Abstract
We have previously identified in the pol gene of human immunodeficiency virus type 1 (HIV-1) a new positive transcriptional regulatory element (nt 4481–4982) containing recognition sites for nuclear proteins (sites B, C, D and a GC-box) [C. Van Lint, J. Ghysdael, P. Paras, Jr, A. Burny and E. Verdin (1994) J. Virol. 68, 2632–2648]. In this study, we have further physically characterized each binding site and have shown that the transcription factors Oct-1, Oct-2, PU.1, Sp1 and Sp3 interact in vitro with the pol region. Chromatin immunoprecipitation assays using HIV-infected cell lines demonstrated in the context of chromatin that Sp1, Sp3, Oct-1 and PU.1 are recruited to the HS7 region in vivo. For each site, we have identified mutations abolishing factor binding to their cognate DNA sequences without altering the underlying amino acid sequence of the integrase. By transient transfection assays, we have demonstrated the involvement of the pol binding sites in the transcriptional enhancing activity of the intragenic region. Our functional results with multimerized wild-type and mutated pol binding sites separately (i.e. in the absence of the other sites) have demonstrated that the PU.1, Sp1, Sp3 and Oct-1 transcription factors regulate the transcriptional activity of a heterologous promoter through their respective HS7 binding sites. Finally, we have investigated the physiological role of the HS7 binding sites in HIV-1 replication and have shown that these sites are important for viral infectivity.
INTRODUCTION
The expression of human immunodeficiency virus type 1 (HIV-1), the etiological agent of AIDS (1–3), is regulated both at the transcriptional and the post-transcriptional levels (4). Control of HIV-1 transcription is mediated by cis-acting elements located in the viral 5′-long terminal repeats (5′-LTRs), by the viral trans-regulatory protein Tat and by cellular transcription factors. In addition to the enhancer located in the 5′-LTR, a 12-O-tetradecanoylphorbol-13-acetate (TPA)-inducible intragenic enhancer has been identified in the pol gene of HIV-1 (5). This element is composed of two functional subdomains encompassing nt 4079–4342 and nt 4781–6026, both exhibiting TPA-inducible enhancing activity, on the herpes simplex virus (HSV) thymidine kinase (TK) promoter in HeLa cells (5), but no significant activity in T-lymphoid and monocyte/macrophage cell lines. Nevertheless, analysis of the chromatin organization of integrated HIV-1 identified a single major nuclease-hypersensitive site in the 8 kb region located between the two LTRs (6,7). This hypersensitive site, centered around nt 4490–4766 [according to the numbering of the HIV-1 NY5 genome (where nt +1 is the start of U3 in the 5′-LTR)], is located in the part of the pol gene encoding the integrase protein, precisely between the two functional domains of the intragenic enhancer identified in HeLa cells (7). This constitutive hypersensitive site is present only in a cell line of monocytic origin (U1) and not in two cell lines of lymphoid origin (8E5 and ACH2), suggesting a cellular specificity associated with this intragenic element (6). A 500 bp fragment (nt 4481–4982) encompassing the pol gene hypersensitive site region (called HS7 region) acts ex vivo as a cis-acting positive regulatory element. Indeed, when cloned downstream and in the sense orientation relative to the HIV-1 promoter, the HS7 region reproducibly increases transcription mediated by the HIV-1 5′-LTR (7).
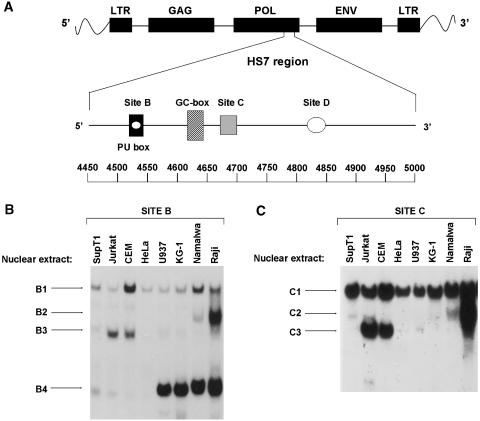
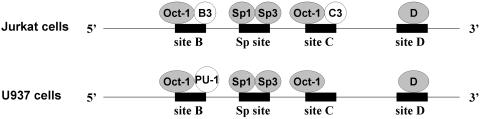
Physical characterization of the HS7 region identified several recognition sites for ubiquitous and cell-type-specific nuclear factors, suggesting their involvement in the control of the transcriptional activity of this region (7). By in vitro binding studies of the HS7 region, four distinct DNA-binding motifs for nuclear proteins (called from 5′ to 3′ site B, GC-box, site C and site D) were physically defined (Figure 1A). Site B (nt 4519–4545) specifically binds four distinct nuclear protein complexes: an ubiquitous factor (B1), a B-cell specific factor (B2), a T-cell specific factor (B3) and a protein(s) related to the transcription factor PU.1 (B4) (Figure 1B). The GC-box (nt 4623–4631) binds purified Sp1 protein in vitro. Site C (nt 4681–4701) specifically binds three nuclear protein complexes: an ubiquitous factor (C1), a B-cell specific factor (C2) and a T-cell specific factor (C3) (Figure 1C). These factors have a DNA-binding specificity similar to that of factors binding to site B (B1, B2 and B3, respectively). Site D (nt 4816–4851) specifically binds an ubiquitously expressed factor(s) (7). However, the physiological role of these HS7 binding sites in HIV-1 transcription and replication remains unknown to date.
Figure 1.
EMSA analysis of nuclear factors binding to site B and to site C. (A) Summary of protein binding sites within the pol regulatory region of HIV-1. (B). The site B oligonucleotide (5′-CAGCATACTTCCTCTTAAAATTAGCAG-3′) was labeled, used as probe and incubated with nuclear extracts from human cell lines of different origins (indicated above the lanes). Retarded DNA–protein complexes (B1, B2, B3 and B4) are indicated by arrows. Adapted with modification from Van Lint et al. (7) with permission. (C) The site C oligonucleotide (5′-TAGAATCTATGAATAAAGAAT-3′) was used as probe and incubated with nuclear extracts from human cell lines of different origins (indicated above the lanes). Retarded DNA–protein complexes (C1, C2 and C3) are indicated by arrows. Taken with modification from Van Lint et al. (7) with permission.
In this study, we have further characterized physically each of these four binding sites (sites B, C, D and the GC-box) located in the HS7 region, examined the functional transcriptional role of each site separately in a heterologous context, and investigated their biological significance in the HIV-1 replication cycle.
MATERIALS AND METHODS
Cell culture
The monocytic cell lines U937 and U1 and the T-lymphoid cell lines JE6-1 (a clonal line of Jurkat cells), A3.01 (a clonal derivative of the CEM cell line), the clonal Jurkat JE6-1 cell lines stably expressing HIV-1 Tat proteins (Tat72 and Tat101) or the empty vector cassette (8), and ACH2 were maintained in RPMI 1640-Glutamax I medium (Invitrogen) supplemented with 10% fetal bovine serum (Myoclone Superplus). The HIV-1-infected U1 and ACH2 cell lines were obtained from AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). The adherent cell line NIH3T3 (a murine fibroblastic cell line) and 293T cells (human embryonic kidney cell line) were cultured in Dulbecco's modified Eagle's-Glutamax I medium containing 10% calf serum. The cell line SL2 (a Drosophila cell line) was cultured in Schneider's Drosophila medium supplemented with l-glutamine and with 10% fetal bovine serum. All media (Invitrogen) also contained 50 U of penicillin/ml and 50 μg of streptomycin/ml (Invitrogen). All cells were grown at 37°C in a 5% CO2 atmosphere, except for SL2 cells which were grown at 28°C without CO2.
Plasmid constructs
An SmaI–XhoI fragment containing the HIV-1 LAI 5′-LTR (nt 1–789) was purified from the previously described pLTR(1-789)-luc construct (9). This fragment was cloned into the unique EcoICRI–XhoI sites of the reporter vector pGL3-Basic (Promega). The resulting plasmid was designated pLTR.
PCR was used to amplify the pol gene fragment (nt 4481–4982) corresponding to the HS7 region from the infectious proviral molecular clone pNL4-3 (reagent no. 114, received from the AIDS Research and Reference Reagent Program, NIAID, NIH). BamHI sites were introduced in the PCR primers, and the BamHI-restricted PCR fragment was cloned in the unique BamHI site of the pLTR, placing the fragment downstream of the 5′-LTR-luc transcriptional unit. The 5′ primer oligonucleotide encompassed the coding strand sequence from nt 4481 to 4505 and contained an added BamHI restriction site (underlined) at the 5′ end (5′-TCCCCCGGGATCC[nt 4481]GAAGCAGAAGTAATTCCAGCAGAG-3′). The 3′ primer oligonucleotide encompassed the complementary sequence of the pol gene from nt 4957 to 4982, and contained an added BamHI site (underlined) at the 5′ end (5′-TCCCCCGGGATCC[nt 4982]TATTACTACTGCCCCTTCACCTTTCC-3′). This plasmid was designated pLTR-HS7wt. Fragments containing the HS7 region mutated in the different binding sites (sites B, C, D and the GC-box) individually or in combination were PCR amplified from the pCV11, pCV14, pCV16, pCV19, pCV25, pCV1069 and pCV1107 plasmids (see below). The 5′ and 3′ primer oligonucleotides were as described above. The different amplified fragments were digested with BamHI and then ligated into the BamHI site of pLTR to generate pLTR-HS7totmut, pLTR-HS7mutB/Oct, pLTR-HS7mutB/PU, pLTR-HS7mutC, pLTR-HS7mutSp, pLTR-HS7mutD and pLTR-HS7mutCmutB/Oct, respectively.
The pTK reporter plasmid contains the HSV TK minimal promoter and was described previously (10).
The p(Cwt)3TK was generated by inserting, upstream of the TK promoter, a double-stranded oligonucleotide corresponding to three direct repeats of the site C sequence (5′-TAGAATCTATGAATAAAGAAT-3′) in the forward orientation into SmaI-digested pTK-luc. Similarly, the p(CmutOct)3TK was generated by inserting, upstream of the TK promoter, a double-stranded oligonucleotide corresponding to three direct repeats of site C mutated in the Oct motif with the sequence 5′-TAGAATCCATGAACAAAGAAT-3′ (the mutations are underlined) in the forward orientation into SmaI-digested pTK.
The p(Bwt)3TK was generated by inserting, upstream of the TK promoter, a double-stranded oligonucleotide corresponding to three direct repeats of the site B sequence (5′-CAGCATACTTCCTCTTAAAATTAGCAG-3′) in the forward orientation into SmaI-digested pTK. Similarly, the p(BmutOct)3TK and p(BmutPU.1)3TK were generated by inserting, upstream of the TK promoter, a double-stranded oligonucleotide corresponding to three direct repeats either of site B mutated in the Oct motif with the sequence 5′-CAGCATACTTCCTCTTGAAGTTGGCAG-3′, or of site B mutated in the PU box with the sequence 5′-CAGCATATTTTCTCTTAAAATTAGCAG-3′ (the mutations are underlined) in the forward orientation into SmaI-digested pTK.
The p(Spwt)3TK was generated by inserting, upstream of the TK promoter, a double-stranded oligonucleotide corresponding to three direct repeats of the Sp site sequence (5′-CCTGTTGGTGGGCGGGGATCAAG-3′) in the forward orientation into SmaI-digested pTK. Similarly, the p(Spmut)3TK was generated by inserting, upstream of the TK promoter, a double-stranded oligonucleotide corresponding to three direct repeats of the Sp site mutated in the GC-box with the sequence 5′-CCTGTTGGTGGGCAGGAATCAAG-3′ (the mutations are underlined) in the forward orientation into SmaI-digested pTK-luc.
The Oct-1 and Oct-2 expression vectors (pCG-Oct-1 and pCG-Oct-2) (kindly provided by Dr Winship Herr) contained the human Oct-1 and Oct-2 cDNAs cloned in the pCG parent plasmid and were described previously (11). The PU.1 expression vector pJ6-PU.1 was described previously (12). The Sp1 and Sp3 expression vectors (pPacSp1 and pPacSp3) (kindly provided by Dr Guntram Suske) contained the human Sp1 and Sp3 cDNAs cloned in the pPac parent plasmid and were described previously (13).
Electrophoretic mobility shift assays
Nuclear extracts were prepared by a rapid method described by Osborn et al. (14). All buffers contained the protease inhibitors antipain (10 μg/ml), aprotinin (2 μg/ml), chymostatin (10 μg/ml), leupeptin (1 μg/ml) and pepstatin (1 μg/ml). Protein concentrations were determined by the method of Bradford (15). The DNA sequences of the coding strand of the double-stranded oligonucleotides used for this study are listed in Figure 9 or in the figure legends. Electrophoretic mobility shift assays (EMSAs) were performed as described previously (7). Briefly, nuclear extract (15 μg of protein) was first incubated on ice for 10 min in the absence of probe and specific competitor DNA in a 16 μl reaction mixture containing 10 μg of DNase-free BSA (Amersham Biosciences), 2 μg of poly(dI–dC) (Amersham Biosciences) as non-specific competitor DNA, 50 μM ZnCl2, 0.25 mM DTT, 20 mM HEPES (pH 7.3), 60 mM KCl, 1 mM MgCl2, 0.1 mM EDTA and 10% (v/v) glycerol. 20 000 c.p.m. of probe (10–40 fmol) was then added to the mixture with or without a molar excess of an unlabeled specific DNA competitor, and the mixture was incubated for 20 min on ice. Samples were subjected to electrophoresis at room temperature on 6% polyacrylamide gels at 150 V for 2–3 h in 1× TGE buffer (25 mM Tris–acetate (pH 8.3), 190 mM glycine and 1 mM EDTA). Gels were dried and autoradiographed for 24–48 h at −70°C. For supershift assays, polyclonal antibodies against Oct-1 (sc-232X), Oct-2 (sc-233X), Sp1 (sc-059X), Sp2 (sc-643X), Sp3 (sc-644X), Sp4 (sc-645X), MEF-2 (sc-10794X) (Santa Cruz Biotechnology), PU.1 (16), MEF2A, -B or -D (kindly provided by Dr Ron Prywes) (17), anti-MEF2C (kindly provided by Dr John Schwarz) (18), or a purified rabbit immunoglobulin (IgG) were added to the reaction mixture and incubated for 30 min on ice before the addition of the radiolabeled probe.
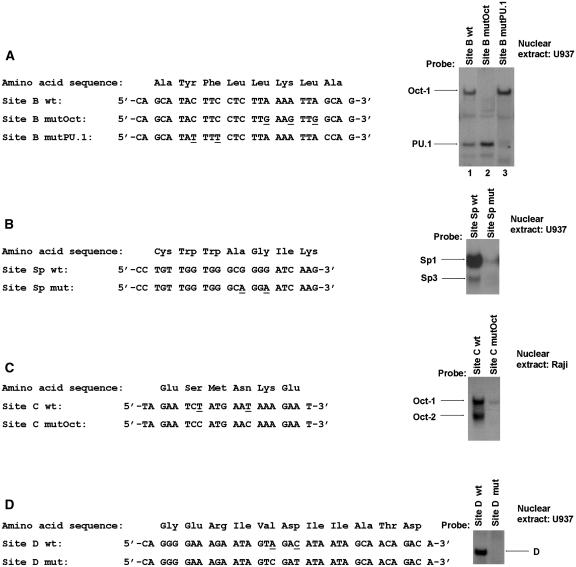
Figure 9.
Mutagenesis of the HS7 binding sites. Left panels: The wild-type and mutated oligonucleotides corresponding to the HS7 site B (A), Sp site (B), site C (C) and site D (D) are shown. The amino acids encoded by these oligonucleotide sequences are indicated. Bases that are changed in the mutated versions of the HS7 binding sites relative to the wild-type version are underlined. Right panels: Probes (indicated at the top of each lane) were incubated with 15 μg of nuclear extracts from U937 (A, B and D) or Raji (C) cells. The figure shows only the specific retarded bands of interest. The retarded complexes corresponding to Oct-1, Oct-2, PU.1, Sp1, Sp3 and D are indicated by arrows.
For analysis of PU.1 binding, whole-cell extracts of Spodoptera frugiperda SF9 cells infected with a recombinant Ac-NPV-PU.1 baculovirus were used as a source for PU.1 protein and extracts from uninfected insect cells were used as a control. Conditions for EMSAs were as described previously (19).
Chromatin immunoprecipitation assays
The Chromatin immunoprecipitation (ChIP) assays were performed by using the ChIP assay kit (Upstate Biotechnology) according to the manufacturer's recommendations. Formaldehyde cross-linking reactions from 5 × 106 HIV-infected U1 cells or ACH2 cells were quenched with 125 mM glycine. Cells were lysed, and chromatin was sonicated to obtain an average DNA length of 500 bp. Following centrifugation, the chromatin was diluted 10-fold and precleared with a protein A–agarose slurry containing salmon sperm DNA and BSA (Upstate Biotechnology). Precleared chromatin was incubated overnight at 4°C with no antibody or with 5 μg of either anti-Oct-1 antibody (sc-232X), anti-Sp1 (sc-59) antibody, anti-Sp3 (sc-644) antibody, anti-PU.1 antibody (sc-352) or normal rabbit IgG control antibody (Upstate Biotechnology, ref. no. 12-370) as control, followed by immunoprecipitation with protein A–agarose. Immunoprecipitated complexes were washed and eluted twice with 200 μl of elution buffer. The protein–DNA cross links were reversed by heating at 65°C overnight, and 10% of the recovered DNA was used for PCR amplification (35 cycles) with a primer set amplifying the HS7 region (nt 4497–4769): 5′-CCAGCAGAGACAGGGCAAGAA-3′/5′-ACTGCCATTTGTACTGCTGTCTT-3′ or with an unrelated primer set amplifying a pol gene region located ∼2 kb upstream of the HS7 region (nt 2326–2573): 5′-TACAGGAGCAGATGATACAG-3′/5′-CCTGGCTTTAATTTTACTGG-3′. PCR products from all reactions were resolved by agarose gel electrophoresis.
Transient transfection and luciferase reporter assays
NIH3T3 and SL2 cells were transfected using FuGENE™-6 (Roche Molecular Biochemicals) according to the manufacturer's protocol. Twenty-four hours before transfection, cells were seeded at a density of 2 × 105 cells/well in 6-well plates. FuGENE™-6 was added directly to serum-free medium 5 min before addition to the DNA. An aliquot of 100 μl of this FuGENE™-6/serum-free medium mixture was added to the DNA mixture in microcentrifuge tubes. The mixture was incubated for 15 min at room temperature and, finally added to each well of a 6-well plate. Transfected cells were grown in 2 ml of supplemented medium for 40–44 h. Cells were then lysed and assayed for luciferase activity (Promega). Luciferase activities derived from TK promoters were normalized with respect to protein concentrations using the Detergent-Compatible Protein Assay (Bio-Rad).
Jurkat and U937 cells were transfected using jetPEI™ (POLYplus) according to the manufacturer's protocol. Briefly, on the day of transfection, cells were harvested at a density of 1 × 106/ml, resuspended in fresh complete medium and seeded at a density of 0.5 × 106 cells/well in 12-well plates. For each well, 1 μg of DNA was diluted into 50 μl of 150 mM NaCl and 4 μl of jetPEI™ were diluted into 50 μl of 150 mM NaCl. Next, 50 μl of this jetPEI™ solution were added to the 50 μl DNA solution. The 100 μl jetPEI™/DNA mixture was then incubated for 15 min at room temperature and, finally added dropwise to each well. Transfected cells were grown in 1.5 ml for 40–44 h, lysed and assayed for luciferase activity (Promega). All DNA solutions also contained 50 ng of pRL-TK (used as an internal control for transfection efficiency) in which a cDNA encoding Renilla luciferase is under the control of the HSV TK promoter region (Promega). Firefly luciferase activities derived from the HIV-1 LTR were normalized with respect to the Renilla luciferase activities by using the dual-luciferase reporter assay system (Promega).
Site-directed mutagenesis of the HS7 binding sites
An ApaI/EcoRI fragment containing nt 2011–5743 of the HIV-1 genome was obtained after digestion with ApaI and EcoRI of pNL4-3. This fragment was cloned in pBluescript II SK (Stratagene) digested with ApaI and EcoRI to generate pCV10. This plasmid was used as a substrate for mutagenesis of the HS7 binding sites by the transformer site-directed mutagenesis method (Clontech). The HS7 region was mutated in all the HS7 sites (site B, Sp site, site C and site D) with the following five mutagenic oligonucleotides (mutations are highlighted in boldface and underlined): CV1 (siteBmutPU.1), 5′-CCTGCTAATTTTAAGAGAAAATATGCTGTTTCTTGCC-3′; CV3 (siteBmutOct), 5′-CCTGCCAACTTCAAGAGGAAGTATGCTGTTTCTTGCC-3′; CV4 (siteBmutOct/PU.1), 5′-CCTGCCAACTTCAAGAGAAAATATGCTGTTTCTTGCC-3′; CV6 (siteCmutOct), 5′-CTTTAATTCTTTGTTCATGGATTCTATTACTCCTTGACTTTG-3′; CV7 (siteSpmut), 5′-CCTGCTTGATTCCTGCCCACCAAC-3′; and CV14 (siteDmut), 5′-GTATGTCTGTTGCTATTATATCGACTATTCTTTCCCCTGC-3′.
In addition, three individual mutations of the HS7 site B PU box, site C or Sp site alone and one double mutation of the octamer motif in both site B and site C were generated with the CV1, CV6 or CV7 and CV6/CV3, respectively. The oligonucleotide 5′-CTTTTGCTCCCATGGTCTTTCCTG-3′, changing a unique AflIII restriction site in pCV10 into a NcoI site (highlighted in boldface) and the reverse oligonucleotide 5′-CTTTTGCTCACATGTTCTTTCCTG-3′, changing the unique NcoI restriction site in a mutated pCV10 into a AflIII site (highlighted in boldface) were successively used for selection during mutagenesis. The five mutant resulting pCV10-derivative plasmids were designated as pCV14 (HS7totmut), pCV11 (HS7mutB/PU), pCV25 (HS7mutC), pCV19 (HS7mutSp) and pCV16 (HS7mutCmutB/Oct), respectively. In addition, two other individual mutations were generated by the QuikChange Site-Directed Mutagenesis Kit (Stratagene) using the pCV10 construct as a substrate with the following two pairs of mutagenic oligonucleotide primers (mutations are highlighted in boldface and underlined on the coding strand primer): CV1141-CV1142 (siteBmutOct), 5′-AGCATACTTCCTCTTGAAGTTGGCAGGAAGATGGCCA-3′ and CV1147-CV1148 (siteDmut), 5′-CAGGGGAAAGAATAGTCGATATAATAGCAACAGAC-3′.
The resulting pCV10-derivative plasmids were designated as pCV1107 (HS7mutB/Oct) and pCV1069 (HS7mutD), respectively.
The mutated clones containing the HS7 binding sites mutated individually or in combination were fully resequenced between ApaI and EcoRI after identification (Applied Biosystems).
Construction of infectious proviruses containing the HS7 mutations
To eliminate the pUC8 MCS EcoRI site (nt 420) and to keep only the pol gene EcoRI site (nt 5743), the HIV-1 circularly permuted single-LTR-containing infectious molecular clone pEV46 (pHIV, a derivative of pILIC) (20) was partially digested with EcoRI, dephosphorylated and gel purified. The resulting DNA fragment was next ligated to a phosphorylated partially double-stranded oligonucleotide. The coding and non-coding sequences of this oligonucleotide were as follows: CV20, 5′-AATTAGTGGACGTCAC-3′ and CV21, 5′-AATTGTGACGTCCACT-3′, respectively. The resulting plasmid was designated pCV1. The ApaI/EcoRI mutagenized fragment from the pCV14 was introduced into the unique ApaI–EcoRI sites of pCV1 to generate pCV426 (named pHIV-1*-HS7totmut). As a control, an unmutated ApaI–EcoRI fragment was purified from pCV10 and cloned into the unique ApaI–EcoRI sites of pCV1. We refer to this construct as pCV422 (named pHIV-1*).
Another set of mutated proviral infectious clones were also constructed. To that end, the ApaI/EcoRI mutagenized fragments from pCV14, pCV1107, pCV11, pCV25, pCV19, pCV1069 and pCV16 were introduced into the unique ApaI–EcoRI sites of the two LTRs containing proviral clone pNL4.3 to generate pCV1102 (named pHIV-1-HS7totmut), pCV1106 (named pHIV-1-HS7mutB/Oct), pCV1101 (named pHIV-1-HS7mutB/PU), pCV1105 (pHIV-1-HS7mutC), pCV1104 (named pHIV-1-HS7mutSp), pCV1082 (named pHIV-1-HS7mutD) and pCV1103 (pHIV-1-HS7mutCmutB/Oct), respectively. As a control, an unmutated ApaI–EcoRI fragment was purified from pCV10 and cloned in an identical manner into the unique ApaI–EcoRI sites of pNL4.3. We refer to this construct as pHIV-1.
Generation of viral stocks
Wild-type and mutant HIV-1 infectious DNAs were generated from the single-LTR-containing proviral constructs pCV422 and pCV426, as described above, by BamHI digestion and self-ligation. These concatemerized proviral DNAs (10 μg) were transfected into 107 Jurkat cells using the DEAE–dextran procedure. At 24 h post-transfection, the cultures were cocultivated with 107 SupT1 cells to allow rapid and efficient recovery of progeny virus. HIV-1 wild-type (HIV-1*) and mutant (HIV-1*-HS7totmut) stocks were prepared at the peak of viral production from supernatants after filtration through a 0.45 μm pore-size membrane. The full-length molecular clones pHIV-1, pHIV-1-HS7totmut, pHIV-1-HS7mutB/Oct, pHIV-1-HS7mutB/PU, pHIV-1-HS7mutC, pHIV-1-HS7mutSp, pHIV-1-HS7mutD and pHIV-1-HS7mutCmutB/Oct described above were used to generate the other stocks of wild-type and mutant viruses (HIV-1, HIV-1-HS7totmut, HIV-1-HS7mutB/Oct, HIV-1-HS7mutB/PU, HIV-1-HS7mutC, HIV-1-HS7mutSp, HIV-1-HS7mutD and HIV-1-HS7mutCmutB/Oct). These DNAs (3 μg) were transfected into 293T cells by Lipofectamine 2000 (Invitrogen). At 48 h post-transfection, HIV-1 stocks were prepared from supernatants after filtration through a 0.45 μm pore-size membrane and stored at [−80°C. All viral stocks were quantified by determining p24 antigen concentration by an enzyme-linked immunosorbent assay (ELISA) (Innogenetics).
Viral infections
Infections were performed by incubating 0.5 × 106 cells with 50 ng of p24 protein of wild-type or mutant viruses (at 37°C for 2 h in 500 μl of culture medium). After infection, the cells were pelleted at 300 g, washed three times with 1 ml of culture medium, resuspended in 1 ml of standard medium, and grown under standard conditions. Every 2 or 3 days, aliquots of 200 μl were removed from the infected cultures and replaced by normal medium. The aliquots were assayed for p24 concentration in order to monitor the kinetics of viral replication.
Sequence analysis of HIV-1 genomic RNA
Viral particles from each stock were pelleted by ultracentrifugation (250 ng of p24 at 20 000 g for 2 h at 4°C) and digested with RNase-free DNase I (60 U/ml for 10 min at 4°C; Roche) in the presence of RNasin (40 U/ml; Promega) to remove contaminating DNA. HIV-1 genomic RNA was purified by using Trizol Ls reagent (Invitrogen). cDNA synthesis was performed by the Titan One Tube RT–PCR kit method (Roche Molecular Biochemicals). cDNAs were amplified by PCR with a 5′ oligonucleotide primer corresponding to nt 4342–4364 (5′-CCAGCTGTGATAAATGTCAGCT-3′) and a 3′ primer corresponding to nt 4982–4961 (5′-TATTACTACTGCCCCTTCACC-3′). PCR fragments were subcloned into the vector pCR4 Blunt-TOPO (Zero Blunt TOPO PCR Cloning Kit; Invitrogen). After identification of recombinant clones, three inserts from each construct were sequenced with the BigDye terminator sequencing kit (Applied Biosystems). The nucleotide sequences of all three clones were identical and confirmed the presence of the original mutations.
RNase protection assays
HIV-1 genomic RNAs were detected by RNase protection analysis. An HIV-1-specific 32P-labeled antisense riboprobe was synthesized in vitro by transcription of pGEM23 (a gift from M. Laspia (21) with SP6 polymerase. For HIV-1 genomic RNA analysis, equivalent amounts of viral particles from each stock (6 μg of p24) were pelleted by ultracentrifugation (at 20 000 g for 2 h at 4°C) and viral RNAs were prepared using Trizol Ls reagent (Invitrogen). The RNase protection assays were performed with the RPA II kit (Ambion) according to the manufacturer's recommendations by using the HIV-1 riboprobe as described above. The protected RNA fragments were analyzed by electrophoresis through 6% urea polyacrylamide gels.
Western immunoblot analysis
HIV-1 lysates were prepared by ultracentrifugation of each virus stock and the pellets were resuspended in Laemmli buffer at a concentration of 37.5 ng of p24/μl. Lysates were heated at 95°C for 5 min, separated by electrophoresis on an 8% polyacrylamide gel, transferred onto a polyvinylene difluoride membrane, and probed with a 1:2000 dilution of purified human anti-HIV-1 IgG (NIH AIDS Research and Reagent Program, reagent no. 192 donated by Alfred Prince). A second antibody, horseradish peroxidase-conjugated goat anti-human IgG (Pierce) (diluted 1:3300) was used for enhanced chemiluminescence detection (Cell Signaling).
RESULTS
Localization of the DNA sequences required for factor binding to the HS7 sites B and C by substitution mutant analysis
In order to identify the nuclear factors leading to the formation of complexes B1/C1, B2/C2 and B3/C3, we performed an in silico search (using the TESS program) for potential transcription factor binding sites in the HS7 sites B and C. The numerous potential sites revealed by this in silico analysis prompted us to perform a systematic substitution-scanning mutational analysis of sites B and C in order to localize by in vitro studies the nucleotides important in these sites for formation of the retarded complexes B1/C1, B2/C2 and B3/C3.
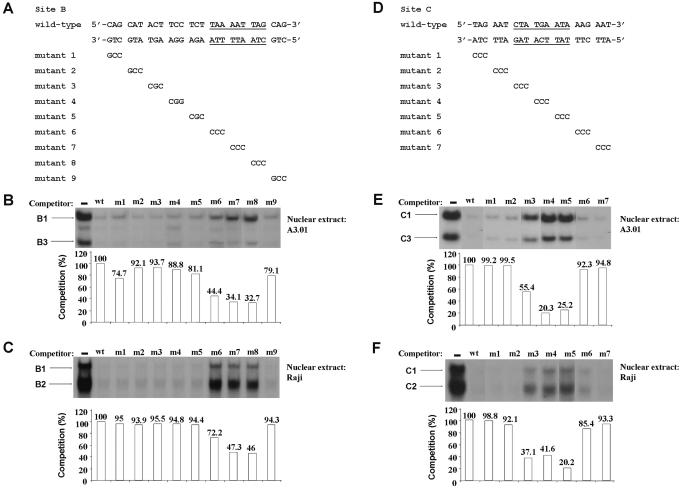
Nine mutant site B oligonucleotides (named m1–m9) were prepared in which consecutive 3 bp regions of the wild-type site B were sequentially replaced with a 3 bp C/G-C/G-C/G sequence (Figure 2A). To examine the effects of these mutations on specific factor binding to site B, we have systematically analyzed the ability of the scanning mutants to inhibit the formation of complexes B1, B2 and B3 in competition EMSAs. If mutations are introduced at critical positions, then a decrease in the ability of the unlabeled mutant oligonucleotide to compete for factor binding will be detected by the increase in band intensity as compared to the competition with the wild-type site B. Mutations at non-essential bases are expected to demonstrate binding competition similar to the wild-type. The competition assays were performed using site B wild-type as probe, nuclear extracts from the A3.01 cell line and unlabeled wild-type and substitution mutant site B double-stranded oligonucleotides as competitors (Figure 2B). Phosphorimaging analyses were used to quantify the percentage of competition for formation of complexes B1 and B3 by each of the unlabeled oligonucleotides. The competition activities of mutant sites B m1–m9 were expressed relative to the competition activity of the wild-type site B, which was arbitrarily assigned a value of 100% of competition. Figure 2B shows a graphic representation of the quantification results for complex B1. Site B mutants m6, m7 and m8 exhibited the lowest percentages of competition (Figure 2B, 44.4, 34.1 and 32.7%, respectively), indicating that the base pairs which were substituted in these three double-stranded oligonucleotides, were the most important for complex B1 formation. Quantification of the bands corresponding to complex B3 indicated the importance of the same base pair (data not shown). Similar results were observed for complexes B1 and B2 when identical experiments were conducted with Raji nuclear extracts (Figure 2C). These results demonstrate that nucleotides between 4534 and 4542 in site B are critical for formation of all three complexes B1, B2 and B3, suggesting the presence of overlapping binding sites in site B or competition among factors for the same site.
Figure 2.
Identification of the DNA sequences required for factor binding to the HS7 sites B and C. (A) Nucleotide sequences of the wild-type and mutant site B oligonucleotides are shown with underlined bases indicating the base pairs required for the formation of complexes B1, B2 and B3. For the mutant site B oligonucleotides, only the bases that are changed compared with the wild-type sequence are indicated. The wild-type site B oligonucleotide was 5′ end labeled and used as probe in EMSA competition experiments with nuclear extracts from A3.01 cells (B) or Raji cells (C) and wild-type and substitution mutant site B double-stranded oligonucleotides as competitors. Binding assays were performed in the absence of competitor or in the presence of a 100-fold molar excess of either the homologous site B oligonucleotide or one of the nine mutated sites B (m1–m9). The competitor used is indicated at the top of each lane. The figure shows only the specific retarded bands of interest. The complexes B1 and B3 (B) or B1 and B2 (C) are indicated by arrows. Quantification of EMSAs was performed with an InstantImager (Packard) and is shown for the B1 complex in both (B and C). The histograms indicate the competition activities of site B m1–m9 mutants expressed relative to the competition activity of the wild-type site B (100%). (D) Nucleotide sequences of the wild-type and mutant site C oligonucleotides are shown with underlined bases indicating the base pairs required for the formation of complexes C1, C2 and C3. The 3 bp region different from the wild-type site C sequence is indicated in the seven mutant site C oligonucleotides (m1–m7). The wild-type site C used as probe was incubated with nuclear extracts from A3.01 cells (E) or Raji cells (F) and wild-type and substitution mutant site C double-stranded oligonucleotides as competitors. Binding assays were performed in the absence of competitor or in the presence of a 100-fold molar excess of either the homologous site C oligonucleotide or one of the seven mutated sites C (m1–m7). The competitor used is indicated at the top of each lane. The figure shows only the specific retarded bands of interest. The complexes C1 and C3 (E) or C1 and C2 (F) are indicated by arrows. Quantification of EMSAs was performed with an InstantImager (Packard) and is shown for the C1 complex in both (E and F). The histograms indicate the competition activities of site C m1–m7 mutants expressed relative to the competition activity of the wild-type site C (100%).
We next prepared seven scanning mutants of site C (named m1–m7), which sequentially replace 3 bp at a time of the entire site C sequence with a CCC sequence (Figure 2D). We tested the ability of this set of substitution mutants to inhibit formation of complexes C1, C2 and C3 in competition EMSAs using wild-type site C as probe, A3.01 nuclear extracts and the unlabeled wild-type and mutant double-stranded oligonucleotides as competitors (Figure 2E). Phosphorimaging analyses were used to quantify the percentage of competition for the formation of complexes C1 and C3 by each of the unlabeled oligonucleotides. The competition activities of mutant sites C m1–m7 were expressed relative to the competition activity of the wild-type site C, which was arbitrarily assigned to a value of 100% of competition. Figure 2E shows a graphic representation of the quantification results for complex C1. Site C mutants m3, m4 and m5 exhibited significantly reduced competition activities (Figure 2B, 55.4, 20.3 and 25.2%, respectively), indicating that the base pairs which were substituted in these three oligonucleotides were the most important for complex C1 formation. Quantification of the bands corresponding to complex C3 indicated the importance of the same base pair (data not shown). Similar results were observed for complexes C1 and C2 when identical experiments were conducted with Raji nuclear extracts (Figure 2F). These results demonstrate that nucleotides between 4687 and 4695 in site C are critical for the formation of all three complexes C1, C2 and C3, suggesting the presence of overlapping binding sites in site C or competition among factors for the same site.
In addition, we confirmed these results by complementary experiments in which a labeled probe containing site B was used and complex formation was competed by unlabeled oligonucleotides containing wild-type and mutant sites C, as well as by reverse experiments in which a labeled probe containing site C was used and complex formation was competed by unlabeled oligonucleotides containing wild-type and mutant sites B (data not shown), confirming that factors binding to site C have a DNA-binding specificity similar to that of factors binding to site B, except for PU.1. These data are in agreement with our previous work (7).
In conclusion, our results indicate that the DNA sequences identified here as critical for factors binding to sites B and C are located between nt 4534 and 4542 and between nt 4687 and 4695, respectively.
The transcription factors Oct-1 and Oct-2 bind to sites B and C
Based on the above systematic mutational analysis, we performed a new computer search for potential transcription factor binding sites, which was focused on the critical nucleotide sequences of sites B and C. This search revealed the presence of a putative octamer regulatory sequence both in site B and in site C (Figure 3A, nt 4532–4539 in site B and nt 4689–4696 in site C). These two putative octamer sequences presented three mismatches with respect to the Oct consensus. Oct-1 and Oct-2 along with Pit-1 and Unc-86 are founding members of the POU family of transcription factors. These members show homology in the domain responsible for specific DNA-binding, the so-called POU domain. While Oct-1 is ubiquitous, Oct-2 is predominantly expressed in B-cells, as well as in activated T-cells and in nervous system. However, B-cells appear to contain the highest amounts of Oct-2 (22,23).
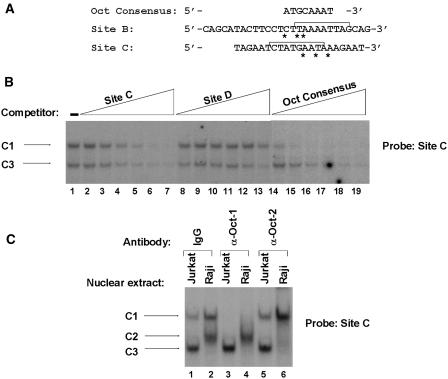
Figure 3.
Oct-1 and Oct-2 specifically interact with site C. (A) The nucleotide sequence of the wild-type sites B and C oligonucleotides are aligned with the octamer consensus motif. The asterisks indicate the mismatches of the potential site B and site C octamer motifs with respect to the octamer consensus. The brackets in site B and site C show the 9 bp (identified in Figure 2) as critical for the formation of complexes B1, B2 and B3 and C1, C2, and C3, respectively. (B) The site C oligonucleotide probe was incubated with nuclear extracts from Jurkat cells. Binding assays were performed in the absence of competitor (lane 1) or in the presence of increasing concentrations (5-, 10-, 25-, 50-, 100- or 200-fold molar excess) of the homologous site C oligonucleotide (lanes 2–7), of the heterologous HS7 site D oligonucleotide (lanes 8–13) or of the Oct consensus oligonucleotide (lanes 14–19). The sequence of the coding strand of the Oct consensus oligonucleotide was as follows: 5′-TGTCGAATGCAAATCACTAGAA-3′. The sequence of the coding strand of the HS7 site D oligonucleotide is shown in Figure 7. The figure shows only the specific retarded bands of interest. The complexes C1 and C3 are indicated by arrows. (C) Antibodies directed against Oct-1 (lanes 3 and 4) and Oct-2 (lanes 5 and 6) or purified rabbit IgG as negative control (lanes 1 and 2) were incubated with 15 μg of nuclear extracts from Jurkat (lanes 1, 3 and 5) or Raji (lanes 2, 4 and 6) cells before the addition of the site C oligonucleotide probe. The figure shows only the specific retarded bands of interest. The retarded DNA–protein complexes C1, C2 and C3 are indicated by arrows.
To determine whether transcription factors binding to sites B and C are related to Oct proteins, unlabeled double-stranded oligonucleotides were prepared and used as competitors in EMSAs. Site C probe was incubated with nuclear extracts from Jurkat cells in the absence or in the presence of different competitor double-stranded oligonucleotides: the unlabeled homologous oligonucleotide, an unlabeled heterologous oligonucleotide corresponding to site D and an unlabeled oligonucleotide corresponding to the octamer consensus sequence (Figure 3B). In the absence of competitor, two retarded complexes were detected: the ubiquitous complex C1 and the T-cell specific complex C3 (Figure 3B, lane 1). Formation of complex C1 was inhibited by a molar excess of unlabeled homologous oligonucleotide (Figure 3B, lanes 2–7), by the Oct consensus oligonucleotide (Figure 3B, lanes 14–19), but not by an oligonucleotide with an unrelated sequence containing site D (Figure 3B, lanes 8–13). Complex C3 was also out-competed by the homologous site C oligonucleotide and by the Oct consensus oligonucleotide (although to a lesser extent than complex C1) but not by the site D oligonucleotide. Similar competition experiments using site C as probe and nuclear extracts from Raji cells confirmed the above results for complex C1 and indicated that complex C2 was inhibited by an excess of unlabeled homologous oligonucleotide and by the Oct consensus, but not by the unrelated site D oligonucleotide (data not shown). We conclude from these experiments that the ubiquitous complex C1, the B-cell-specific complex C2 and the T-cell-specific complex C3 could contain a member(s) of the Oct proteins. Similar competition experiments performed with site B as probe supported the notion that formation of complexes B1, B2 and to a lesser extent B3 also resulted from binding of proteins belonging to the Oct family (data not shown).
To identify directly the factors present in the specific complexes B1/C1, B2/C2 and B3/C3, we performed supershift assays using antibodies directed against two individual members of the Oct family of transcription factors: the ubiquitously expressed Oct-1 protein and the lymphoid-specific Oct-2 protein. Labeled site C oligonucleotide probe was incubated with nuclear extracts from Jurkat or Raji cells in the presence of either an anti-Oct-1 antibody or an anti-Oct-2 antibody or purified rabbit IgG as control (Figure 3C). With Jurkat nuclear extracts, we observed, in the presence of the IgG control, the formation of the ubiquitous complex C1 and of the T-cell-specific complex C3 (Figure 3C, lane 1). Addition of the anti-Oct-1 antibody resulted in the complete disappearance of complex C1 (Figure 3C, lane 3), whereas the addition of the anti-Oct-2 antibody did not modify the migration profile of complexes C1 and C3 (Figure 3C, lane 5). With Raji nuclear extracts, we observed, in the presence of the IgG control, the formation of the ubiquitous complex C1 and the B-cell-specific complex C2 (Figure 3B, lane 2). The anti-Oct-1 antibody completely eliminated complex C1 (Figure 3C, lane 4) and complex C2 was eliminated by the anti-Oct-2 antibody (Figure 3C, lane 6). Similar results were obtained for complexes B1, B2 and B3 when identical experiments were conducted with site B as probe (data not shown).
In order to identify the protein(s) present in complexes B3/C3, we first tested, by supershift experiments, a series of protein candidates for potential transcription factor binding sites revealed by our in silico analysis: among others, the members of the MEF-2 transcription factor family. However, the latter assays did not provide any clue about the identity of the complexes B3/C3. Of note, the validity and/or the specificity of the different α-MEF-2 (α-MEF-2A, -2B, -2C and -2D) antibodies were established in separate experiments (data not shown). We also tried to purify complexes B3/C3 by affinity chromatography. However, despite many attempts, we did not succeed in the identification of these B3/C3 complexes. Additional purification and microsequencing studies are currently underway in our laboratory to identify the nature of the protein(s) present in complexes B3/C3.
From these data, we conclude that the ubiquitous complexes B1/C1 contain the Oct-1 transcription factor and that the B-cell-specific complexes B2/C2 contain the Oct-2 transcription factor. The transcription factor(s) present in the T-cell-specific complexes B3/C3 still remains (remain) to be identified.
The transcription factor PU.1 specifically interacts with site B of the HS7 region to form complex B4
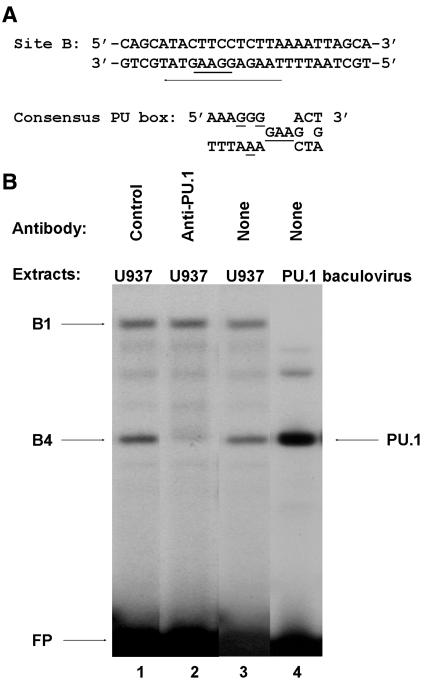
Complex B4 was previously demonstrated by our laboratory to contain a protein related to the transcription factor PU.1 (7). Indeed, our laboratory has previously reported that a SV40 PU.1 consensus oligonucleotide inhibits specifically the formation of complex B4, whereas no competition of binding in complexes B1 and B2 was observed. We have also previously shown that the PU.1 protein, derived from lysates of Cos-1 cells transfected with a PU.1 expression vector, binds to site B in a PU box-dependent manner. Nevertheless, because proteins that bind DNA by means of an Ets domain share DNA recognition properties, we could not rule out the possibility that other Ets family members also bind to the PU box present in the HS7 site B (Figure 4A).
Figure 4.
The complex B4 of the HS7 site B contains PU.1. (A) Nucleotide sequence of the wild-type site B oligonucleotide with the PU box indicated by an arrow on the non-coding strand. The core sequence 5′-GGAA-3′ of the PU box is indicated by a thicker bar. (B) Nuclear extracts from U937 cells (15 μg) were incubated in the absence of antibody (lane 3) or in the presence of a PU.1-specific antiserum (lane 2) or the preimmune rabbit serum as a negative control (lane 1) before the addition of the site B oligonucleotide probe. The same probe was also incubated with whole-cell extracts from SF9 cells infected with an Ac-NVP-PU.1 recombinant baculovirus (PU.1 baculovirus, lane 4). The DNA–protein complexes B1 and B4, the PU.1 complex and the free probe (FP) are indicated by arrows.
To determine directly the presence of PU.1 in complex B4, we performed supershift assays. Preimmune serum or antiserum specific for PU.1 was incubated with nuclear extracts from U937 cells before the addition of the labeled probe corresponding to site B (Figure 4B). We observed that the addition of the anti-PU.1 antibody interfered with the formation of complex B4, leading to its disappearance (Figure 4B, lane 2). In contrast, the preimmune serum did not affect complex B4 formation (Figure 4B, lane 1), indicating the specificity of the protein–antibody interaction. These results indicated that the B4 complex contained PU.1. In addition, we also demonstrated that PU.1 protein produced in baculovirus bound to site B and induced the formation of a complex comigrating with complex B4 (Figure 4B, compare lanes 3 and 4).
In conclusion, the macrophage- and B-cell-specific transcription factor PU.1 specifically interacts with site B in the HIV-1 HS7 region. Moreover, the comigration of complex B4 with purified PU.1 protein and the complete disappearance of complex B4 when using the anti-PU.1 antiserum suggest that this complex does not contain other Ets proteins.
The transcription factors Sp1 and Sp3 specifically interact with the GC-box located in the HIV-1 HS7 region
We have previously identified a GC-box with close homology to the Sp1 consensus sequence at position nt 4623–4631 in the HS7 region and we have shown, by in vitro DNase I footprinting analysis, that this site binds affinity-purified human Sp1 protein (7). In order to further assess the presence of Sp1 and/or other Sp family members on this GC-box, we designed a double-stranded oligonucleotide, and designed site Sp wt (nt 4616–4638) encompassing the potential Sp site (Figure 5A). This oligonucleotide was radiolabeled and tested in EMSAs for DNA–protein interactions with nuclear extracts from Jurkat cells (Figure 5B). Two retarded protein–DNA complexes were observed. Similar results were obtained with nuclear extracts from the U937, Raji and A30.1 cell lines (data not shown). To evaluate the sequence specificity of the binding to the Sp wt probe, we performed competition EMSAs using increasing concentrations of different unlabeled double-stranded competitor oligonucleotides (Figure 5B). The specificity of the protein binding was demonstrated because their formation was inhibited by competition with molar excesses of the unlabeled homologous Sp wt oligonucleotide (Figure 5B, lanes 2–5), but not by the same molar excesses of a heterologous oligonucleotide corresponding to site B (Figure 5B, lanes 14–17). A Sp1 consensus oligonucleotide (named Sp1 cons) inhibited the formation of the retarded complexes even at lower concentrations than the homologous oligonucleotide (Figure 5B, lanes 6–9). In contrast, these complexes were not competed by a mutated version of the Sp1 consensus oligonucleotide (named Sp1 cons mut) containing a GG to TT substitution, thereby demonstrating the specificity of the retarded complexes to the HS7 Sp motif (Figure 5B, lanes 10–13). These results support the hypothesis that both complexes contain Sp family members.
Figure 5.
Sp proteins bind to the GC-box located in the HS7 region. (A) The nucleotide sequence of the wild-type Sp site oligonucleotide is aligned with the Sp1 consensus sequence. The GC-box present in the Sp site is indicated by an arrow and the recognition core sequence 5′-GG-3′ is indicated by a thicker bar. (B) The HS7 Sp wt site oligonucleotide probe was incubated with 15 μg of nuclear extracts from Jurkat cells in the absence of competitor (lane 1) or in the presence of increasing concentrations (25-, 50-, 100- or 200-fold molar excess) of the homologous HS7 Sp site wt oligonucleotide (lanes 2–5), of the Sp1 consensus oligonucleotide (lanes 6–9), of a mutated Sp1 consensus oligonucleotide (lanes 10–13) or of the heterologous HS7 site B oligonucleotide (lanes 14–17). The sequence of the coding strand of the Sp1 consensus oligonucleotide and the sequence of the mutated Sp1 consensus oligonucleotide were as follows: 5′-ATTCGATCGGGGCGGGGCGAG-3′ and 5′-ATTCGATCGGTTCGGGGCGAGC-3′, respectively. The figure shows only the specific retarded bands of interest. The two retarded DNA–protein complexes are indicated by arrows. (C) Nuclear extracts from Jurkat cells (15 μg) were incubated in the absence of antibody or in the presence of antibodies directed against MEF-2, Sp1 and/or Sp3 (as indicated at the top of each lane) before the addition of the HS7 Sp wt site oligonucleotide probe. The figure shows only the specific retarded bands. The retarded DNA–protein complexes and the supershifted complexes are indicated by arrows.
To identify directly the Sp family members within the two retarded complexes observed with the HS7 Sp wt probe, we performed supershift assays using specific antibodies directed against individual members of the Sp family of transcription factors (Figure 5C). The HS7 Sp wt probe was incubated with nuclear extracts from Jurkat cells and polyclonal antibodies directed against Sp1, Sp2, Sp3 or Sp4 were added to the binding reaction mixture (Figure 5C and data not shown). The α-Sp1 antibody selectively supershifted the major slower migrating complex (Figure 5C, lane 3) and the α-Sp3 antibody resulted in the appearance of a supershifted complex and the corresponding disappearance of the faster migrating complex (Figure 5C, lane 4). We confirmed these results when both anti-Sp1 and anti-Sp3 antibodies were included in the same binding reaction (Figure 5C, lane 5). Similar relative mobilities of Sp1 and Sp3 EMSA complexes were reported in previous studies (24–27). In contrast, the binding pattern was not affected by the addition of the antibodies directed against the other Sp proteins (Sp2 and Sp4) (data not shown), showing that the two complexes did not seem to involve these other proteins. Moreover, the binding pattern was not affected by the addition of an unrelated antibody against MEF-2, used as a negative control (Figure 5C, lane 2).
Overall, these results demonstrate that Sp1 and Sp3 transcription factors interact with the GC-box (renamed Sp site hereafter in the manuscript) located in the intragenic HS7 region of HIV-1.
Transcription factor(s) binding to site D seems (seem) to contain a zinc finger DNA-binding domain
Our laboratory has previously reported that an ubiquitously expressed factor(s) interacts (interact) specifically with site D (7). This site is well conserved among many HIV-1 isolates, but computer analysis has revealed no relevant homology between this binding site and the recognition sequences for known transcription factors (7). Methylation interference analysis of site D has identified the guanine residues that are important for binding: methylation of two guanine residues at positions 4830 and 4833 on the coding strand and of one guanine residue at position 4835 on the non-coding strand has been shown to strongly interfere with binding to site D (7).
In this study, we further characterized site D by EMSAs to identify the protein(s) present in complex D. Since many transcription factors are zinc finger proteins, the probe corresponding to site D was incubated with nuclear extracts from Jurkat cells in the absence or in the presence of increasing concentrations of a chelator of zinc, the 1,10-phenanthroline. Incubation of this site D probe resulted in one major ubiquitous retarded complex (labeled D) (Figure 6, lane 1) as previously reported by our laboratory (7). We observed that complex D disappeared in the presence of phenanthroline (Figure 6, lanes 2–5). The presence of increasing concentrations of ZnCl2 caused the restoration of complex D (Figure 6, lanes 6–9), whereas the presence of increasing concentrations of another ion (MgCl2) did not allow the reappearance of complex D (Figure 6, lanes 10–13).
Figure 6.
Binding of nuclear factors to the HS7 site D is Zn2+-dependent. The site D oligonucleotide probe was incubated with 15 μg of nuclear extracts from Jurkat cells in the absence (lane 1) or in the presence of increasing concentrations (0.5, 1, 1.5 and 2 mM) of 1,10-phenanthroline (lanes 2–13). Increasing amounts of ZnCl2 (0.4, 0.8, 1 and 1.2 mM) or of MgCl2 (0.4, 0.8, 1 and 1.2 mM) were added to the binding reactions (lanes 6–9 or lanes 10–13, respectively). The figure shows only the specific retarded bands of interest. The complex D is indicated by an arrow.
We conclude that the binding of the ubiquitously expressed protein(s) to site D is Zn2+-dependent. This observation suggests that this (these) protein(s) could contain zinc finger domain(s). However, despite many purification and protein sequencing attempts, we did not succeed to identify this (these) protein(s).
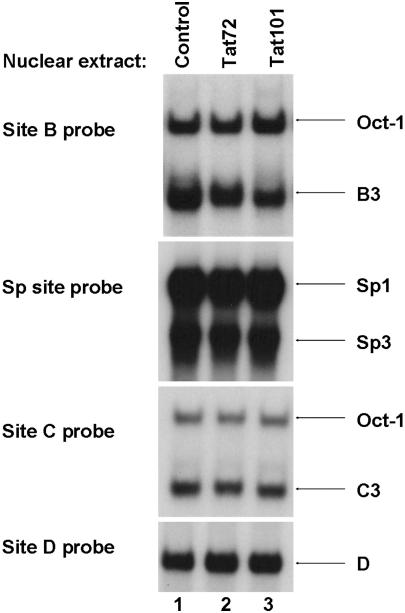
Tat expression does not affect the binding of the nuclear factors to the HS7 binding sites
To determine whether Tat can influence the binding of factors to the HS7 binding sites, we performed EMSAs, using nuclear extracts prepared from clonal Jurkat cell lines that stably expressed either the one-exon form of Tat [72 amino acids (Tat72)] (Figure 7, lane 2) or the two-exon form of Tat [101 amino acids (Tat101)] (Figure 7, lane 3) (8). Nuclear extracts from a clone transfected with the empty expression cassette vector were used as controls (Figure 7, lane 1). Both forms of Tat were used because Tat101 is expressed both early and late in the virus life cycle, while Tat72 is expressed solely in the late phase (28). Probes corresponding to HS7 site B, Sp site, site C and site D were incubated with these nuclear extracts. No difference in binding activity between the Tat72 and the Tat101 clones and the control clone was noted when the four probes were used (Figure 7, compare lanes 2 and 3 with lane 1).
Figure 7.
Analysis of Tat effect on factors binding to the HS7 sites. Probes corresponding to the HS7 site B, Sp site, site C or site D were incubated with 15 μg of nuclear extracts from clonal Jurkat cell lines expressing either Tat72 (lane 2) or Tat101 (lane 3) or the empty vector cassette as control (lane 1). The figure shows only the specific retarded bands of interest. The retarded complexes corresponding to Oct-1, C3, Sp1, Sp3 and D are indicated by arrows.
We conclude from these in vitro experiments that Tat has no effect on nuclear factor binding to the HS7 intragenic region.
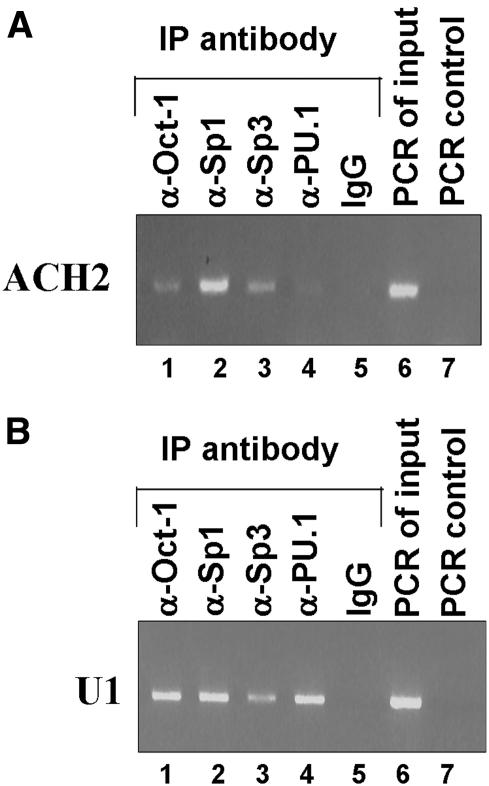
Sp1, Sp3, Oct-1 and PU.1 are recruited to the HS7 regulatory region in vivo
To demonstrate in vivo in the context of chromatin the relevance of our in vitro binding studies, we performed chromatin ChIP assays using both the lymphoid HIV-infected ACH2 cell line and the monocytic HIV-infected U1 cell line. Formaldehyde cross-linked chromatin from these cell lines was used for immunoprecipitation with antibodies directed against Sp1, Sp3, Oct-1 or PU.1 or a purified rabbit IgG as negative control. Following reverse of the cross-link, the purified DNA was subjected to PCR analysis using a set of primers flanking the HS7 region (Figure 8). In ACH2 cells, Oct-1, Sp1 and Sp3 binding to the HS7 region was detected (Figure 8A, lanes 1, 2 and 3, respectively), whereas, as expected, immunoprecipitations of the cross-linked chromatin with the anti-PU.1 antibody or with the purified rabbit IgG gave no signal (Figure 8A, lanes 4 or 5, respectively). In U1 cells, analysis of PCR products from immunoprecipitated DNA showed significant enrichment of the HS7 region when immunoprecipitation was carried out with the anti-Oct-1, anti-Sp1, anti-Sp3 or anti-PU.1 antibodies (Figure 8B, lanes 1, 2, 3 or 4, respectively). In contrast, no such enrichment was observed following immunoprecipitation of the cross-linked chromatin with the purified rabbit IgG (Figure 8B, lane 5). As a control, we used another set of primers flanking a region of the pol gene located 2 kb upstream of the HS7 region that has not been reported so far as binding any of these transcription factors. Immunoprecipitations with all the antibodies did not enrich eluates with DNA from this control region in both ACH2 and U1 cell chromatin, demonstrating the specificity of the HS7 interactions (data not shown).
Figure 8.
Recruitment of Oct-1, Sp1, Sp3 and PU.1 to the HS7 region in vivo. ChIP assays were used to detect binding of transcription factors to the HS7 region in the chromosomal context of proviruses integrated in (A) ACH2 T-lymphoid cells and (B) U1 monocytic cells. DNA and protein were cross-linked with formaldehyde for 10 min, and DNA was sheared. The cross-linked protein–DNA complexes were immunoprecipitated with an anti-Oct1 antibody (lane 1), an anti-Sp1 antibody (lane 2), an anti-Sp3 antibody (lane 3), an anti-PU.1 antibody (lane 4) or with a purified rabbit IgG as negative control (lane 5). The protein–DNA cross-links were reversed and the purified DNA was amplified by PCR using primers amplifying the HS7 region. PCR of the inputs (samples representing amplification from 1:100 dilution of total input chromatin from the ChIP experiments) are shown in lane 6. The PCR control represents the PCR amplification in the absence of DNA (lane 7).
These data, thus, demonstrate the occupancy in vivo of the HS7 region by Oct-1, Sp1 and Sp3 in both ACH2 and U1 cell lines as well as, consistently, the binding of PU.1 to the HS7 only in the U1 cells.
Identification of point mutations abolishing factor binding to the DNA motifs in the HIV-1 HS7 region
To further characterize physically the DNA motifs located in the HS7 region of the pol gene, we studied by using EMSA the effect of selected mutations on binding affinity. Point mutations were designed to abolish binding of factors to their respective sites without modifying the underlying amino acid sequence of the integrase.
Site B: We demonstrated that Oct-1, Oct-2 and PU.1 bound to site B. To abolish binding of Oct-1 and Oct-2, three adenine residues at positions 4535, 4538 and 4541 were substituted with guanine residues (Figure 9A). The oligonucleotide corresponding to this mutation was designated site B mutOct. To abolish PU.1 binding to site B, two cytosine residues at positions 4526 and 4529 were substituted with thymine residues in the site B PU box (Figure 9A). The oligonucleotide corresponding to this mutation was designated site B mutPU.1. The effect of these selected mutations was analyzed on binding affinity by using EMSAs. The wild-type and the two mutated site B oligonucleotides were used as probes and were incubated with nuclear extracts from U937 (Figure 9A), Raji and Jurkat (data not shown) cells. Figure 9A demonstrated the lack of Oct-1 binding to the site B mutOct oligonucleotide probe (lane 2) and the lack of PU.1 binding to the site B mutPU.1 oligonucleotide probe (lane 3). Similar experiments using Raji nuclear extracts showed the lack of Oct-2 binding to the site B mutOct oligonucleotide and confirmed the lack of PU.1 binding to site B mutPU.1 oligonucleotide probe (data not shown). Similar EMSAs using Jurkat nuclear extracts confirmed the lack of Oct-1 binding and showed the lack of T-cell-specific binding to the site B mutOct oligonucleotide probe (data not shown). Taken together, our results identify an octamer motif and a PU box in the HS7 site B. The octamer motif specifically binds the octamer proteins Oct-1 and Oct-2 in vitro. The PU box specifically binds the Ets protein PU.1. We report a 3 bp mutation, referred to as site B mutOct, that abrogates the binding of Oct-1 and Oct-2 and binding of the T-cell-specific factor to site B and a 2 bp point mutation, referred to as site B mutPU.1, that abrogates PU.1 binding to site B.
Sp site: We showed that Sp1 and Sp3 bound to the HS7 GC-box. Two guanine residues at positions 4629 and 4632 were substituted with two adenine residues in this motif (Figure 9B). The wild-type and the mutated oligonucleotide (referred to as site Sp wt and site Sp mut, respectively) were used as probes and incubated with nuclear extracts from U937 cells. Figure 9B showed the lack of Sp1 and Sp3 binding to the site Sp mut oligonucleotide probe, thereby demonstrating that the selected 2 bp mutation abolished Sp binding to the HS7 Sp site.
Site C: We demonstrated that Oct-1, Oct-2 and a T-cell factor(s) bound to the HS7 site C. To abolish factor binding to this site, two thymine residues at positions 4688 and 4694 were substituted with cytosine residues in the octamer sequence of site C (Figure 9C). The effect of this 2 bp mutation on binding affinity was analyzed by EMSAs. The wild-type and the mutated oligonucleotides (referred to as site C wt and site C mutOct, respectively) were used as probes and incubated with Raji (Figure 9C) or Jurkat nuclear extracts (data not shown). Figure 9C showed the lack of Oct-1 and Oct-2 binding to the site C mutOct oligonucleotide probe. Similar EMSAs using Jurkat nuclear extracts confirmed the lack of Oct-1 binding and showed the lack of T-cell-specific binding to the same mutated probe (data not shown). These results demonstrated that the selected point mutations introduced in site C abolished both the binding of Oct-1 and Oct-2 and the binding of the T-cell-specific factor.
Site D: To abolish binding of nuclear factors to site D, we substituted one adenine residue at position 4832 for a cytosine residue and one cytosine at position 4835 for a thymine residue (Figure 9D). The adenine residue (nt 4832) is located between two guanine residues on the coding strand (nt 4830 and 4833), which were previously demonstrated by our laboratory to be critical for complex D formation by methylation interference (7). The cytosine residue (nt 4835) corresponds to the guanine residue previously identified on the non-coding strand by the same technique as critical for complex D formation. The wild-type and the mutated oligonucleotides (referred to as site D mut) were used as probes in EMSAs with U937 nuclear extracts. Figure 9D demonstrated that the selected 2 bp mutation abolished formation of complex D.
The HS7 binding sites are involved in the transcriptional activity of the pol gene region
Our laboratory has previously shown using chloramphenicol acetyltransferase (CAT) transient expression plasmids that a 500 bp fragment encompassing the HS7 region (nt 4481–4982) exhibited transcription enhancing activity (∼2-fold) when it was cloned in its natural position with respect to the HIV-1 promoter in U937 and CEM cells in the presence of Tat. However, in the absence of Tat, the CAT values were too low to detect any enhancing activity of the HS7 region.
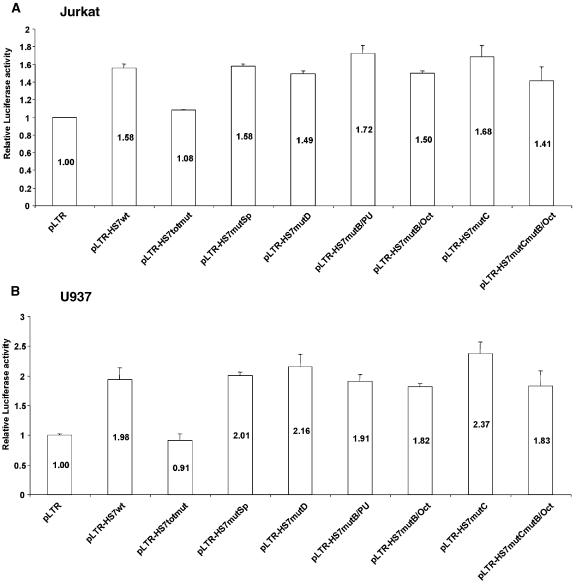
In order to address the potential functional role of the HS7 region in the basal (Tat-independent) activity of the HIV-1 promoter, we used the more sensitive luciferase transient expression system. We subcloned the 500 bp HS7 fragment into the construct pLTR in the sense orientation with respect to the transcriptional unit, downstream of the luciferase reporter gene, thereby generating the pLTR-HS7wt. The pLTR construct contains the complete HIV-1 LAI 5′-LTR (plus the leader sequence up to the ATG of gag) driving the expression of the luciferase gene. The constructs pLTR and pLTR-HS7wt were transiently transfected into Jurkat cells (Figure 10A) or U937 cells (Figure 10B). The reporter constructs were cotransfected into the cells with pRL-TK, and used as an internal control to measure the transfection efficiency. At 44 h post-transfection, cells were lysed and assayed for luciferase activity. In Jurkat cells, transfection of plasmid pLTR-HS7wt caused a 1.58-fold increase in luciferase activity compared with that of the control plasmid pLTR tested under the same conditions (Figure 10A). In U937 cells, transfection of pLTR-HS7wt caused a 1.98-fold increase in luciferase activity compared with that of pLTR (Figure 10B). These results indicate that, in the absence of Tat, the region associated with the intragenic HS7 shows a weak transcriptional enhancing activity when it is cloned in a position similar to that observed within the viral genome (downstream and in sense orientation relative to the 5′-LTR-luc transcriptional unit).
Figure 10.
Functional significance of the HS7 binding sites. The transcriptional enhancing activity of the wild-type and mutated pol gene HS7 regions was tested after cloning of these regions in pLTR and transfection of the plasmids pLTR, pLTR-HS7wt, pLTR-HS7totmut pLTR-HS7mutB/Oct, pLTR-HS7mutB/PU, pLTR-HS7mutC, pLTR-HS7mutSp, pLTR-HS7mutD and pLTR-HS7mutCmutB/Oct in Jurkat (A) or U937 (B) cells. Cells were cotransfected with 50 ng of pRL-TK in which the HSV TK promoter is driving the Renilla luciferase gene expression. Luciferase activities (Firefly and Renilla) were measured in cell lysates 44 h after transfection. Results are expressed as LuciferaseFirefly/LuciferaseRenilla and are presented as histograms indicating luciferase activities relative to that of the control vector pLTR, which was assigned a value of 1. Means and standard errors of the means from 6 to 8 independent transfections performed with three different DNA preparations are indicated.
In order to determine the relative contribution of each factor binding sites (site B, site C, Sp site and site D) to the HS7 enhancing activity, the point mutations identified in EMSAs (Figure 9) were introduced individually or in combination in the context of the pLTR-HS7wt plasmid. The mutated plasmids were designed pLTR-HS7totmut, pLTR-HS7mutSp, pLTR-HS7mutD, pLTR-HS7mutB/PU, pLTR-HS7mutB/Oct, pLTR-HS7mutC and pLTR-HS7mutCmutB/Oct. These plasmids were assayed for luciferase activity after transient transfection in Jurkat or U937 cells (Figure 10A or B, respectively). Remarkably, in both cell lines, transfection of plasmid pLTR-HS7totmut exhibited luciferase activity similar to that obtained with the control vector pLTR, thereby demonstrating that the enhancing effect observed with pLTR-HS7wt required intact site B, site C, Sp site and site D motifs. Moreover, in both cell lines, transfection of the plasmids containing the individual mutations presented luciferase activities similar to that obtained with the wild-type pLTR-HS7wt construct. In conclusion, mutations in the HS7 binding sites in combination abolish the transcription-enhancing activity of the pol HS7 region, suggesting that these sites are responsible for most of this activity. However, we could not demonstrate the functional role played by one individual site in the HS7 enhancing activity (Figure 10A and B).
We next studied the effect of mutating the HS7 binding sites on the response of the HIV-1 promoter to Tat. We observed that the pol HS7 region exhibited transcriptional enhancing activity in the presence of Tat, in agreement with our previous results (7), and that mutations in all the HS7 binding sites in combination abolished this activity (data not shown).
Our functional results, thus, demonstrate a positive regulatory role of the pol HS7 region in both Tat-independent and Tat-dependent HIV-1 promoter-driven gene expression. These data indicate that the loss of transcriptional enhancing activity caused by mutations in the HS7 binding sites correlated with the loss in factor binding to these sites. Moreover, mutation of each HS7 binding site individually did not affect the enhancing activity of the intragenic positive regulatory region.
Functional analysis of the individual HS7 region binding sites by overexpression assays
We next wanted to further characterize the functionality of each HS7 site individually (sites for which we were able to identify by the above experiments the bound transcription factors) and to examine whether these transcription factors (PU.1, Sp1, Sp3, Oct-1 and Oct-2) act through the HS7 binding sites. Because these transcription factors present distinct cell-specific expression, we decided to study each site separately by overexpression assays using for each individual factor a cell line which does not express the factor considered. To this end, we produced a series of artificial luciferase reporter constructs in which multimerized copies of each wild-type and mutated HS7 binding sites were inserted upstream of the HSV TK minimal promoter into the pTK construct. The resulting constructs were cotransfected with expression vectors for the various transcription factors (PU.1, Sp1, Sp3, Oct-1 and Oct-2) and assayed for luciferase activity. The minimal TK promoter was used instead of the HIV-1 promoter (LTR) because the latter is known to contain binding sites for Oct, Ets and Sp family members (29) and could have therefore complicated the interpretation of the transfection results. In these experiments, we used multimerized HS7 binding sites because multimerization of a transcription factor binding site allows the amplification of its transcriptional effect.
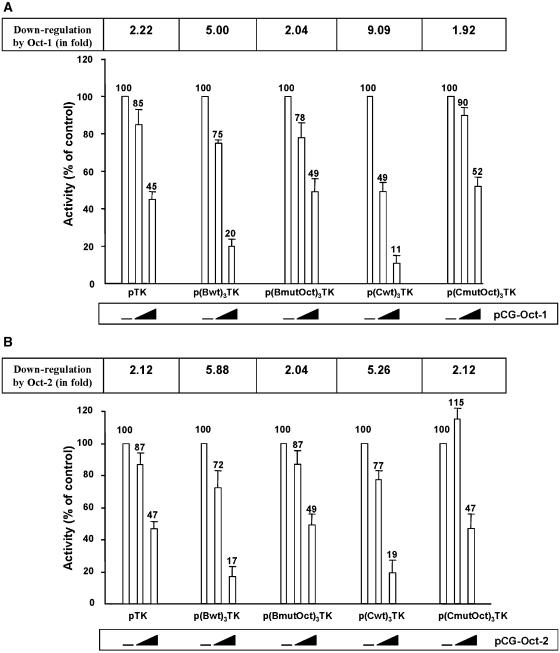
Ectopic expression of the Oct-1 and Oct-2 transcription factors down-regulates TK promoter activity through multimerized sites B and C
To determine whether the HS7 sites B and C can act as independent regulators of transcription, we constructed four luciferase reporter plasmids driven by the HSV TK minimal promoter with three tandem repeats of the wild-type and Oct-mutated site B or site C inserted upstream of the TK promoter. These four plasmids were referred to as p(Bwt)3TK, p(BmutOct)3TK, p(Cwt)3TK and p(CmutOct)3TK, respectively. To examine the response of these reporter constructs to the POU-homeodomain transcription factors Oct-1 and Oct-2, murine NIH3T3 fibroblats were transiently cotransfected with each of them and increasing amounts of either the Oct-1 or the Oct-2 expression vector (Figure 11A and B, respectively) and then assayed for luciferase activity. The murine NIH3T3 cell line was used in these experiments because our EMSAs performed with NIH3T3 nuclear extracts and with probes corresponding to site B or site C revealed no binding activity corresponding to complexes B1/C1, B2/C2, B3/C3 and even no binding at all (data not shown).
Figure 11.
Multimerized copies of the HS7 sites B and C confer Oct-1 and Oct-2 down-regulation to a heterologous minimal promoter. NIH3T3 cells were transiently cotransfected with 500 ng of the pTK, p(Bwt)3TK, p(BmutOct)3TK, p(Cwt)3TK or p(CmutOct)3TK reporter construct and with increasing amounts (0, 100 and 500 ng) of either pCG-Oct-1 (A) or pCG-Oct-2 (B). To maintain the same amount of transfected DNA and to avoid squelching artifacts, the different amounts of Oct-1/Oct-2 expression vectors cotransfected were complemented to 500 ng of DNA by using the empty pCG vector. Luciferase activities were measured in cell lysates 44 h after transfection and were normalized with respect to protein concentrations of the lysates. Results are presented as histograms indicating the luciferase activity of each reporter construct in the absence of ectopically expressed Oct-1 or Oct-2, which was arbitrarily assigned a value of 100% of activity. The down-regulation of the TK promoter constructs by Oct-1 (A) and Oct-2 (B) is also indicated (in fold). Means of triplicate samples and standard errors of the means are shown. An experiment representative of three independent transfections performed with at least two different DNA preparations is shown.
As shown in Figure 11, the control pTK construct was repressed by Oct-1 up to 2.22-fold (i.e. by 55%) and by Oct-2 up to 2.12-fold (i.e. by 53%). Cotransfection of the reporter construct p(Bwt)3TK with the Oct-1 or Oct-2 expression vectors resulted in a dose-dependent decrease in luciferase activity by ectopically expressed Oct-1 (up to 5.00-fold) and Oct-2 (up to 5.88-fold); thus, representing a 2.3- and 2.8-fold down-regulation when compared with the Oct-1 and Oct-2 responses of the control pTK construct devoid of upstream HS7 sites B. This effect required intact Oct motifs in site B, because mutations in these motifs [p(BmutOct)3TK] resulted in levels of Oct-1- and Oct-2-mediated repression similar to those obtained with the control pTK (Figure 11A and B, respectively). Similar results were obtained when examining site C. Indeed, addition of three copies of the HS7 site C upstream of the TK promoter [p(Cwt)3-TK] resulted in a dose-dependent decrease in luciferase activity by ectopically expressed Oct-1 (up to 9.09-fold) and Oct-2 (up to 5.26-fold), thus representing a 4.1-fold and 2.5-fold down-regulation when compared with the Oct-1 and Oct-2 responses of the control pTK devoid of upstream HS7 sites C (Figure 11A and B, respectively). This effect also required intact Oct motifs in site C [see p(CmutOct)3-TK in Figure 11A and B].
We conclude from these experiments that ectopic Oct-1 and Oct-2 proteins have a site B- or site C-dependent inhibitory effect on the heterologous TK promoter containing multiple upstream site B or site C, respectively. These results, thus, demonstrate that the HS7 sites B and C function as negative regulatory elements in response to ectopic Oct-1 or Oct-2 proteins in a heterologous context, suggesting that Oct-1 and Oct-2 might be negatively involved in the HS7 transcriptional activity.
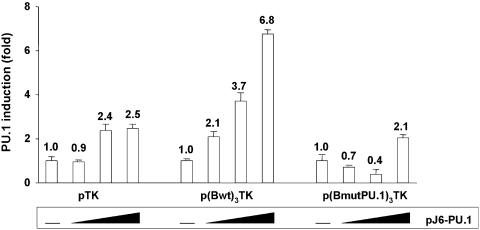
Ectopic expression of the PU.1 transcription factor up-regulates TK promoter activity through multimerized site B
In order to define the potential role of PU.1 through the PU box of the HS7 site B, we generated a mutant derivative of the p(Bwt)3TK construct with the upstream insertion of three tandem repeats of a site B mutated in the PU box sequence. This derivative was designated p(BmutPU.1)3TK. The constructs pTK, p(Bwt)3TK and p(BmutPU.1)3TK were cotransfected with increasing amounts of the PU.1 expression vector pJ6-PU.1 into PU.1-negative NIH3T3 cells (Figure 12). Transfected cells were assayed for luciferase activity. Results presented in Figure 12 show that the control pTK (lacking specific PU boxes) construct was moderately transactivated by PU.1 (up to 2.5-fold). This PU box-independent activation could be attributed to stimulation by PU.1 through other DNA sequences present in the pTK vector. Cotransfection of the PU.1 expression vector with the reporter construct containing three wild-type sites B [p(Bwt)3TK] resulted in a dose-dependent stimulation of the luciferase activity (up to 6.8-fold); thus, representing a 2.7-fold up-regulation when compared with the PU.1 response of the control pTK devoid of upstream site B PU boxes. This effect required an intact PU box in site B, because mutations in this motif [p(BmutPU.1)3TK] resulted in levels of PU.1-mediated transactivation similar to those obtained with the control pTK.
Figure 12.
Ability of multimerized HS7 site B motifs to confer PU.1 stimulation to a TK minimal promoter. NIH3T3 cells were transiently cotransfected with 500 ng of either pTK, or p(Bwt)3TK or p(BmutPU.1)3TK and with increasing amounts (0, 100, 250 and 500 ng) of the PU.1 expression vector, pJ6-PU.1. To maintain the same amount of transfected DNA and to avoid squelching artifacts, the different amounts of PU.1 expression vector cotransfected were complemented to 500 ng of DNA by using the empty pJ6 vector. Luciferase activities were measured in cell lysates 44 h after transfection and were normalized with respect to protein concentrations of the lysates. Results are presented as histograms indicating the induction by PU.1 (in fold) with respect to the activity of each TK reporter construct in the absence of PU.1, which was assigned a value of 1. Means of triplicate samples and standard errors of the means are shown. An experiment representative of three independent transfections performed with at least two different DNA preparations is shown.
We conclude from these experiments that ectopic PU.1 protein has a site B PU box-dependent stimulatory effect on the heterologous TK promoter containing multiple upstream sites B. These results thus establish the functional significance of PU.1 through the PU box present in the HS7 site B and suggest that the transcriptional activity of the HS7 region is positively regulated by PU.1.
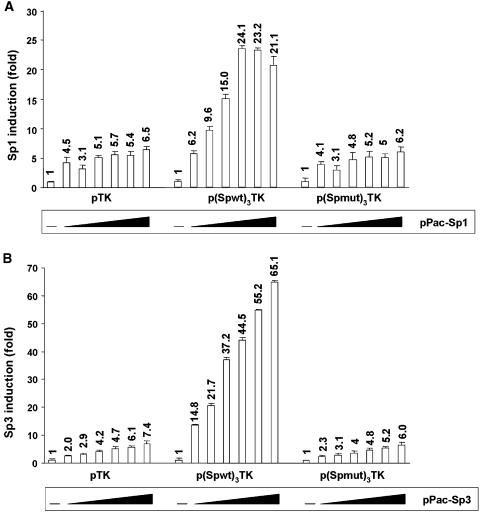
Ectopic expression of the Sp1 or Sp3 transcription factors up-regulates TK promoter activity through the HS7 Sp binding site
To examine whether Sp1 and/or Sp3 act through the HS7 GC-box, we produced synthetic reporter constructs in which multimerized copies of the wild-type or mutated HS7 Sp site were inserted upstream of the TK promoter into the pTK construct. The resulting plasmids were designated p(Spwt)3TK or p(Spmut)3TK, respectively. To examine the response of these reporter constructs to Sp1 and/or Sp3, Drosophila Schneider (SL2) cells (which unlike most mammalian cells lack endogenous Sp factors) were transiently cotransfected with each of them and increasing amounts of either the Sp1 or the Sp3 expression vector (Figure 13A and B, respectively), and then assayed for luciferase activity. The control pTK construct was moderately transactivated by Sp1 (up to 6.5-fold) and by Sp3 (up to 7.4-fold). Addition of three copies of the HS7 Sp site upstream of the TK promoter resulted in a dose-dependent increase in luciferase activity by ectopically expressed Sp1 (up to 24.1-fold) and Sp3 (up to 65.1-fold); thus, representing a 3.7- and 8.8-fold up-regulation when compared with the Sp response of the control pTK devoid of upstream HS7 Sp sites. Mutations in the HS7 Sp sites reversed the activating response to Sp1 and Sp3 to the luciferase activities obtained with the control vector pTK [see p(Spmut)3TK in Figure 13A and B]. Moreover, the coexpression of Sp1 and Sp3 together in equal amounts on each TK reporter constructs had no more effect than either factor transfected alone (data not shown).
Figure 13.
Ability of multimerized HS7 Sp motifs to confer Sp1 and Sp3 stimulation to a TK minimal promoter. SL2 cells were transiently cotransfected with 700 ng of either pTK, or p(Spwt)3TK or p(Spmut)3TK and with increasing amounts (0, 5, 10, 25, 50, 100 and 250 ng) of either the Sp1 or the Sp3 expression vector, pPac-Sp1 or pPac-Sp3 [(A) or (B), respectively]. To maintain the same amount of transfected DNA and to avoid squelching artifacts, the different amounts of Sp expression vector cotransfected were complemented to 250 ng of DNA by using the empty pPac vector. Luciferase activities were measured in cell lysates 44 h after transfection and were normalized with respect to protein concentrations of the lysates. Results are presented as histograms indicating the induction by Sp1 or Sp3 (in fold) with respect to the activity of each TK reporter construct in the absence of Sp, which was assigned a value of 1. Means of triplicate samples and standard errors of the means are shown. An experiment representative of three independent transfections performed with at least two different DNA preparations is shown.
We conclude from these experiments that ectopic Sp1 and Sp3 proteins have a HS7 Sp site-dependent stimulatory effect on the heterologous TK promoter containing multiple upstream HS7 Sp sites. The ability of the concatemerized HS7 Sp site to confer transactivation by ectopically expressed Sp1 and Sp3 in a heterologous context suggests that Sp1 and Sp3 are direct contributors to the transcriptional enhancing activity of the HS7 region.
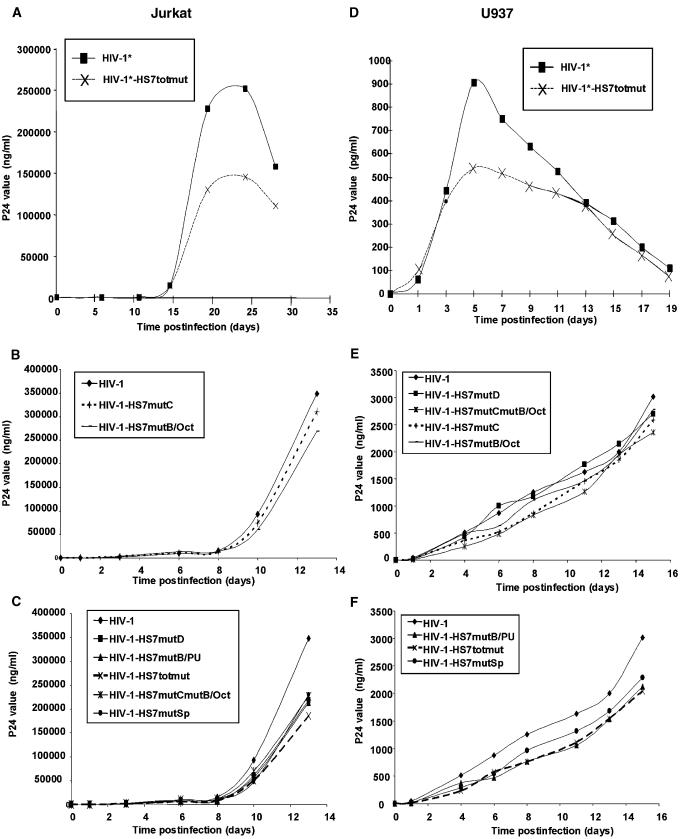
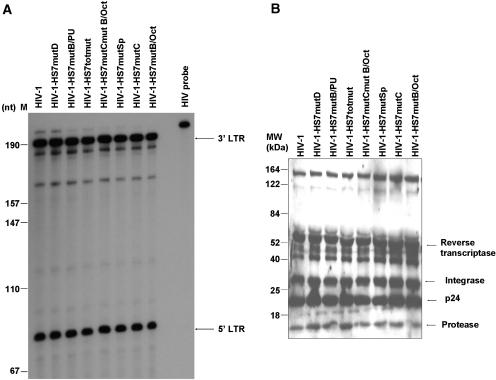
The HS7 binding sites are critical for HIV-1 infectivity
To address the biological significance of the HS7 binding sites in the HIV-1 life cycle, we tested the effect of mutations abolishing factor binding to the different sites (sites B, C, Sp and D) on HIV-1 replication in infection experiments with wild-type and mutant HIV-1 infectious viruses. To this end, mutations of each site were introduced in combination into an infectious clone of HIV-1. In vitro site-directed mutagenesis of the DNA-binding sites was performed with a plasmid (pCV10) containing the ApaI (nt 2011)/EcoRI (nt 5743) fragment of the NL4-3 HIV-1 genome and oligonucleotides containing the mutated sites as described in Materials and Methods. After site-directed mutagenesis and confirmation of the mutations by sequencing, the fragment (ApaI/EcoRI) containing the mutations was subcloned back into the corresponding restriction sites of the pCV1 plasmid. The resulting mutant plasmid was designated pCV426 (pHIV-1*-HS7totmut). As a control, the corresponding wild-type ApaI/EcoRI fragment was similarly cloned into pCV1 to generate pCV422 (pHIV-1*).
Wild-type and mutant HIV-1 infectious proviruses were generated from the single-LTR-containing constructs pCV422 and pCV426 by BamHI digestion and self-ligation. To obtain stocks of infectious viruses, these proviruses were transfected into Jurkat cells. Transfected cells were cocultivated with SupT1 cells for 1 day after transfection. Progeny virus production in coculture supernatants was then monitored by measuring the level of p24 gag antigen over a 15 day period. Cell-free supernatants were harvested at the peak of viral production to generate virus stocks for subsequent infectivity studies. To assess the possibility of reversion to a wild-type phenotype as an explanation for the growth of the mutant virus, HIV-1 genomic RNA from HIV-1*-HS7totmut virus stock was purified and analyzed by RT–PCR with a primer pair that amplified a 640 bp intragenic portion of the genome (nt 4342–4982) encompassing the mutated HS7 binding sites. PCR fragments were subcloned, and three individual clones were resequenced. This analysis confirmed the presence of the original mutations in the HS7 region (data not shown).
To study the effect of the HS7 mutations on HIV-1 growth kinetics, we infected either human CD4+ T-lymphoid Jurkat or human promonocytic U937 cells with the wild-type (HIV-1*) and mutant (HIV-1*-HS7totmut) viral stocks. We performed at least three replication assays with the Jurkat T-cell line and with the U937 monocytic cell line by measuring p24 production over time. Figure 14 shows a representative replication curve for each cell line, which revealed differences in replication rate between the wild-type and the HS7 mutant viruses. Infection of Jurkat cells with the wild-type virus resulted in rapid and vigorous viral production, with p24 concentration reaching a peak on days 20–25 post-infection, followed by a rapid decrease reflecting a rapid reduction in viable cell numbers (Figure 14A). Mutant virus HIV-1*-HS7totmut replicated with a replication kinetics that was similar to that of the wild-type control virus. However, infection of Jurkat cells with the mutant HIV-1*-HS7totmut virus produced lower p24 release (about two times less than the amount released from cultures infected with the wild-type HIV-1* at the peak of infection), demonstrating reduced growth properties. Similar results were obtained when the growth properties of the mutant HIV-1*-HS7totmut were assayed on the human promonocytic U937 cell line (Figure 14B). These data are consistent with those obtained after transient expression assays (Figure 10).
Figure 14.
Mutations in the HS7 binding sites cause a reduced efficiency of HIV-1 replication. Jurkat (A–C) or U937 (D–F) cells were infected with equivalent amounts of p24 concentration of wild-type (HIV-1 or HIV-1*), totally mutated (HIV-1-HS7totmut or HIV-1*-HS7totmut) or individually mutated (HIV-1-HS7mutB/PU, HIV-1-HS7mutB/Oct, HIV-1-HS7mutC, HIV-1-HS7mutCmutB/Oct, HIV-1-HS7mutSp and HIV-1-HS7mutD) viral infectious stocks as described in Materials and Methods. Results show viral production, which was estimated by measuring p24 antigen concentration in culture supernatants at different times following infection. The experiment shown is representative of at least nine independent infection experiments and the variation for a given mutant between different experiments was <15% in most cases.
In order to further evaluate the effect of the HS7 mutations on HIV-1 replicative properties, we next introduced individually the mutations into the two-LTRs-containing infectious HIV-1 molecular clone pNL4-3. The wild-type and totally mutated proviruses were also generated in this context. The resulting proviruses were designated as pHIV-1, pHIV-1-HS7totmut, pHIV-1-HS7mutB/Oct, pHIV-1-HS7mutB/PU, pHIV-1-HS7mutC, pHIV-1-HS7mutSp, pHIV-1-HS7mutD and pHIV-1-HS7mutCmutB/Oct. Stocks of infectious viruses wild-type, totally mutated or containing the individual mutations were generated by transfection of 293T cells, were tested for the presence of the original muations by RT–PCR and sequencing analyses, and were used in infection studies of Jurkat and U937 cells (see Materials and Methods). HIV-1 replication was monitored by measuring the production of p24 in the cell supernatants over a ∼15 day period. On the basis of their growth characteristics, the five individual and the double HS7 mutant viruses were classified into two replicative phenotypes.
In Jurkat cells, (i) mutant viruses HIV-1-HS7mutB/Oct and HIV-1-HS7mutC replicated efficiently with replication kinetics similar to that of the wild-type control virus HIV-1, but with levels of viral production slightly lower than that of the control virus, indicating that the individual mutations of the octamer sequence either in site B or in site C slightly affected HIV-1 replication but not as strongly as the combination of all the HS7 mutations (compare with HIV-1-HS7totmut) (Figure 14B and C); and (ii) mutant viruses HIV-1-HS7mutB/PU, HIV-1-HS7mutCmutB/Oct, HIV-1-HS7mutSp and HIV-1-HS7mutD also replicated with a replication kinetics that was similar to that of the wild-type control virus. However, infection with these mutant viruses produced p24 release as low as the totally mutated virus, demonstrating reduced growth properties similar to those observed with HIV-1-HS7totmut (Figure 14C).
In U937 cells, (i) mutant viruses HIV-1-HS7mutB/Oct, HIV-1-HS7mutC, HIV-1-HS7mutCmutB/Oct and HIV-1-HS7mutD replicated efficiently with replication kinetics and levels of viral production that were similar to those of the wild-type control virus HIV-1, indicating that the individual mutations of the octamer sequence either in site B or in site C, the individual mutation of site D and the double mutation of the octamer sequences in both site B and site C did not affect HIV-1 replication (Figure 14E); and (ii) mutant viruses HIV-1-HS7mutB/PU and HIV-1-HS7mutSp also replicated with a replication kinetics that was similar to that of the wild-type HIV-1. However, infection with these mutant viruses produced lower p24 release than the wild-type HIV-1, demonstrating reduced growth properties similar to those observed with HIV-1-HS7totmut (Figure 14F).
Further infection of target cells in a second round with supernatants collected 15 days after the first infection indicated that all the HS7 mutant viruses were not impaired in terms of infectivity (data not shown).
Thus, the integrity of the HS7 DNA-binding sites located 5 kb downstream of the HIV-1 transcription start site is important for HIV-1 replication in human CD4+ cell lines, indicating a positive regulatory function for this region. Our findings strongly suggest a crucial role of the HS7 Sp and PU.1 sites on HIV-1 replication.
Mutations in the HS7 binding sites do not affect the integrity of the viral particles
Although mutations in the HS7 region are not expected to impair HIV-1 RNA packaging, we performed RNase protection assays to quantify viral RNA from equal amounts of viral particles from the wild-type and each HS7 mutant virus stocks (Figure 15A). This experiment showed that the wild-type and mutant viruses contained the same amount of packaged genomic RNA (Figure 15A). Moreover, we also analyzed the amount of HIV-1-specific proteins in the wild-type and each HS7 mutant virus stock (Figure 15B). To this end, lysates from equal amounts of p24 from the wild-type and mutant virus stocks were prepared, and western blot analyses were performed with purified human anti-HIV-1 IgG as a source of antibody. Similar levels of the HIV-1 proteins (including the integrase which is partially encoded by the HS7 region) were detected in all lysates (Figure 15B).
Figure 15.
Mutations in the HS7 binding sites do not affect viral particle formation. (A) Equivalent amounts of viral particles (assessed by p24 ELISA assay) from the wild-type and each HS7 mutant virus stocks were pelleted by ultracentrifugation. After lysis of viral particles, HIV-1 RNA was detected by RNase protection analysis with an antisense riboprobe which protects two bands (200 and 83 nt) corresponding to the HIV-1 3′- and 5′-LTR, respectively. The undigested HIV probe is shown for reference. Lane Marker (M) contains pSK-/ApaII markers, whose sizes (in nt) are indicated on the left-hand side. Intensities of RNA bands were quantified by radioimaging (InstantImager). (B) Equivalent amounts of viral particles (assessed by p24 ELISA assay) from the wild-type and each HS7 mutant virus stocks were pelleted by ultracentrifugation. The ultracentrifuged viral stocks were lysed in Laemmli buffer, analyzed by western blotting with anti-HIV-1 immunoglobulin, and detected by enhanced chemiluminescence with a horseradish peroxidase-conjugated goat anti-human IgG. The bands corresponding to the HIV-1 reverse transcriptase, integrase, p24 protein and protease are indicated. MW, molecular mass (indicated in kDa).
These results demonstrate that the reduced replication phenotypes we observed with the HIV-1 HS7 mutant viruses are due neither to a defect in RNA packaging nor to a defect in the protein content of viral particles. Thus, mutant and wild-type HIV-1 particles from the viral stocks used in the infection studies are structurally undistinguishable at both the genomic RNA and the protein levels, suggesting an effect of the mutations in the HS7 cis-acting elements at the level of HIV-1 transcription.
DISCUSSION
We have previously identified and physically characterized a new positive transcriptional regulatory element associated with a DNase I-hypersensitive site (HS7) present in the pol gene of HIV-1 (7). A fragment encompassing this hypersensitive site positively regulates transcription from the HIV-1 5′-LTR in transient transfection experiments. Several recognition sites for nuclear proteins (sites B, C, D and a GC-box) have been identified by in vitro binding studies (7). In this study, we have further physically characterized each of these four binding sites located in the HS7 region and have shown that the transcription factors Oct-1, Oct-2, PU.1, Sp1 and Sp3 interact in vitro with this region. Other in vitro binding studies have also shown that Tat expression does not affect the binding of the nuclear factors to the HS7 binding sites. Furthermore, ChIP assays have revealed that Sp1, Sp3, Oct-1 and PU.1 are recruited in vivo to the HS7 intragenic region of HIV-1 proviruses integrated in several infected cell lines (Figure 16). For each of the four sites (sites B, Sp, C and D), we have identified point mutations abolishing binding of the nuclear factors to their cognate DNA sites without altering the underlying amino acid sequence of the integrase. By transient transfection assays, we have demonstrated the functional involvement of the HS7 binding sites in the transcriptional enhancing activity of the pol gene region. We have next examined the functional transcriptional role of each site separately in a heterologous context. Our results with multimerized wild-type and mutated HS7 binding sites in isolation (i.e. in the absence of the other DNA-binding sites) have demonstrated that the PU.1, Sp1 and Sp3 transcription factors, on one hand, and the Oct-1 and Oct-2 transcription factors, on the other hand, up- and down-regulate, respectively, the transcriptional activity of the minimal TK promoter. Finally, we have addressed the biological significance of the HS7 binding sites in the HIV-1 replication cycle by mutating these sites individually or in combination in the context of infectious clones of HIV-1. These mutations do not affect the integrity of the viral particles both at the genomic RNA and the protein levels. Our infection studies have demonstrated that the HS7 sites located 5 kb downstream of the HIV-1 transcription start site are important for HIV-1 replication in human CD4+ cell lines, indicating a positive regulatory function for pol intragenic region.
Figure 16.
Putative schematic representation of the nuclear factors binding to their respective DNA sites in the intragenic HS7 region of HIV-1. This assignment is based on in vitro binding studies. Moreover, the transcription factors Oct-1, Sp1, Sp3 and PU.1 have also been demonstrated by ChIP assays to be recruited to the HS7 region in vivo in the chromosomal context of integrated proviruses. The DNA-binding sites are represented as closed rectangles. The ubiquitous nuclear factors are represented as gray-shaded ovals, whereas factors specific to a given cell line appear as open circles. Our experiments do not establish whether the nuclear factors indeed bind together, successively or alternatively to their cognate DNA sequences.
Functional role of the HS7 transcription factor binding sites
Octamer binding sites
By in vitro binding studies, we have demonstrated that the HS7 sites B and C specifically bind the octamer proteins Oct-1 and Oct-2. In addition, we have shown by ChIP assays the binding of the Oct-1 protein to the HS7 region in vivo both in the HIV-1 latently infected T-lymphoid cell line ACH2 and in the monocytic cell line U1. Octamer binding proteins belong to the POU homeodomain transcription factor family (30). Two of the best characterized octamer binding proteins are Oct-1 and Oct-2. Whereas the Oct-1 protein is ubiquitously expressed, Oct-2 is expressed predominantly in the B-cell lineage (31–35). However, Oct-2 expression can be induced by antigenic stimulation of T-cells (36), suggesting that the Oct-2 protein may play a role in gene expression during T-cell activation. Oct-1 is a broadly expressed and versatile transcription factor performing many divergent roles in cellular and viral transcriptional regulation and acting both as a repressor and as an activator of transcription [reviewed in (37)].
Using transient transfection assays, we have shown that ectopically expressed Oct-1 and Oct-2 repress the transcriptional activity of the minimal TK promoter through the HS7 sites B and C. However, our infection experiments using viruses mutated in the octamer sequences of site B and/or site C are not consistent with a repressive effect mediated by these sites. Indeed, in U937 cells, the individual mutations of the octamer sequences either in site B or in site C as well as the double mutation of the octamer sequences in both site B and site C do not affect HIV-1 replication. In Jurkat cells, the individual Oct mutations slightly reduce the levels of viral production relative to those observed with the wild-type virus, whereas the mutant virus containing the double Oct mutation demonstrates more severely reduced growth properties similar to those observed with the totally mutated virus HIV-1-HS7totmut. Different hypotheses can be drawn to explain these discrepancies between our ex vivo transfection and in vivo infection results. First, assays using synthetic reporter constructs with multimerized binding sites may not be representative of the regulation, which takes place in the complete regulatory HS7 region and involves interactions between proteins binding to adjacent elements. A second possible explanation is that, in our ex vivo studies, the vectors containing the mutations in the octamer sequences are transfected in cells which do not express Oct-1/Oct-2, the nuclear T-lymphoid factors B3/C3 or the PU.1 protein. In contrast, the Jurkat or U937 cells used in the infection studies contain Oct-1 and B3/C3 or Oct-1 and PU.1, respectively, thereby representing a more complex biological system. Third, transient transfection experiments may not reflect the regulation found in vivo with the intact provirus since transiently transfected DNA is not assembled into physiological chromatin. Similar discrepancies between transient transfection studies and in vivo functional studies have been previously reported for HIV by different groups including our laboratory (20,38–40). More specifically, regarding the HS7 Oct binding sites, the functional effect of the individual mutations in site B/Oct or site C/Oct could compensate each other; and the functional effect of the combined mutation site B/Oct+site C/Oct could be compensated by other factors binding to the HS7 region and still unidentified. In Jurkat cells, mutation of the octamer sequence of site B or site C does abolish not only the binding of Oct-1, but also the binding of the T-cell-specific factor B3/C3, which makes impossible to functionally distinguish in infection studies between the role of Oct-1 and the role of the T-cell factor. In U937 cells, mutation in the octamer sequence of site B abolishes the binding of Oct-1 but not the binding of PU.1. However, we can not exclude the possibility that the mutation of the Oct sequence might affect the binding of PU.1 in vivo. Indeed, our in vitro binding studies suggest that the binding of these two proteins to site B is mutually exclusive.
We have demonstrated that the PU.1 protein, which binds to site B, activates the transcriptional activity of the TK promoter through the HS7 site B. Therefore, in monocytes/macophages, the competition between Oct-1 and PU.1 both expressed in these cells for binding to their respective overlapping cognate sequences in the HS7 site B could determine the global contribution of site B to the transcriptional activity of the pol intragenic region. In T-cells, in which PU.1 is not expressed, such a competition between Oct-1 and the T-cell-specific factor (B3/C3) could similarly takes place in both site B and site C and could therefore determine the contribution of these two sites to the HS7 transcriptional activity. Previous publications have reported links between the Oct-1 protein and the HIV-1 transcriptional regulation. Indeed, Liu and Latchman (41) have previously identified four potential Oct binding sites in the HIV-1 LTR. In this latter study, Oct-1 has been shown to repress HIV-1 LTR promoter activity and its transactivation by Tat (41). A more recent study contrasts with the latter one and demonstrates that Oct-1 and Oct-2 fail to bind the HIV-1 LTR, and that overexpression of Oct proteins has no effect on HIV-1 transcription or replication in primary human CD4+ T-cells (42). Interestingly, the role of Oct-1 in HIV-1 transcription has also been indirectly highlighted by Bahr and co-workers. They have demonstrated that murabutide, which is a clinically acceptable immunomodulator presenting the capacity to decrease HIV-1 transcription (43,44), presents the ability to induce Oct-1 expression and DNA-binding activity in HIV-1 infected macrophages (45). These results suggest that the murabutide-induced suppression of HIV-1 transcription could be mediated, at least in part, by its capacity to increase Oct-1 expression (45). In the present study, we have demonstrated that the HIV-1 HS7 sites B and C bind Oct-1 and Oct-2 and confer transrepression by ectopically expressed Oct-1 and Oct-2 in reporter gene assays. Therefore, these results suggest that the murabutide-induced repression of HIV-1 transcription reported by Darcissac et al. (43) could be correlated to its capacity to increase the expression of Oct-1 and its subsequent binding to the HS7 sites B and C. Thus, further exploration of the role of Oct transcription factors in the activity of the HIV-1 LTR are warranted. Moreover, it would be interesting to study the relative contribution of the debated LTR Oct sites and of the HS7 Oct sites identified in this report to HIV-1 transcriptional regulation.
PU box binding site
We have demonstrated in vitro and in vivo that the HS7 site B is specifically bound by the PU.1 protein. PU.1 is an ETS family transcription factor that is highly expressed in B-lymphocytes, myeloid cells and immature erythrocytes (46,47). PU.1 binding motifs are found in a large number of B-lymphoid, myeloid and erythroid cell specific transcription elements (48). Using transient transfection assays, we have demonstrated that ectopic PU.1 protein has a site B PU box-dependent stimulatory effect on the heterologous TK promoter containing multiple upstream sites B. Consistently, we have demonstrated by infection studies in U937 cells that virus mutated in the HS7 site B PU box presents reduced growth kinetics when compared with a wild-type virus, thereby supporting the notion that PU.1 positively regulates the transcriptional activity of the HS7 region. The individual mutation of the site B PU box also reduces the efficiency of HIV-1 replication in the PU.1-negative Jurkat T cell line. It is worth noting that, although our EMSA experiments did not allow us to demonstrate in T-cells the binding of proteins to the PU box, we cannot exclude the possibility that this motif might be occupied by another protein in vivo. Another possible explanation is that some bases in the PU box could be important for the recruitment of co-factors for the Oct-1 protein whose binding sequence in site B is located just next to the PU box motif.
DNA regulatory regions in eukaryotic genomes frequently adopt a nuclease-hypersensitive configuration (49). This hypersensitivity is thought to result from the disruption of the packaging of DNA into nucleosomes by DNA-bound regulatory proteins. The absence of a hypersensitive site in the pol gene of HIV-1 integrated in two chronically infected T-cell lines (ACH2 and 8E5) could be due to differences in nucleotide sequence among HIV-1 isolates or to the presence of different regulatory factors in different cell types (lymphoid versus monocyte/macrophage cells). It is also possible that the pol region is nucleosome free in ACH2 and 8E5 cells but is occupied by bound factors in such a way as to adopt a configuration resistant to nuclease digestion. The ChIP assays reported here provide additional information on this issue and support the latter possibility. Indeed, these assays demonstrate the recruitment of Oct-1, Sp1 and Sp3 to the HS7 region in vivo both in T-lymphoid (ACH2) and in monocytic (U1) HIV-1-infected cells. Moreover, we have shown by ChIP assays that, in addition to Oct-1, Sp1 and Sp3, PU.1 also occupies the HS7 region in U1 cells and could therefore be involved in the nuclease hypersensitivity of the intragenic region in these cells. Indeed, studies on the B-cell-specific μ and κ3′ enhancers have shown that PU.1 is involved in chromatin remodeling events by functioning in some contexts as an accessibility factor (i.e. a protein capable of binding its site in the repressive context of chromatin, thereby increasing accessibility of the region) (50–52). Therefore, the presence in the pol gene of HIV-1 of a regulatory region containing a binding site for the macrophage and B-cell-specific factor PU.1 and associated with a monocyte-specific nuclease hypersensitive site suggests that PU.1 could act through the HS7 site B as an accessibility factor triggering the open chromatin configuration in the pol region. Further work including mutation and in vitro chromatin reconstruction studies is needed to investigate this hypothesis.
Although previous studies have demonstrated that the HIV-1 macrophage tropism is primarily determined at the level of the entry, several reports have highlighted additional determinants of macrophage tropism present at the level of transcription. Indeed, two motifs located in the HIV-1 LTR bind members of the C/EBP family of transcription factors and are required for virus replication in macrophages, but not T-cells (53–55). The tropism of other viruses for their target cells is determined in part at the transcriptional level. In murine leukemia viruses, a switch in tropism to T-cells was found to be caused by alterations in the U3 region of the LTR (56). The enhancer element of lymphotropic papovavirus, which contains a PU box required for its activity, contributes to the restricted tropism of the virus for primate B lymphocytes (57). The LTR enhancer of all known virulent strains of the equine infectious anemia virus (EIAV) contains three Ets binding motifs that interact with the transcription factor PU.1 and that are necessary for viral transcription in primary macrophages (58–60). Finally, our laboratory has identified a functional PU box in the bovine leukemia virus LTR (61).
Sp binding site
We have demonstrated by in vitro binding experiments that the HS7 GC-box is specifically bound by the Sp1 and Sp3 proteins. The recruitment of both proteins to the pol region was also demonstrated in vivo in the ACH2 and U1 cell lines. Using transient transfection assays, we have shown that ectopically expressed Sp1 and Sp3 have a positive effect on a luciferase reporter plasmid containing three copies of the HS7 Sp site cloned upstream of the heterologous promoter TK. In vivo, we have shown that the mutant virus containing the mutation in the HS7 Sp site demonstrates a reduced replicative phenotype similar to that observed with the totally mutated virus HIV-1-HS7totmut, thereby indicating a positive regulatory function of the Sp site.
The Sp family of transcription factors comprises eight members (Sp1–Sp8) [reviewed in (62,63)]. The most studied members of the Sp family, Sp1 and Sp3, are ubiquitously expressed but also fulfil distinct functions, as has been indicated by gene ablation studies (64,65). Sp1 is often described as a general activator of transcription, whereas Sp3 can act as an activator or as a repressor of Sp1-mediated activation, depending on the sequence context, the number of Sp binding sites and the availability of specific coactivators, corepressors or other transcription factors. However, the interplay between Sp1 and Sp3 in the regulation of specific promoters and the way the cells used the combination of these two related transcription factors to regulate gene expression is not clear. In this study, we have demonstrated that both Sp1 and Sp3 proteins have a HS7 Sp site-dependent stimulatory effect in the context of a heterologous promoter. In some systems including the HIV-1 LTR (13,66), Sp3 has been shown to cause repression of Sp1-mediated transcriptional activation. However, we could not observe such an inhibitory activity for TK transcription as shown in Drosophila SL2 cells transfected with both the Sp1 and Sp3 expression vectors (data not shown). Such a lack of repression of Sp1-mediated transcriptional activation by Sp3 has been previously reported for the IL-10 promoter regulation (67).
The Sp1 protein has been shown to mediate the formation of DNA loops between Sp1 proteins bound at two different sites on a DNA molecule (68–70). This DNA bending induced by Sp1 may play a direct role in the activation of transcription by bringing together factors bound at non-adjacent sites or facilitating binding of factors involved in the formation of an initiation complex. The presence of three tandemly arranged Spl sites in the HIV-1 5′-LTR and of one Spl site in the pol nuclease-hypersensitive site suggests a putative interaction between these elements. Such an association could bring in close proximity other factors bound to the 5′-LTR and to the pol regulatory element, and this arrangement could play a role in transcriptional regulation of the HIV-1 promoter. Similarly, Sp protein interactions could take place between the pol Sp site and the two juxtaposed Sp1 binding sites previously identified by our laboratory in the leader region positive transcriptional regulatory element (20).
Site D
We have previously published that site D specifically binds an ubiquitously expressed factor (7). In this report, despite many in silico and experimental attempts, we were unable to identify the factor(s) binding to site D. However, we have demonstrated that the binding of the ubiquitously expressed protein(s) to site D is Zn2+-dependent. This observation suggests that this (these) protein(s) could contain zinc finger domain(s). Our infection studies have demonstrated that a virus mutated in the HS7 site D replicates as efficiently as the wild-type control virus in the monocytic cell line U937. In contrast, in Jurkat T cells, the same mutant virus demonstrates reduced growth properties similar to those of the totally mutated virus HIV-1-HS7totmut, thereby demonstrating in vivo the biological significance of this site in the HIV-1 life cycle.
Since complex D is ubiquitous, this difference in terms of replication between the two cell types cannot be attributed to the presence or the absence of the D factor(s) in one cell line. However, numerous explanations have been proposed to explain how a ubiquitously expressed factor can participate in lineage-specific gene regulation. For instance, limited access of the ubiquitous factor to its cognate DNA sequence or post-translational modifications of the factors, such as phosphorylation, acetylation and glycosylation, have been reported as important for tissue-specific regulation. Moreover, protein–protein interactions between ubiquitous and cell-specific transcription factors could also contribute to cell type-specific transcriptional control.
We have thus demonstrated an important physiological role of the pol region in HIV-1 replication. The growth properties of a virus containing combined mutations abolishing nuclear factor binding to all the HS7 sites (site B, Sp site, site C and site D) are reduced when compared with those of a wild-type HIV-1. This reduced replicative phenotype is observed both in the Jurkat T-cell line and in the U937 monocyte/macrophage cell line and is indicative of a positive regulatory function for the pol intragenic region. No defect in RNA-packaging and no defect in the protein content of viral particles could be measured for any of the mutant viruses as determined by quantification of their HIV genomic RNA (by RNAse protection assay) and as determined by western blotting analysis, respectively. Therefore, our results suggest that the reduced replicative phenotypes of the HS7 mutant viruses are due to a decreased level of transcription as the result of point mutations in the HS7 region and that the cis-acting DNA elements we identified here, within this region, are required for optimal LTR transcriptional activity. Consistent with this hypothesis, our findings indicate that the loss of transcriptional enhancing activity of the pol regulatory region and the reduced replicative phenotype of HIV-1 caused by mutations in the HS7 binding sites correlate with the loss in factor binding to these sites.
Why does HIV-1 need two enhancers? During the course of infection, HIV-1 has to replicate in different cell types which can present various activation states. The presence of different transcriptional regulatory elements in the viral genome may suggest a viral strategy to be productive across several cellular environments with large differences in the pool of transcription factors. Indeed, it is estimated that more than 2000 transcription factors are encoded by the human genome (71,72). These factors are not equally expressed in all cell types, and many can be activated at specific stages of the cell developmental programme. The transcriptional environment of HIV-1 is therefore multidimensional, as the pool of transcription factors varies both temporally and spatially. Therefore, the redundancy and overlap in the transcription factor binding sites found in the HIV-1 genome could represent a solution for the virus to ensure its replication in several cellular environments. Our results demonstrate that the HS7 region is composed of multiple factor binding sites; some of these factors are ubiquitously expressed (Oct-1, Sp1/Sp3 and complex D), whereas others are cell-specific factors (PU.1, T-cell complexes B3/C3), supporting the notion that the HS7 transcriptional activity is determined by a combinatorial control: the combined action of tissue-restricted and ubiquitously expressed proteins. The HS7 regulatory region described in this report could either bring additional cellular specificity, or increase the strength of the promoter/enhancer unit located in the HIV-1 LTR, or allow viral responses to a broader variety of exogenous stimuli. Other viruses have been shown to possess intragenic regulatory regions. For example, the presence of two enhancer regions contributing to transcriptional regulation by adding a cellular specificity has been previously reported for the human hepatitis B virus and the closely related woodchuck hepatitis virus (73,74).
In conclusion, the pol positive cis-regulatory element located in the transcribed region of the HIV-1 genome brings an additional factor in an already complex network of regulators affecting the level of HIV-1 replication. Such a complexity could allow a finer-tuned regulation than a simple ‘On/Off’ switching mechanism; this fine-tuning might find its purpose when HIV-1 transcription needs to be moderately and/or transiently modified.
Acknowledgments
We thank Dr Guntram Suske (Philipps-Universitat, Marburg, Germany), Dr Winship Herr (Cold Spring Harbor Laboratory, New York, USA), Dr Ron Prywes (Columbia University, New York, USA) and Dr John Schwarz (University of Texas Medical School, Houston, USA) for reagents used in this study. We thank T. Folks, Malcolm Martin and Alfred Prince for reagents obtained through the AIDS Research and Reference Reagent Program. This work was supported by grants to C.V.L. from the ‘Fonds National de la Recherche Scientifique’ (FNRS, Belgium), the Télévie-Program, the ‘Université Libre de Bruxelles’ (ULB, ARC program no. 04/09-309), the Internationale Brachet Stiftung, the CGRI-INSERM cooperation, the Région Wallonne-Commission Européenne FEDER (Project Intergenes, program Interreg III), the Agence Nationale de Recherches sur le SIDA (ANRS, France), the Theyskens-Mineur Foundation, the ‘Fortis Banque Assurance’ and the ‘Fédération Belge contre le Cancer’. C.V.L. is ‘Maître de Recherches’ of the FNRS. V.G. is a fellow of the Belgian ‘Fonds pour la Recherche dans l'Industrie et l'Agriculture (FRIA)’. D.D. and S.dW. are supported by post-doctoral fellowships from the ‘Région Wallonne’ (Programs WALEO 021/5110 and 021/5347, respectively). Funding to pay the Open Access publication charges for this article was provided by the FNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barre-Sinoussi F., Chermann J.C., Rey F., Nugeyre M.T., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vezinet-Brun F., Rouzioux C., et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Gallo R.C., Salahuddin S.Z., Popovic M., Shearer G.M., Kaplan M., Haynes B.F., Palker T.J., Redfield R., Oleske J., Safai B., et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 3.Levy J.A., Hoffman A.D., Kramer S.M., Landis J.A., Shimabukuro J.M., Oshiro L.S. Isolation of lymphocytopathic retroviruses from San Francisco patients with AIDS. Science. 1984;225:840–842. doi: 10.1126/science.6206563. [DOI] [PubMed] [Google Scholar]
- 4.Varmus H. Regulation of HIV and HTLV gene expression. Genes Dev. 1988;2:1055–1062. doi: 10.1101/gad.2.9.1055. [DOI] [PubMed] [Google Scholar]
- 5.Verdin E., Becker N., Bex F., Droogmans L., Burny A. Identification and characterization of an enhancer in the coding region of the genome of human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1990;87:4874–4878. doi: 10.1073/pnas.87.12.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verdin E. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J. Virol. 1991;65:6790–6799. doi: 10.1128/jvi.65.12.6790-6799.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Lint C., Ghysdael J., Paras P., Jr, Burny A., Verdin E. A transcriptional regulatory element is associated with a nuclease-hypersensitive site in the pol gene of human immunodeficiency virus type 1. J. Virol. 1994;68:2632–2648. doi: 10.1128/jvi.68.4.2632-2648.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ott M., Emiliani S., Van Lint C., Herbein G., Lovett J., Chirmule N., McCloskey T., Pahwa S., Verdin E. Immune hyperactivation of HIV-1-infected T cells mediated by Tat and the CD28 pathway. Science. 1997;275:1481–1485. doi: 10.1126/science.275.5305.1481. [DOI] [PubMed] [Google Scholar]
- 9.Quivy V., Adam E., Collette Y., Demonte D., Chariot A., Vanhulle C., Berkhout B., Castellano R., de Launoit Y., Burny A., et al. Synergistic activation of human immunodeficiency virus type 1 promoter activity by NF-kappaB and inhibitors of deacetylases: potential perspectives for the development of therapeutic strategies. J. Virol. 2002;76:11091–11103. doi: 10.1128/JVI.76.21.11091-11103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calomme C., Nguyen T.L., de Launoit Y., Kiermer V., Droogmans L., Burny A., Van Lint C. Upstream stimulatory factors binding to an E box motif in the R region of the bovine leukemia virus long terminal repeat stimulates viral gene expression. J. Biol. Chem. 2002;277:8775–8789. doi: 10.1074/jbc.M107441200. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka M., Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 12.Klemsz M.J., McKercher S.R., Celada A., Van Beveren C., Maki R.A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 13.Hagen G., Muller S., Beato M., Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osborn L., Kunkel S., Nabel G.J. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc. Natl Acad. Sci. USA. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Quang C.T., Pironin M., von Lindern M., Beug H., Ghysdael J. Spi-1 and mutant p53 regulate different aspects of the proliferation and differentiation control of primary erythroid progenitors. Oncogene. 1995;11:1229–1239. [PubMed] [Google Scholar]
- 17.Han T.H., Prywes R. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol. Cell. Biol. 1995;15:2907–2915. doi: 10.1128/mcb.15.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firulli A.B., Miano J.M., Bi W., Johnson A.D., Casscells W., Olson E.N., Schwarz J.J. Myocyte enhancer binding factor-2 expression and activity in vascular smooth muscle cells. Association with the activated phenotype. Circ. Res. 1996;78:196–204. doi: 10.1161/01.res.78.2.196. [DOI] [PubMed] [Google Scholar]
- 19.Bosselut R., Duvall J.F., Gegonne A., Bailly M., Hemar A., Brady J., Ghysdael J. The product of the c-ets-1 proto-oncogene and the related Ets2 protein act as transcriptional activators of the long terminal repeat of human T cell leukemia virus HTLV-1. EMBO J. 1990;9:3137–3144. doi: 10.1002/j.1460-2075.1990.tb07511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Lint C., Amella C.A., Emiliani S., John M., Jie T., Verdin E. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J. Virol. 1997;71:6113–6127. doi: 10.1128/jvi.71.8.6113-6127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laspia M.F., Wendel P., Mathews M.B. HIV-1 Tat overcomes inefficient transcriptional elongation in vitro. J. Mol. Biol. 1993;232:732–746. doi: 10.1006/jmbi.1993.1427. [DOI] [PubMed] [Google Scholar]
- 22.Hatzopoulos A.K., Stoykova A.S., Erselius J.R., Goulding M., Neuman T., Gruss P. Structure and expression of the mouse Oct2a and Oct2b, two differentially spliced products of the same gene. Development. 1990;109:349–362. doi: 10.1242/dev.109.2.349. [DOI] [PubMed] [Google Scholar]
- 23.Stoykova A.S., Sterrer S., Erselius J.R., Hatzopoulos A.K., Gruss P. Mini-Oct and Oct-2c: two novel, functionally diverse murine Oct-2 gene products are differentially expressed in the CNS. Neuron. 1992;8:541–558. doi: 10.1016/0896-6273(92)90282-i. [DOI] [PubMed] [Google Scholar]
- 24.Ihn H., LeRoy E.C., Trojanowska M. Oncostatin M stimulates transcription of the human alpha2(I) collagen gene via the Sp1/Sp3-binding site. J. Biol. Chem. 1997;272:24666–24672. doi: 10.1074/jbc.272.39.24666. [DOI] [PubMed] [Google Scholar]
- 25.Ihn H., Trojanowska M. Sp3 is a transcriptional activator of the human alpha2(I) collagen gene. Nucleic Acids Res. 1997;25:3712–3717. doi: 10.1093/nar/25.18.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J.M., Datto M.B., Shen X., Hu P.P., Yu Y., Wang X.F. Sp1, but not Sp3, functions to mediate promoter activation by TGF-beta through canonical Sp1 binding sites. Nucleic Acids Res. 1998;26:2449–2456. doi: 10.1093/nar/26.10.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brightbill H.D., Plevy S.E., Modlin R.L., Smale S.T. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J. Immunol. 2000;164:1940–1951. doi: 10.4049/jimmunol.164.4.1940. [DOI] [PubMed] [Google Scholar]
- 28.Malim M.H., Hauber J., Fenrick R., Cullen B.R. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature. 1988;335:181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- 29.Pereira L.A., Bentley K., Peeters A., Churchill M.J., Deacon N.J. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000;28:663–668. doi: 10.1093/nar/28.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herr W., Sturm R.A., Clerc R.G., Corcoran L.M., Baltimore D., Sharp P.A., Ingraham H.A., Rosenfeld M.G., Finney M., Ruvkun G., et al. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 31.Scholer H.R., Hatzopoulos A.K., Balling R., Suzuki N., Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 1989;8:2543–2550. doi: 10.1002/j.1460-2075.1989.tb08392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cockerill P.N., Klinken S.P. Octamer-binding proteins in diverse hemopoietic cells. Mol. Cell. Biol. 1990;10:1293–1296. doi: 10.1128/mcb.10.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheidereit C., Cromlish J.A., Gerster T., Kawakami K., Balmaceda C.G., Currie R.A., Roeder R.G. A human lymphoid-specific transcription factor that activates immunoglobulin genes is a homoeobox protein. Nature. 1988;336:551–557. doi: 10.1038/336551a0. [DOI] [PubMed] [Google Scholar]
- 34.Staudt L.M., Clerc R.G., Singh H., LeBowitz J.H., Sharp P.A., Baltimore D. Cloning of a lymphoid-specific cDNA encoding a protein binding the regulatory octamer DNA motif. Science. 1988;241:577–580. doi: 10.1126/science.3399892. [DOI] [PubMed] [Google Scholar]
- 35.Sturm R.A., Das G., Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 36.Zwilling S., Dieckmann A., Pfisterer P., Angel P., Wirth T. Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science. 1997;277:221–225. doi: 10.1126/science.277.5323.221. [DOI] [PubMed] [Google Scholar]
- 37.Wysocka J., Herr W. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 2003;28:294–304. doi: 10.1016/S0968-0004(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 38.Harrich D., Hsu C., Race E., Gaynor R.B. Differential growth kinetics are exhibited by human immunodeficiency virus type 1 TAR mutants. J. Virol. 1994;68:5899–5910. doi: 10.1128/jvi.68.9.5899-5910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J.Y., Gonzalez-Scarano F., Zeichner S.L., Alwine J.C. Replication of type 1 human immunodeficiency viruses containing linker substitution mutations in the −201 to −130 region of the long terminal repeat. J. Virol. 1993;67:1658–1662. doi: 10.1128/jvi.67.3.1658-1662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeichner S.L., Kim J.Y., Alwine J.C. Linker-scanning mutational analysis of the transcriptional activity of the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 1991;65:2436–2444. doi: 10.1128/jvi.65.5.2436-2444.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y.Z., Latchman D.S. The octamer-binding proteins Oct-1 and Oct-2 repress the HIV long terminal repeat promoter and its transactivation by Tat. Biochem. J. 1997;322:155–158. doi: 10.1042/bj3220155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M., Genin A., Cron R.Q. Overexpression of octamer transcription factors 1 or 2 alone has no effect on HIV-1 transcription in primary human CD4 T cells. Virology. 2004;321:323–331. doi: 10.1016/j.virol.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Darcissac E.C., Truong M.J., Dewulf J., Mouton Y., Capron A., Bahr G.M. The synthetic immunomodulator murabutide controls human immunodeficiency virus type 1 replication at multiple levels in macrophages and dendritic cells. J. Virol. 2000;74:7794–7802. doi: 10.1128/jvi.74.17.7794-7802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bahr G.M., Darcissac E.C., Casteran N., Amiel C., Cocude C., Truong M.J., Dewulf J., Capron A., Mouton Y. Selective regulation of human immunodeficiency virus-infected CD4+ lymphocytes by a synthetic immunomodulator leads to potent virus suppression in vitro and in hu-PBL-SCID mice. J. Virol. 2001;75:6941–6952. doi: 10.1128/JVI.75.15.6941-6952.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truong M.J., Delsart V., Bahr G.M. Differentially expressed genes in HIV-1-infected macrophages following treatment with the virus-suppressive immunomodulator murabutide. Virus Res. 2004;99:25–33. doi: 10.1016/j.virusres.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 46.McKercher S.R., Torbett B.E., Anderson K.L., Henkel G.W., Vestal D.J., Baribault H., Klemsz M., Feeney A.J., Wu G.E., Paige C.J., et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 47.Scott E.W., Simon M.C., Anastasi J., Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 48.Tenen D.G., Hromas R., Licht J.D., Zhang D.E. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 49.Wolffe A. Chromatin: Structure and Function. San Diego, CA: Academic Press; 2005. [Google Scholar]
- 50.Nelsen B., Tian G., Erman B., Gregoire J., Maki R., Graves B., Sen R. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- 51.Pongubala J.M., Nagulapalli S., Klemsz M.J., McKercher S.R., Maki R.A., Atchison M.L. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3′ enhancer activity. Mol. Cell. Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marecki S., McCarthy K.M., Nikolajczyk B.S. PU.1 as a chromatin accessibility factor for immunoglobulin genes. Mol. Immunol. 2004;40:723–731. doi: 10.1016/j.molimm.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Henderson A.J., Zou X., Calame K.L. C/EBP proteins activate transcription from the human immunodeficiency virus type 1 long terminal repeat in macrophages/monocytes. J. Virol. 1995;69:5337–5344. doi: 10.1128/jvi.69.9.5337-5344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henderson A.J., Connor R.I., Calame K.L. C/EBP activators are required for HIV-1 replication and proviral induction in monocytic cell lines. Immunity. 1996;5:91–101. doi: 10.1016/s1074-7613(00)80313-1. [DOI] [PubMed] [Google Scholar]
- 55.Henderson A.J., Calame K.L. CCAAT/enhancer binding protein (C/EBP) sites are required for HIV-1 replication in primary macrophages but not CD4+ T cells. Proc. Natl Acad. Sci. USA. 1997;94:8714–8719. doi: 10.1073/pnas.94.16.8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Celander D., Haseltine W.A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984;312:159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- 57.Erselius J.R., Jostes B., Hatzopoulos A.K., Mosthaf L., Gruss P. Cell-type-specific control elements of the lymphotropic papovavirus enhancer. J. Virol. 1990;64:1657–1666. doi: 10.1128/jvi.64.4.1657-1666.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hines R., Sorensen B.R., Shea M.A., Maury W. PU.1 binding to ets motifs within the equine infectious anemia virus long terminal repeat (LTR) enhancer: regulation of LTR activity and virus replication in macrophages. J. Virol. 2004;78:3407–3418. doi: 10.1128/JVI.78.7.3407-3418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maury W. Monocyte maturation controls expression of equine infectious anemia virus. J. Virol. 1994;68:6270–6279. doi: 10.1128/jvi.68.10.6270-6279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maury W., Perryman S., Oaks J.L., Seid B.K., Crawford T., McGuire T., Carpenter S. Localized sequence heterogeneity in the long terminal repeats of in vivo isolates of equine infectious anemia virus. J. Virol. 1997;71:4929–4937. doi: 10.1128/jvi.71.7.4929-4937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dekoninck A., Calomme C., Nizet S., de Launoit Y., Burny A., Ghysdael J., Van Lint C. Identification and characterization of a PU.1/Spi-B binding site in the bovine leukemia virus long terminal repeat. Oncogene. 2003;22:2882–2896. doi: 10.1038/sj.onc.1206392. [DOI] [PubMed] [Google Scholar]
- 62.Li L., He S., Sun J.M., Davie J.R. Gene regulation by Sp1 and Sp3. Biochem. Cell Biol. 2004;82:460–471. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- 63.Bouwman P., Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol. Cell. Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- 64.Gollner H., Dani C., Phillips B., Philipsen S., Suske G. Impaired ossification in mice lacking the transcription factor Sp3. Mech. Dev. 2001;106:77–83. doi: 10.1016/s0925-4773(01)00420-8. [DOI] [PubMed] [Google Scholar]
- 65.Bouwman P., Gollner H., Elsasser H.P., Eckhoff G., Karis A., Grosveld F., Philipsen S., Suske G. Transcription factor Sp3 is essential for post-natal survival and late tooth development. EMBO J. 2000;19:655–661. doi: 10.1093/emboj/19.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rohr O., Aunis D., Schaeffer E. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J. Biol. Chem. 1997;272:31149–31155. doi: 10.1074/jbc.272.49.31149. [DOI] [PubMed] [Google Scholar]
- 67.Tone M., Powell M.J., Tone Y., Thompson S.A., Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J. Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 68.Sjottem E., Andersen C., Johansen T. Structural and functional analyses of DNA bending induced by Sp1 family transcription factors. J. Mol. Biol. 1997;267:490–504. doi: 10.1006/jmbi.1997.0893. [DOI] [PubMed] [Google Scholar]
- 69.Mastrangelo I.A., Courey A.J., Wall J.S., Jackson S.P., Hough P.V. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc. Natl Acad. Sci. USA. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su W., Jackson S., Tjian R., Echols H. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 71.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 72.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 73.Moolla N., Kew M., Arbuthnot P. Regulatory elements of hepatitis B virus transcription. J. Viral Hepat. 2002;9:323–331. doi: 10.1046/j.1365-2893.2002.00381.x. [DOI] [PubMed] [Google Scholar]
- 74.Ueda K., Wei Y., Ganem D. Cellular factors controlling the activity of woodchuck hepatitis virus enhancer II. J. Virol. 1996;70:4714–4723. doi: 10.1128/jvi.70.7.4714-4723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]