] > 2.0) at markers D4S2367 and D4S2689 (−log [P value] = 3.0 and 2.25, respectively), at 5q33.3 at D5S404 and D5S1505 (−log [P value] = 2.03 and 2.02, respectively), at 8q11.23 at D8S1113 (−log [P value] = 2.21), at 11q22 at D11S908 and D11S968 (−log [P value] = 2.52 and 2.16, respectively), and at 17p11 at D17S1308 and D17S1876 (−log [P value] = 2.33 and 2.70, respectively). Other significant results for PDT and genoPDT (−log [P value] > 1.31) can be accessed at the ADHD Genetic Research Study Web site.
Linkage-heterogeneity analysis for the five regions subject to fine mapping (4q13.2, 5q33.3, 8q11.23, 11q22, and 17p11) strongly rejected the H0 hypothesis (homogeneity and no linkage) in favor of the H2 hypothesis (linkage heterogeneity) for every region analyzed. By contrasting the H1 hypothesis (linkage homogeneity) with H2, we found that H1 was rejected only at chromosome 17 (χ21=6.952; P<.008; α=0.16). Testing for the presence of any number (m) of trait loci (m=5 in the present study) showed that the posterior-probability assignment of each family to every region did correlate with the preliminary results. For example, family F8 showed a 0.993 probability of linkage to 11q22, family F9 showed a 0.995 probability of linkage to 4q13.2, and family F14 showed a 0.993 probability of linkage to 17p11. Haplotypes cosegregating with ADHD at these regions are shown in figure 5A–5C. It is interesting that the fraction of unlinked families was only 0.241 after the presence of these five loci was considered.
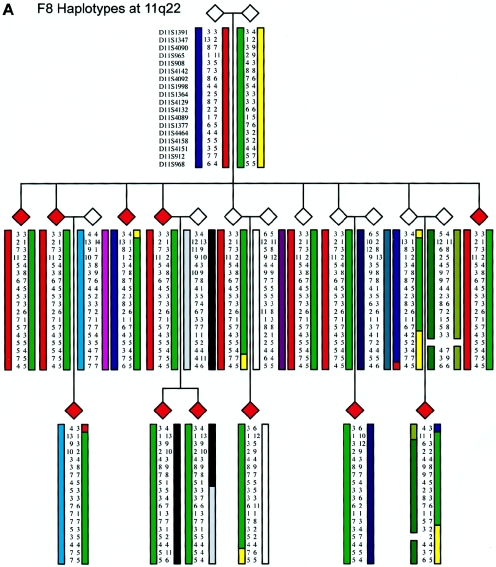
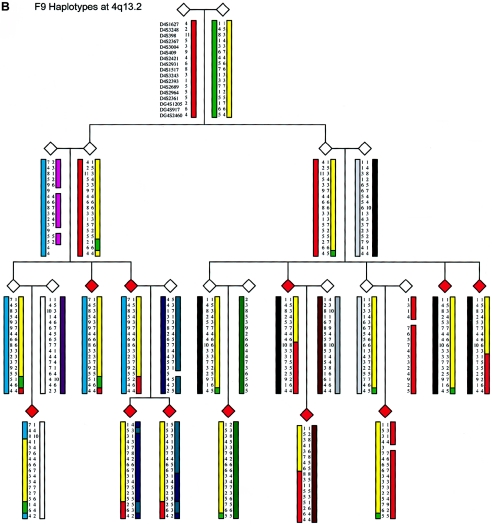
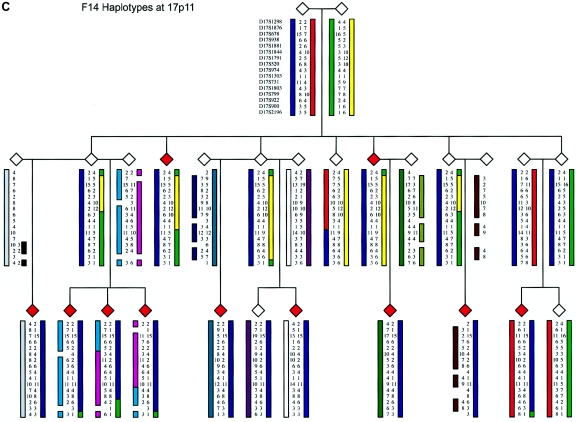
Figure 5.
Family structure haplotypes of three informative pedigrees that show those chromosomal regions segregating with ADHD at chromosomes 4q13.2, 11q22, and 17p11. Haplotypes were reconstructed using SIMWALK2, and they are shown for family F8 at 11q22, family F9 at 4q13.2, and family F14 at 17p11, with markers listed close to the top left symbol.
Discussion
We have studied ADHD in large multigenerational families from a genetic isolate and have identified linkage with a number of chromosomal regions. Several of these regions identified in our study are novel (4q13.2, 5q33.3, and 8q11.23) and have not been described in the previous genomewide scans (Fisher et al. 2002; Bakker et al. 2003; Ogdie et al. 2003). We found two regions (11q22 and 17p11) that overlapped with regions that were reported by previous ADHD genomewide scans to be suggestive of linkage, and these findings were replicated in our set of families, with significant linkage (Fisher et al. 2002; Bakker et al. 2003; Ogdie et al. 2003). It is noteworthy to emphasize that each of our significant regions exhibits a strong concordance of positive results, no matter which method of linkage analysis was used: model-based, model-free, or FBAT. Although multipoint linkage statistics are limited in use here, the sparse inheritance trees for pedigree analysis, as implemented in MERLIN (Abecasis et al. 2002), resulted in strongly significant multipoint NPLall Z scores, which were replicated by use of the conditional logistic model (Olson 1999) in a two-point NPL analysis without trimming individuals.
Although these results appear to be consistent among different analytical methods, specific differences and appropriate caveats related to each method need to be noted. The first difference is the strikingly significant results obtained for the combined set of families at chromosomes 4, 5, 11, and 17 by use of the conditional logistic model (Olson 1999) for ASPs (Risch 1990), as implemented in LODPAL. In contrast, only one significant result was obtained, for chromosome 17, by use of a parametric approach. Although it is well known that model-free linkage has an advantage in robustness when used to test complex traits, where the model of inheritance may vary throughout the sample of families (Kruglyak et al. 1996), we consider that the cause of this difference could lie in the fact that single-marker IBD-based statistics—instead of multipoint IBD-probability estimates—were calculated, a consequence of the restrictions generated by the size of the families. Single-marker statistics are highly variable, and the highest (among several) single-marker statistic in a region often greatly overestimates the linkage evidence (Elston et al. 2002). In fact, the most important benefit of multipoint analysis is an increase in the accuracy of the linkage statistic and of the estimate of trait-locus location by reduction of the statistic’s point-to-point variability (i.e., by smoothing the curve of the observed statistic along the chromosome) (Elston et al. 2002). However, the argument against the possibility of overestimation of linkage is supported by the fact that multipoint IBD probabilities were estimated using the sparse inheritance trees for pedigree analysis (in MERLIN), and equivalent values of significance were reported.
We also considered the possibility that the difference was obtained because an incorrect inheritance model was specified when we applied the parametrical approach. This effect has been well described elsewhere (Greenberg and Berger 1994, 1998; Durner et al. 1999). However, two previous linkage analyses of simulated and real data have considered different models of inheritance that are based on variations of dominance, penetrance, trait-allelic genetic frequencies, and marker-allelic frequencies. These detailed studies have produced no significant variations in the results (Arcos-Burgos et al. 2002, 2004). Additionally, this model was derived from the results disclosed by a complex segregation analysis of samples from patients with ADHD from the same isolated population (Lopera et al. 1999).
Additional caution is necessary when interpreting results of the model-free linkage approaches. Most of these model-free methods were developed under the assumption of independent affected pairs. In the case of the present study, affected relative pairs came from large families, and the statistics we computed are subject to correlations, despite the assumption of independence. However, several authors claim that the absence of independence does not increase the type I error rate, provided that IBD-sharing estimates are computed using all marker data in the pedigree.
As a consequence of the large number of markers used for scanning the genome, multiple analytical methods for contrasting the hypothesis of linkage with the analysis of the individual family data, with adjustment for multiple comparisons, must to be taken into account. We have estimated the empirical probability of the generation of a type I error by randomly simulating, by use of SIMLINK (Ploughman and Boehnke 1989), an unlinked marker in 10,000 replicates of the families and then determining the probability of obtaining LODs as high as the ones identified by our current analysis. It is noteworthy that the empiric probability of a LOD score or MLS that was >1.0, 2.0, or 3.0, for a true recombination fraction of 0.50, was 0.001, 0.0000, and 0.0000, respectively, when families were analyzed under homogeneity and heterogeneity models. Similar results were obtained for individual families F8, F9, and F14, which were the ones who accounted for the significant results. As described by Morton (1998), all but a small proportion of results significant at α=0.001 are true. This corresponds to a LOD score of 3.0 in a sequential test but to a LOD score of only 2.07 in large-sample theory. The procedure of ascertainment for this set of families has followed a strategy that is similar to sequential ascertainment, since the sample size (N) is varied, and the parameters—α, β, and effect size (D)—have remained fixed (Elston et al. 2002). (For example, the genomewide search was initiated once we demonstrated that only a portion of the data from 40 families that were originally estimated had a power of 80% [α=0.05] for the detection of a gene with a major effect by use of standard criteria for determining the presence of linkage [Arcos-Burgos et al. 2002]; we did increase the N of individuals belonging to the whole set of families to perform the fine mapping and to improve the attempt to narrow our minimal critical region.)
Furthermore, as described by Risch (1991), the posterior probability that a significant linkage finding is false (posterior false-positive rate [Φ]) increases with decreasing sample size. However, as demonstrated by Risch (1991), an N of 50 fully informative gametes is required to maintain Φ<5%, a value that is far exceeded in any of the individual families with ADHD who showed significant results of linkage under the homogeneity or the heterogeneity model. Thus far, for the linkage results concerning this set of families with ADHD—taking into account their empiric probability of generating a type I error for a LOD score of 2.0, the strategy of ascertainment, and the size of fully informative gametes—we consider that the guidelines by Lander and Kruglyak (1995) can be used in establishing the level of significance for this linkage study. We agree that, because of the observed linkage heterogeneity, one way to adjust the significance values would be to correct by the number of families involved in the analysis; therefore, applying a method such as the Bonferroni correction (critical P value/16) would provide a more conservative and accurate decision.
We are aware of the importance of replicating results of genomic regions that cosegregate with ADHD in different populations and that are ascertained under different schemes. Here, we demonstrate linkage in Paisa families to markers in regions on chromosomes 11q22, 17p11, and 20q13.33. These regions overlap with (1) the regions reported in a previous genomewide scan performed in ASPs with ADHD who were ascertained in the United States (Fisher et al. 2002; Ogdie et al. 2003, 2004); (2) nominal regions at chromosomes 3q13.33 and 9q33.3, which suggests an overlap with regions reported by the genomewide scan in the Dutch ASPs with ADHD (Bakker et al. 2003); and (3) the nominal region located at chromosome 5p13.3 that overlaps with both the U.S. and Dutch genomewide scans (Fisher et al. 2002; Bakker et al. 2003; Ogdie et al. 2003). The overlap of these regions among different studies is independent of the ascertainment conditions, analytical units (sib pairs, case-control, extended, and multigenerational pedigrees), population, and phenotype-definition methods, and it supports the contention that ADHD is a real biological entity amenable to the study of its underlying genetic influences.
Although all 16 pedigrees in our study were recruited from a population isolate with presumed reduced genetic heterogeneity, different pedigrees appear to be linked to different genomic regions. The test of heterogeneity demonstrated that, for the five densely analyzed regions, the alternative hypothesis of the presence of both linkage and heterogeneity was significantly better than the null hypothesis of absence of both linkage and homogeneity. Additionally, the hypothesis of linkage homogeneity was rejected in favor of the hypothesis of linkage heterogeneity when chromosome 17 was tested and probabilities of assignment of a family to each one of the analyzed regions exceeded 0.99 and were consistent with the homogeneity analysis.
More interesting, two pedigrees (F9 and F14) are linked to one region, 4q13.2. In some cases (e.g., families F8, F9, and F14), several regions cosegregate with ADHD in one family, suggesting that genetic heterogeneity and potential interaction between loci may underlie the susceptibility to develop ADHD. In fact, with the exception of 19p13.2, which has a heterogeneity α value of ∼20%, those regions exhibiting MLS peaks >1.0 all had an α value of ∼10%, which suggests that one family pointed to each region. This strong pattern of heterogeneity in ADHD is not unexpected for a condition that does not compromise reproductive fitness. The significant and suggestive linkage to different genomic regions in individual families (i.e., family F9: 4q13.2, 5q33.3, and 8q11.23; family F8: 11q22 and 8p23.1; and family F14: 4q13.2, 5q33.3, and 17p11) is compatible with the presence of epistasis (i.e., the interaction between different loci). We recognize that our results represent only indirect evidence, but epistasis is biologically plausible when one considers the size of the pedigrees and the fact that they came from an isolated population. However, we also recognize that some of these regions may represent false positives; therefore, we are currently performing an additional fine-mapping study that uses SNPs at a spacing of ∼200 kb in this set of families and in 100 additional nuclear families from the same isolated community. We believe that this study will provide more information about linkage in several of these interesting regions and that it may help detect the presence of interacting loci. Furthermore, we plan to combine this set of families with sib pairs from other communities to aid in the identification of ADHD susceptibility loci and to gain a better understanding of how epistasis of several genes may underlie the phenotypic spectrum of ADHD.
Acknowledgments
We are grateful to the Colombian families who participated in this research. We thank Dr. Maria Teresa Acosta, for bringing together the collaborators from Colombia and the United States, and Dr. R. Arlen Price, for critically reading this manuscript. This research was supported in part by COLCIENCIAS grant 1115-04-12010.
Electronic-Database Information
The URLs for data presented herein are as follows:
- ADHD Genetic Research Study, http://www.nhgri.nih.gov/10004331 (for additional data regarding results obtained by different methods of linkage)
- Center for Inherited Disease Research, http://www.cidr.jhmi.edu/
- deCODE, http://www.decode.com/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ADHD) [PubMed]
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 [DOI] [PubMed] [Google Scholar]
- Acosta MT, Arcos-Burgos M, Muenke M (2004) Attention deficit/hyperactivity disorder (ADHD): complex phenotype, simple genotype? Genet Med 6:1–15 [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders, 4th ed. American Psychiatric Association, Washington, DC [Google Scholar]
- Arcos-Burgos M, Castellanos FX, Konecki D, Lopera F, Pineda D, Palacio JD, Rapoport JL, Berg K, Bailey-Wilson J, Muenke M (2004) Pedigree disequilibrium test (PDT) replicates association and linkage between DRD4 and ADHD in multigenerational and extended pedigrees from a genetic isolate. Mol Psychiatry 9:252–259 [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Castellanos FX, Lopera F, Pineda D, Palacio JD, Garcia M, Henao GC, Palacio LG, Berg K, Bailey-Wilson JE, Muenke M (2002) Attention-deficit/hyperactivity disorder (ADHD): Feasibility of linkage analysis in a genetic isolate using extended and multigenerational pedigrees. Clin Genet 61:335–343 [DOI] [PubMed] [Google Scholar]
- Arcos-Burgos M, Muenke M (2002) Genetics of population isolates. Clin Genet 61:233–247 [DOI] [PubMed] [Google Scholar]
- Bakker SC, van der Meulen EM, Buitelaar JK, Sandkuijl LA, Pauls DL, Monsuur AJ, van ’t SR, Minderaa RB, Gunning WB, Pearson PL, Sinke RJ (2003) A whole-genome scan in 164 Dutch sib pairs with attention-deficit/hyperactivity disorder: suggestive evidence for linkage on chromosomes 7p and 15q. Am J Hum Genet 72:1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat A, Heath SC, Ott J (1999) Heterogeneity for multiple disease loci in linkage analysis. Hum Hered 49:229–231 [DOI] [PubMed] [Google Scholar]
- Biederman J, Newcorn J, Sprich S (1991) Comorbidity of attention deficit hyperactivity disorder with conduct, depressive, anxiety, and other disorders. Am J Psychiatry 148:564–577 [DOI] [PubMed] [Google Scholar]
- Bravo ML, Valenzuela CY, Arcos-Burgos OM (1996) Polymorphisms and phyletic relationships of the Paisa community from Antioquia (Colombia). Gene Geogr 10:11–17 [PubMed] [Google Scholar]
- Castellanos FX, Tannock R (2002) Neuroscience of attention-deficit/hyperactivity disorder: the search for endophenotypes. Nat Rev Neurosci 3:617–628 [DOI] [PubMed] [Google Scholar]
- Chang KKD, Steiner H, Ketter TA (2000) Psychiatric phenomenology of child and adolescent bipolar offspring. J Am Acad Child Adolesc Psychiatry 39:453–460 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schaffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- Durner M, Vieland VJ, Greenberg DA (1999) Further evidence for the increased power of LOD scores compared with nonparametric methods. Am J Hum Genet 64:281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston RC, Olson JM, Palmer L (eds) (2002) Biostatistical genetics and genetic epidemiology. John Wiley and Sons, Chichester, United Kingdom [Google Scholar]
- Faraone SV, Biederman J, Chen WJ, Krifcher B, Keenan K, Moore C, Sprich S, Tsuang MT (1992) Segregation analysis of attention deficit hyperactivity disorder. Psychiatr Genet 2:257–275 [Google Scholar]
- Faraone SV, Biederman J, Mennin D, Gershon J, Tsuang MT (1996) A prospective four-year follow-up study of children at risk for ADHD: psychiatric, neuropsychological, and psychosocial outcome. J Am Acad Child Adolesc Psychiatry 35:1449–1459 [DOI] [PubMed] [Google Scholar]
- Faraone SV, Biederman J, Monuteaux MC (2000) Toward guidelines for pedigree selection in genetic studies of attention deficit hyperactivity disorder. Genet Epidemiol 18:1–16 [DOI] [PubMed] [Google Scholar]
- ——— (2001) Attention deficit hyperactivity disorder with bipolar disorder in girls: further evidence for a familial subtype? J Affect Disord 64:19–26 [DOI] [PubMed] [Google Scholar]
- Fisher SE, Francks C, McCracken JT, McGough JJ, Marlow AJ, MacPhie IL, Newbury DF, Crawford LR, Palmer CG, Woodward JA, Del’Homme M, Cantwell DP, Nelson SF, Monaco AP, Smalley SL (2002) A genomewide scan for loci involved in attention-deficit/hyperactivity disorder. Am J Hum Genet 70:1183–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley HA, Carlton CO, Howell RJ (1996) The relationship of attention deficit hyperactivity disorder and conduct disorder to juvenile delinquency: legal implications. Bull Am Acad Psychiatry Law 24:333–345 [PubMed] [Google Scholar]
- Geller B, Zimerman B, Williams M, DelBello MP, Frazier J, Beringer L (2002) Phenomenology of prepubertal and early adolescent bipolar disorder: examples of elated mood, grandiose behaviors, decreased need for sleep, racing thoughts and hypersexuality. J Child Adolesc Psychopharmacol 12:3–9 [DOI] [PubMed] [Google Scholar]
- Giedd JN (2000) Bipolar disorder and attention-deficit/hyperactivity disorder in children and adolescents. J Clin Psychiatry 61:31–34 [PubMed] [Google Scholar]
- Gillis JJ, Gilger JW, Pennington BF, DeFries JC (1992) Attention deficit disorder in reading-disabled twins: evidence for a genetic etiology. J Abnorm Child Psychol 20:303–314 [DOI] [PubMed] [Google Scholar]
- Goddard KA, Witte JS, Suarez BK, Catalona WJ, Olson JM (2001) Model-free linkage analysis with covariates confirms linkage of prostate cancer to chromosomes 1 and 4. Am J Hum Genet 68:1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Abreu P, Hodge SE (1998) The power to detect linkage in complex disease by means of simple LOD-score analyses. Am J Hum Genet 63:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Berger B (1994) Using LOD-score differences to determine mode of inheritance: a simple, robust method even in the presence of heterogeneity and reduced penetrance. Am J Hum Genet 55:834–840 [PMC free article] [PubMed] [Google Scholar]
- Greenfield B, Hechtman L, Weiss G (1988) Two subgroups of hyperactives as adults: correlations of outcome. Can J Psychiatry 33:505–508 [DOI] [PubMed] [Google Scholar]
- Harada Y, Satoh Y, Sakuma A, Imai J, Tamaru T, Takahashi T, Amano N (2002a) Behavioral and developmental disorders among conduct disorder. Psychiatry Clin Neurosci 56:621–625 [DOI] [PubMed] [Google Scholar]
- Harada Y, Yamazaki T, Saitoh K (2002b) Psychosocial problems in attention-deficit hyperactivity disorder with oppositional defiant disorder. Psychiatry Clin Neurosci 56:365–369 [DOI] [PubMed] [Google Scholar]
- Jimenez I, Mora O, Lopez G, Jimenez ME, Zuluga L, Isaza R, Sanchez JL, Uribe CS, Valenzuela CY, Blanco R, Arcos-Burgos M (1996) Idiopathic epilepsy with generalized tonic clonic seizures in Antioquia, Colombia: is the joint Amerindian and Negroid racial admixture the cause of its high prevalence? Biol Res 29:297–304 [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, DeMeo DL, Laird NM (2002) Power and design considerations for a general class of family-based association tests: quantitative traits. Am J Hum Genet 71:1330–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Laird NM (2002a) On a general class of conditional tests for family-based association studies in genetics: the asymptotic distribution, the conditional power, and optimality considerations. Genet Epidemiol 23:165–180 [DOI] [PubMed] [Google Scholar]
- ——— (2002b) Power calculations for a general class of family-based association tests: dichotomous traits. Am J Hum Genet 71:575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckman JF, Sholomskas D, Thompson WD, Belanger A, Weissman MM (1982) Best estimate of lifetime psychiatric diagnosis: a methodological study. Arch Gen Psychiatry 39:879–883 [DOI] [PubMed] [Google Scholar]
- Levy F, Hay DA, McStephen M, Wood C, Waldman I (1997) Attention-deficit hyperactivity disorder: a category or a continuum? Genetic analysis of a large-scale twin study. J Am Acad Child Adolesc Psychiatry 36:737–744 [DOI] [PubMed] [Google Scholar]
- Lopera F, Palacio LG, Jimenez I, Villegas P, Puerta IC, Pineda D, Jimenez M, Arcos-Burgos M (1999) Genetic and environmental factors discrimination in attention deficit hyperactivity disorder. Rev Neurol 28:660–664 [PubMed] [Google Scholar]
- Lopera F, Rivera N, Arboleda J, Restrepo T, Arcos-Burgos M (2001) Analysis of complex segregation in a large family with hereditary cerebrovascular disease in Antioquia, Colombia. Rev Neurol 32:222–225 [PubMed] [Google Scholar]
- Maher BS, Marazita ML, Moss HB, Vanyukov MM (1999) Segregation analysis of attention deficit hyperactivity disorder. Am J Med Genet 88:71–78 [PubMed] [Google Scholar]
- Martin ER, Bass MP, Gilbert JR, Pericak-Vance MA, Hauser ER (2003) Genotype-based association test for general pedigrees: the genotype-PDT. Genet Epidemiol 25:203–213 [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL (2000) A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JR, Stewart MA (1973) The psychiatric status of the legal families of adopted hyperactive children. Arch Gen Psychiatry 28:888–891 [DOI] [PubMed] [Google Scholar]
- Morton NE (1998) Significance levels in complex inheritance. Am J Hum Genet 62:690–697 (erratum 63:1252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogdie MN, Fisher SE, Yang M, Ishii J, Francks C, Loo SK, Cantor RM, McCracken JT, McGough JJ, Smalley SL, Nelson SF (2004) Attention deficit hyperactivity disorder: fine mapping supports linkage to 5p13, 6q12, 16p13, and 17p11. Am J Hum Genet 75:661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogdie MN, MacPhie IL, Minassian SL, Yang M, Fisher SE, Francks C, Cantor RM, McCracken JT, McGough JJ, Nelson SF, Monaco AP, Smalley SL (2003) A genomewide scan for attention-deficit/hyperactivity disorder in an extended sample: suggestive linkage on 17p11. Am J Hum Genet 72:1268–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JM (1999) A general conditional-logistic model for affected-relative-pair linkage studies. Am J Hum Genet 65:1760–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J (1983) Linkage analysis and family classification under heterogeneity. Ann Hum Genet 47:311–320 [DOI] [PubMed] [Google Scholar]
- Ott J, Bhat A (1999) Linkage analysis in heterogeneous and complex traits. Eur Child Adolesc Psychiatry 8 Suppl 3:43–46 [DOI] [PubMed] [Google Scholar]
- Pineda D, Ardila A, Rosselli M, Arias BE, Henao GC, Gomez LF, Mejia SE, Miranda ML (1999) Prevalence of attention-deficit/hyperactivity disorder symptoms in 4- to 17-year-old children in the general population. J Abnorm Child Psychol 27:455–462 [DOI] [PubMed] [Google Scholar]
- Ploughman LM, Boehnke M (1989) Estimating the power of a proposed linkage study for a complex genetic trait. Am J Hum Genet 44:543–551 [PMC free article] [PubMed] [Google Scholar]
- Reich W (2000) Diagnostic interview for children and adolescents (DICA). J Am Acad Child Adolesc Psychiatry 39:59–66 [DOI] [PubMed] [Google Scholar]
- Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- ——— (1991) A note on multiple testing procedures in linkage analysis. Am J Hum Genet 48:1058–1064 [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- ——— (1997) Genetic analysis of complex diseases. Science 275:1329–1330 [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Tacchini G, Coppola MT, Musazzi A, Altamura AC, Invernizzi G (1994) Multinational validation of the Composite International Diagnostic Interview (CIDI). Minerva Psichiatr 35:63–80 [PubMed] [Google Scholar]
- van den Oord EJ, Boomsma DI, Verhulst FC (1994) A study of problem behaviors in 10- to 15-year-old biologically related and unrelated international adoptees. Behav Genet 24:193–205 [DOI] [PubMed] [Google Scholar]