Abstract
Many approaches were made in recent years to establish urine PCR as a diagnostic tool for Lyme borreliosis, but results are contradictory. In the present study, a standardized protocol spiking urine from healthy donors with a defined amount of whole Borrelia or Borrelia DNA was established. The development of a nested real-time PCR targeting ospA enabled a highly sensitive and quantitative analysis of these samples. We show the following. (i) Storage of spiked urine samples for up to 6 months at −20°C had no negative effect on spike recovery. (ii) Centrifugation of 10 ml of urine at 40,000 × g for 30 min resulted in a concentration of both spikes, i.e., whole Borrelia and DNA. (iii) The inhibition of DNA spike recovery in 48% (11 of 23 samples) of urine samples tested could be attributed to nuclease activity. This was abrogated by alkalizing the urine or by working with the samples on ice. Despite optimized conditions, analysis of urine samples of 12 patients with erythema migrans, the clinical stage considered to be associated with the highest bacterial load, revealed a positive result in only one sample. All 12 samples were negative by an alternative PCR targeting flagellin. The results of our study support doubts that urine is a suitable material for diagnosis of Lyme borreliosis.
Diagnosis of Lyme borreliosis (LB) is currently based on clinical criteria confirmed by serological tests. However, serology has a number of limitations, such as serological cross-reactivity, the delayed production of antibodies in early stages of LB, and an inability to distinguish between ongoing and previous infections, due to the persistence of antibodies. In patients with ongoing or recurring symptoms after antibiotic therapy, it is often impossible to definitely attribute clinical symptoms to a persistent infection with Borrelia burgdorferi sensu lato, the causative agent of LB. A way out of this dilemma might be the direct detection of this pathogen in clinical samples, such as the detection of Borrelia DNA by PCR. The use of urine as a sample material is an attractive approach, since urine is obtained without invasive procedures.
Since 1991, when the detection of Borrelia DNA in urine was shown for the first time by Goodman et al. (14), many studies have been carried out (for reviews, see references 11, 19, and 29). A comparison between the different reports is nearly impossible because of significant differences in patient selection, study design, and DNA extraction and PCR methods. Nevertheless, a meta-analysis of urine PCR assays for diagnosis of LB showed an overall sensitivity of 68% (range, 13 to 100%) and a specificity of 99% (range, 95 to 100%) (11).
Even when comparing only reports on urine of patients with erythema migrans (EM) who clearly have an active infection, detection of Borrelia DNA in urine ranged between 13% and >90% (1, 5, 20, 25, 28, 30). A caution was issued by Brettschneider et al., who were not able to confirm the presence of Borrelia DNA in the 27% PCR-positive urine specimens by hybridization or sequencing (7). Thus, despite all efforts, the value of PCR as a diagnostic tool for LB and of urine as a sample material remains unclear and is controversial (7, 12, 30).
Until now, it has not been clear whether urine of LB patients contains whole Borrelia organisms or Borrelia DNA. There is accumulating evidence that the urinary tract is a site of persistent infection in most animals, since various studies have demonstrated the presence of spirochetes in the urinary bladder tissue in experimentally and naturally infected animals (13, 15, 21, 32). However, culture of spirochetes from animal urine failed (3, 16, 32) or was successful only very rarely (13). Maiwald et al. postulated that soluble or protein-bound DNA is excreted in the urine, not whole Borrelia organisms (24), as the Geneclean kit worked best for sample preparation. However, this conclusion was based on the false premise that this procedure involves no protein denaturing, but the chaotrophic salt NaI was added for sample preparation.
Only few studies quantified Borrelia burgdorferi sensu lato DNA in clinical specimens: the number of spirochetes in a 2-mm skin biopsy of 50 patients with EM ranged from 10 to 10,000, with a median of 1,450 spirochetes (22); in five synovial fluid samples, the number varied from 20 to 41,000 per ml (31). For urine, no proper quantification is available, but Goodman et al. calculated an equivalent of 50 to 5,000 organisms per ml of urine by extrapolation of signal intensities (14). Hence, the number of spirochetes in clinical specimens seems to be rather low, and procedures that concentrate the number of organisms in a given sample and that display high sensitivity are imperative.
Furthermore, urine appears to contain inhibitory factors. Urea inhibits PCR in concentrations of ≥50 mM (4, 18). Given that the normal concentration of urea in adult urine is about 330 mM, it is clear that the method chosen for DNA extraction plays an important role. Until now, this inhibitory effect has only been analyzed on a qualitative, not a quantitative, basis. In the present study, we establish an optimized, quantitative, nested real-time PCR to determine the diagnostic potential of PCR with urine samples spiked with whole Borrelia organisms and Borrelia DNA and apply this method to patient samples.
MATERIALS AND METHODS
Isolation of DNA.
DNA extraction was performed as previously described (5). Briefly, 1 ml of DNAzol (Invitrogen, Karlsruhe, Germany) and 6 μl of a polyacryl carrier (Fermentas, St. Leon-Rot, Germany) were added to each sample. DNA was precipitated with ethanol, centrifuged at 17,500 × g for 20 min, and washed twice with 70% ethanol, according to the manufacturer's instructions. The final pellet was dissolved in 100 μl Tris-EDTA (TE) buffer. A total of 10 μl was used as a DNA template for PCR.
Nested real-time PCR.
For amplification of B. burgdorferi sensu lato DNA, a nested PCR system with primer pairs targeting the outer surface protein A (ospA) was established as a standard PCR, followed by a real-time PCR. The first PCR amplification was performed in a volume of 50 μl containing 2 mM MgCl2, 0.2 μM each primer, 1× PCR buffer, and 0.2 mM desoxynucleoside triphosphate (all from PE Applied Biosystems, Weiterstadt, Germany); 1 U of Taq DNA polymerase (Eppendorf, Hamburg, Germany); and 10 μl of extracted DNA. The sequence of the outer primer pair (Thermo Hybaid, Ulm, Germany) was 5′ ATGAAAAAATATTTATTGGG and 3′ CTGTTGATGACTTGTCTTT, resulting in a 347-bp product. PCR was performed with a GeneAmp PCR System 2400 (PE Applied Biosystems) using the following protocol: initial denaturation at 95°C for 2 min, followed by 20 cycles of denaturation, each cycle consisting of 95°C for 30 s, annealing at 50°C for 40 s, elongation at 72°C for 18 s; there was a final extension at 72°C for 10 min. After amplification, 1 μl of the first PCR served as template for the second PCR, which was performed as real-time PCR with a LightCycler rapid thermal cycler system (Roche Diagnostics GmbH, Mannheim, Germany). Real-time PCR was performed as described previously (27). The use of the LightCycler software 3.5 enabled quantification by import of an external standard curve. An alternative PCR was performed according to Schmidt et al., with primers targeting the B. burgdorferi sensu lato flagellin gene and a detection limit of <5 Borrelia per PCR (i.e., <0.5 Borrelia/μl) (28).
In each step (DNA extraction and outer and inner PCR), negative controls containing water instead of DNA were included to exclude cross-contamination. Borrelia afzelii strain VS461 was used as a positive control. Positive controls contained 5 Borrelia organisms per μl of template in the first PCR and 100 Borrelia organisms per μl of template in the real-time PCR. To avoid cross-contamination and sample carryover, pre- and post-PCR sample processing and PCR amplification were performed in separate rooms; cotton-plugged pipette tips were generally used.
Treatment of patient urine samples.
Midstream urine samples from 12 patients (Table 1) with clinically diagnosed EM were collected by a general practitioner, working in a region endemic for borreliosis. EM was present at the time of urine collection. Urine samples were obtained before antibiotic therapy and frozen immediately at −20°C. The samples were transported frozen and stored at −20°C until use. Urine samples were thawed at 4°C overnight, and 10 ml was centrifuged at 40,000 × g and 4°C for 30 min. DNA extraction was performed with the pellet, as described above.
TABLE 1.
Patient characteristics
| Patient no. | Age (yr) | Sexb | Tick bite recall | Size of EM (cm) | IgM ELISA | Urine
|
|
|---|---|---|---|---|---|---|---|
| pH | Specific density | ||||||
| 1 | 48 | F | No | 8 | − | 6.5 | 1.010 |
| 2 | 67 | M | Yes | 10 | − | 7.0 | 1.015 |
| 3 | 10 | M | Yes | 10 | − | 7.5 | 1.010 |
| 4 | 30 | M | Yes | 8 | − | 8.0 | 1.010 |
| 5 | 38 | F | Yes | 6 | − | 7.0 | 1.005 |
| 6 | 49 | M | Yes | 10 | + | 6.0 | 1.030 |
| 7a | 56 | F | No | 4 × 6 to 8 | + | 6.5 | 1.015 |
| 8 | 59 | F | No | 15 | + | 5.5 | 1.020 |
| 9a | 21 | M | No | 5 × 5 to 8 | + | 6.0 | 1.020 |
| 10 | 55 | F | Yes | 10 | − | 5.5 | 1.015 |
| 11 | 59 | F | No | 50 | + | 5.5 | 1.005 |
| 12 | 27 | F | No | 12 | + | 6.5 | 1.005 |
These patients showed four and five multiple EM lesions, respectively.
F, female; M, male.
EM was diagnosed based on the following criteria. (i) EM had to be an erythema of the skin (without epidermal involvement) of at least 5 cm in size. (ii) The time interval between tick bite (if remembered) and appearance of the EM at the site of the tick bite lasted at least 7 days.
Inhibition assay.
To exclude inhibition of the PCR, all patient samples were analyzed as follows. A total of 100 copies of Chlamydia pneumoniae DNA were added to the extracted DNA of the patient urine samples. PCR was performed according to Mueller et al. (26).
Spiked urine samples.
The efficacy of the DNA preparation was evaluated by extraction and amplification of experimentally spiked urine or water. Midstream urine specimens were collected from healthy donors. If not explicitly mentioned, 500 whole Borrelia organisms or 1 pg of DNA (equivalent to 500 Borrelia organisms) were added to 50 μl of urine or water. The extracted DNA was eluted in a volume of 100 μl TE, resulting in a theoretical concentration of 5 Borrelia equivalents per μl of eluate.
For spike experiments, B. afzelii strain VS461 was used. It was cultured as described previously (10) and was kindly provided by T. Kamradt (Berlin, Germany). Culture density was determined by microscopy using a modified Thoma counting chamber (Merck Eurolab, Ismaning, Germany). DNA from the Borrelia culture was extracted with the DNeasy tissue kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. DNA was quantified photometrically (BioPhotometer, Eppendorf, Germany). A total of 2 fg of DNA was regarded as equivalent to one Borrelia organism (20).
Detection of nuclease activity.
For detection of nuclease activity in urine, 500 ng of the plasmid DNA (pJuFo [8]) was incubated at room temperature with urine or water. After different incubation times, the DNA was resolved on 1% agarose gels by electrophoresis and visualized under UV light with ethidium bromide.
Chemical characterization of urine.
Urine samples were characterized with test strips (Multistix 10SG, Bayer, Fernwald, Germany), a urea kit (Urea Nitrogen, Sigma Diagnostics, Deisenhofen, Germany), and a magnesium kit (Magnesium Merckotest, Merck, Darmstadt, Germany) according to the manufacturer's instructions.
RESULTS
Nested PCR of Borrelia.
We previously described a real-time PCR, which offers the advantage of quantification and differentiation of Borrelia species (27). As only small amounts of Borrelia DNA are expected in clinical samples, the real-time PCR was modified to a nested PCR, by preceding it with a 20-cycle standard PCR. With this modification, we were still able (i) to distinguish between the different Borrelia species (B. afzelii, Borrelia garinii, and B. burgdorferi sensu stricto) by melting curve analysis (data not shown) and (ii) to carry out a quantitative analysis. Starting with 31.6 Borrelia organisms per μl, DNA was diluted in steps of 1:3.16 in triplicates. The nested PCR decreased the detection limit of our original real-time PCR (27) by a factor of 10 to 0.1 to 1 Borrelia per μl (i.e., 1 to 10 Borrelia per 10 μl of template employed in the standard PCR) (data not shown).
Since it is not clear whether urine of LB patients contains Borrelia DNA or whole Borrelia organisms, most of the following experiments were carried out with both. To eliminate possible inhibitors of PCR from the urine, we performed DNA extraction using a guanidin-detergent lysing solution (DNAzol) combined with a polyacrylamide carrier, as recently described by Bergmann et al. as the only DNA extraction method resulting in positive results in urine samples from LB patients (5).
To determine the variance of our nested real-time PCR between different experiments, we determined the standard deviation of the threshold cycle numbers (CT values) of positive controls from 16 independent experiments. Positive controls of DNAzol extraction, which ran through the entire procedure, showed a standard deviation of CT values of 0.81.
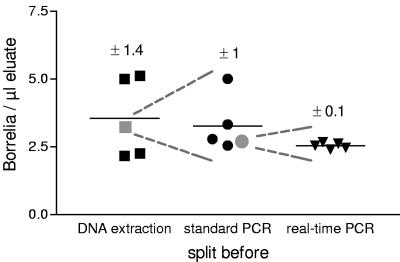
Variance within one experiment was determined by fivefold preparation of a positive control (containing 500 Borrelia organisms). Before each step (DNAzol, block PCR, and LC PCR, respectively) one of the five samples was split into five replicates and treated as separate samples in the next steps (Fig. 1). The coefficients of variation of the samples of each step were 40, 32, and 4% for DNA extraction, standard PCR, and real-time PCR, respectively. These data suggest that DNA extraction contributes more to the variance of results than PCR.
FIG. 1.
Variance within one experiment. DNA of five identical samples, each containing 500 Borrelia equivalents, was extracted with DNAzol. After DNA extraction, one of the five samples was split into five parts, and the resulting nine samples were inserted in the standard PCR. Before real-time PCR was performed, one of the samples was again split into five parts. All 13 samples were quantified in the same real-time PCR run. The means of the split samples are depicted as gray symbols.
Spike experiments.
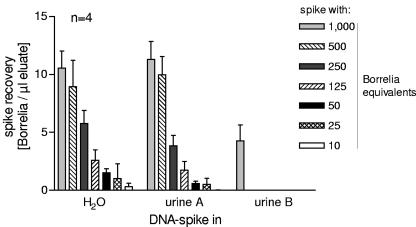
To investigate whether there is inhibition of PCR by urine components, dilution series of Borrelia and Borrelia DNA in water were compared to dilution series in urine samples of two healthy donors (urine samples A and B). A total of 50 μl water or urine was spiked with different amounts of DNA. DNAzol was added after an incubation time of 10 min at room temperature. After extraction with DNAzol, DNA was eluted in 100 μl of TE. Nested real-time PCR was performed, and the spike recovery was determined. No difference was seen between the detection limit of Borrelia DNA in water and urine sample A. In both cases, the recovery of a spike of 10 Borrelia equivalents (i.e., 0.1 Borrelia organisms per μl eluate) was possible. However, in urine sample B, only the highest spike (i.e., 1,000 Borrelia equivalents or 10 Borrelia organisms per μl eluate) could be detected (Fig. 2). This inhibitory effect was not seen when urine B was spiked with whole Borrelia organisms instead of DNA (data not shown), indicating that inhibition was not due to direct inhibition of the PCR by components of the urine transferred into the PCR.
FIG. 2.
Dilution series of Borrelia DNA in water or urine samples (A and B) from two healthy donors. DNA equivalents of 1,000, 500, 250, 100, 25, and 10 Borrelia organisms were added to 50 μl of water or urine, which should result in concentrations of 10 to 0.1 Borrelia equivalents per μl of eluate. Data are depicted as means ± standard errors of the mean from quadruplicate experiments.
To investigate whether the inhibitory effect on DNA recovery could also be seen in other urine samples, we spiked urine from 23 healthy donors with 500 Borrelia equivalents of DNA each. The extracted DNA was eluted in a volume of 100 μl, resulting in a theoretical concentration of five Borrelia organisms per μl eluate. Three aliquots of each urine sample were spiked, and DNA was extracted independently. Definition of one Borrelia organism per μl eluate as the cutoff (corresponding to a 20% recovery of the spike) revealed inhibitory effects in 11 of 23 (48%) urine samples (data not shown).
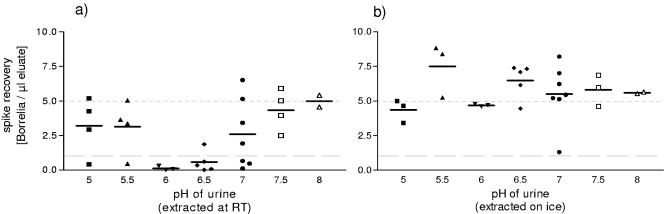
Since it is well known that one of the main inhibitors of PCR in urine is urea (4, 18), we measured the urea concentration of every urine sample and compared it to the respective spike recovery rate, but no correlation was evident (data not shown). This result was not surprising, as DNA extraction with DNAzol could eliminate even high concentrations of urea, as shown by spiking urea solutions (0 mM to 1 M) with Borrelia DNA. Either these solutions were employed directly in the PCR or the DNA was extracted with DNAzol before PCR. Whereas in the first case an inhibitory effect of the urea was already seen at 50 mM, extraction with DNAzol enabled spike recovery even at 1 M urea (data not shown). In contrast to other characteristics of urine, such as high amounts of phosphate, magnesium, or protein or a high osmolality, which did not correlate with disturbed spike recovery (data not shown), the inhibitory effects of urine were strongest at a pH between 6 and 6.5 (Fig. 3a).
FIG. 3.
(a) pH dependence of spike recovery. A total of 29 urine samples from 23 donors were spiked with DNA of 500 Borrelia organisms, and the spike recovery was correlated to the pH of the respective urine. (b) The same approach as in panel a is shown, with 27 urine samples; the exception is that the urine samples were kept on ice until DNAzol was added.
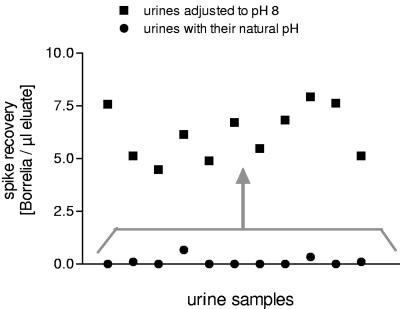
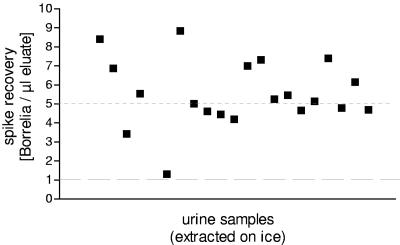
The inhibitory influence of pH 6 or 6.5 could be abrogated by adjusting its pH to 8 with NaOH before the spike was added. In each of the 11 samples tested, this resulted in a positive spike recovery (Fig. 4). In addition, the inhibitory effect was no longer observed if urine samples were kept on ice until DNAzol was added (Fig. 5). Under these conditions, the influence of pH disappeared (Fig. 3b).
FIG. 4.
pH influence on spike recovery. The spike recovery of 11 inhibitory urine samples was compared to the recovery in the same urine samples adjusted to a pH of 8.
FIG. 5.
Spike recovery in 20 of the 29 urine samples shown in Fig. 3a. The experiment was performed as described in the legend to Fig. 3a, with the exception that urine was left on ice until DNAzol was added.
These results indicated that active nucleases, known to exist in urine (17), were responsible for the inhibitory effect on DNA spike recovery. To prove this, purified plasmid DNA was incubated with an inhibitory urine sample for 1 h at room temperature. Complete degradation of the plasmid DNA occurred as seen by ethidium bromide staining, which did not occur if the pH of the urine was shifted to 8 or if bivalent ions were chelated with 10 mM EDTA before the plasmid DNA was added (data not shown). Both experiments substantiate nuclease activity in the original urine sample. The intensity of degradation of plasmid DNA could be correlated with the level of spike recovery. This was demonstrated in three different inhibitory urines and three urines with good spike recovery (data not shown).
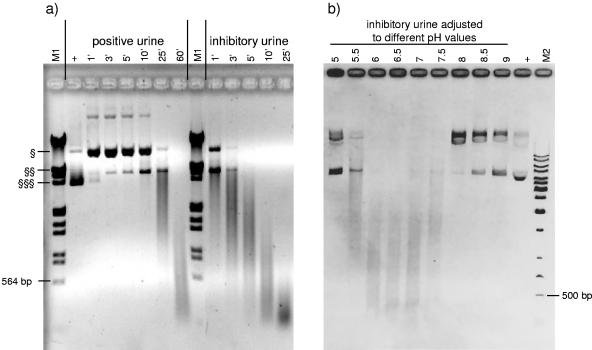
Figure 6 demonstrates the time (Fig. 6a) and pH (Fig. 6b) dependency of nuclease activity. In experiments shown in Fig. 6a, plasmid DNA was incubated with two urine samples (one urine with a good spike recovery and one strongly inhibitory urine) for different times at room temperature. The degradation of plasmid DNA incubated with a urine sample with good spike recovery started after 25 min, whereas degradation of DNA incubated with the inhibitory urine sample could be seen immediately. To demonstrate pH dependency, the inhibitory urine sample was adjusted to pH values between 5 and 9 before plasmid DNA was added and incubated for 10 min at room temperature. The gel shows that the greatest degradation effect occurred in urine with a pH between 6 and 7. No degradation was seen in urine with pH 5 or pH ≥8 (Fig. 6b).
FIG. 6.
(a) Time dependency of nuclease activity. pJuFo plasmid was incubated with a previously tested urine with excellent spike recovery (positive urine) and an inhibitory urine sample for different times (given in minutes) at room temperature. EcoRI/HindIII-digested lambda DNA was used as a marker (M1). (b) pH dependency of nuclease activity. pJuFo plasmid was incubated for 10 min at room temperature with an inhibitory urine which was adjusted to different pH values. The marker is a 1-kb ladder (M2). As a positive control (+), water instead of urine was spiked with plasmid. §, relaxed circular plasmid; §§, linearized plasmid; and §§§, supercoiled form of plasmid.
Storage of urine.
Bergmann et al. (5) reported that storage of frozen urine samples for more than 3 months increased the inhibitory nature of the urine. We tried to assess this observation quantitatively. We froze triplicates of spiked urine samples (50 μl urine, which was inhibitory in previous tests, spiked with 500 Borrelia DNA or whole Borrelia organisms) at different time points over a 6-month period. All aliquots were thawed on ice to avoid nuclease activity, extracted, and amplified in parallel. No significant difference in spike recovery was seen (i) between the spiked water and urine, (ii) over time, or (iii) between Borrelia DNA and whole Borrelia organisms (data not shown). Thus, there is no indication for a decline in Borrelia organism or Borrelia DNA recovery within a 6-month observation period in our experimental setting.
Effect of centrifugation.
Many publications addressing urine PCR try to concentrate samples before DNA extraction by centrifugation of several milliliters of urine (5, 9, 23, 30), but so far it has not been proven whether centrifugation really improves the sensitivity. To examine a possible effect of centrifugation quantitatively, two 10-ml aliquots, each of different urine samples or water, were spiked with 5,000 Borrelia equivalents (Fig. 7a) or 5,000 whole Borrelia organisms (Fig. 7b), respectively.
FIG. 7.
Effect of urine centrifugation. A total of 10 ml of urine or water was spiked with 5,000 equivalents of Borrelia DNA (16 different urines) or 5,000 whole Borrelia organisms (11 different urine samples). Either 50 μl of 10 ml spiked urine or water was submitted to DNA extraction (A) or 10 ml urine or water was centrifuged at 40,000 × g for 30 min; the pellet was then submitted to DNA extraction (B).
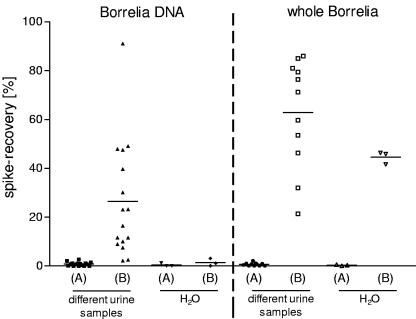
One of the two aliquots was centrifuged for 30 min at 40,000 × g and at 4°C. The supernatant was discarded as completely as possible, and the pellet was submitted to DNA extraction. The pellet was stored on ice until the addition of DNAzol. Assuming a yield of 100% of the initial value, one should find 50 Borrelia equivalents per μl of eluate after DNA extraction. From the second aliquot, 50 μl was submitted directly to DNA extraction. These aliqiots should contain 25 Borrelia equivalents, resulting in 0.25 Borrelia equivalents/μl of eluate. Figure 7 shows the positive effect of centrifugation on all urine samples tested, for detection of both DNA and whole Borrelia organisms. No such concentration effect was seen when water instead of urine was spiked with DNA. It is remarkable that the concentration effect was highly different in the various urine samples. By repeating the experiment with similar results (same rank order), we showed that this effect did not stem from the sample preparation but rather from variations in the different urine samples. Compared to samples spiked with DNA, centrifugation of samples spiked with whole Borrelia organisms showed a greater concentration effect in all cases.
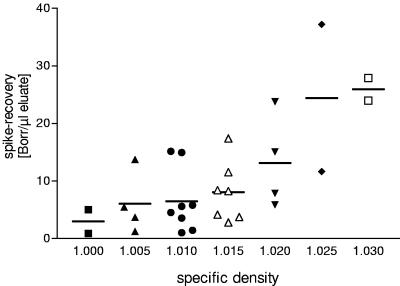
Comparison of the spike recovery with the specific density of the respective urine showed a positive correlation (Fig. 8). No such correlation was seen for recovery of whole Borrelia organisms (data not shown).
FIG. 8.
A total of 10 ml of 29 urine samples from 23 donors was spiked with DNA from 5,000 Borrelia organisms, and the spike recovery rate was set in relation to the respective specific density of each urine sample.
Urine samples from borreliosis patients.
Urine from 12 EM patients was collected by a practitioner and stored at −20°C. To prevent potential nuclease activity, urine was thawed on ice and kept on ice until addition of DNAzol. DNA was extracted from 10 ml of urine of each patient and tested by real-time nested PCR (data not shown). Borrelia DNA was only detected in one patient sample (Table 1, patient number 7), and the Borrelia genospecies was B. afzelii. Since Schmidt et al. were able to detect Borrelia DNA in the urine of >90% of patients with EM, we repeated DNA amplification with the nested PCR based on Borrelia flagellin according to Schmidt et al. (28). With this PCR, no Borrelia DNA was detected in any of the 12 urine samples, although the positive control was detected readily. The characterization of the urine samples from the patients resulted in pH values of 5.5 to 7.5 (Table 1). The urine which was positive by the ospA PCR had a pH of 6.5. None of the samples showed inhibitory activity with the Chlamydia DNA. Additionally, all patient urine samples were tested for their nuclease activity analogous to the spiked urine samples. A total of 7 of the 12 urines (58%) revealed nuclease activity, including the positive urine sample.
DISCUSSION
To evaluate a urine-based PCR assay as a diagnostic tool for LB, we established a standardized protocol, spiking urine samples from healthy donors with a defined amount of whole Borrelia organisms or Borrelia DNA. The use of a nested real-time PCR enabled a quantitative analysis of these spiked samples with a sensitivity of 0.1 to 1 Borrelia organisms per 10 μl of template.
As it is still unclear whether whole Borrelia organisms or Borrelia DNA is present in the urine of LB patients, we performed spike experiments with both. The reported increase of inhibitory qualities of urine samples from healthy donors which were stored for a period of >3 months (5) could not be confirmed in our study, regardless whether we spiked urine with whole Borrelia organisms or DNA. In contrast to Bergmann et al., we spiked urine samples before freezing them and used the same urine sample for every time point. Since Bergmann et al. used different urine samples for the different storage times, it appears that the increase in inhibition was not due to long-term storage but rather to variations of individual samples.
Centrifugation of spiked urine samples resulted in improved detection of both whole Borrelia organisms and DNA and is therefore advisable. It was surprising that the concentration effect was also seen for the DNA spike, as soluble DNA does not sediment in aqueous solution at 40,000 × g. Probably, the DNA binds to or is dragged down by particles in the urine (e.g., crystals or cell debris). In line with this assumption, urine samples with a higher specific density also showed a higher concentration effect upon centrifugation, pointing to a coprecipitation effect. Therefore, centrifugation of greater volumes of urine with low specific density might increase the amount of Borrelia DNA recovered.
Spike recovery was inhibited in nearly 50% of urine samples from healthy donors spiked with Borrelia DNA. Addition of plasmid DNA and visualization on agarose gels demonstrated that the inhibitory effect was due to degradation of the DNA. This implied an enzymatic activity, as no apparent degradation of the plasmid took place in water as expected. DNase I, which is the major nuclease present in urine, is a Ca2+- and Mg2+-dependent endonuclease, with an optimal activity at pH 6.5 (17). By adjusting inhibitory urine samples to pH 8, adding EDTA, or keeping them on ice, we could abrogate the degradation and demonstrate that it was due to nuclease activity in the urine.
We transferred the optimized protocol to the detection of Borrelia DNA in samples from patients with an EM. This patient group has an acute infection; therefore, detection of Borrelia DNA in the urine should be most likely. The urine samples were frozen immediately and thawed on ice to prevent degradation of possible Borrelia DNA. To concentrate the amount of DNA or Borrelia, 10 ml of urine was centrifuged, and nested real-time PCR was performed to increase sensitivity. Despite these optimized conditions, only 1 of the urine samples from the 12 tested LB patients revealed a positive PCR result. The PCR based on flagellin was not even positive for this donor. It could be possible that a large volume of urine dilutes the amount of Borrelia DNA below detectable limits; however, this is an unlikely explanation for all 11 negative results.
The positive patient urine had a pH of 6.5 and showed nuclease activity in the respective experiment—a result which could be used as an argument against free DNA in the urine of borreliosis patients.
Assuming that whole Borrelia organisms are present in the urine of patients, a negative result of urine PCR could be only due to an extremely low count or absence of Borrelia organisms in the given specimen. With the nested real-time PCR, we were able to detect 1 Borrelia organism in 10 μl of template. Hence, the concentration in the eluate was 10 Borrelia organisms per 100 μl eluate. In our spike experiments with whole Borrelia organisms, the average yield by centrifugation of 10 ml of urine was about 63% (Fig. 7). Therefore, 2 Borrelia organisms per ml of urine would be required to result in a positive PCR. This sensitivity thus exceeds the reported estimate of 50 to 5,000 Borrelia organisms per ml of urine (14).
An average of 1,450 spirochetes were detected in 2-mm biopsies (22). Therefore, the number of Borrelia organisms in an EM with a diameter of 10 cm and an equal distribution of spirochetes would be around 4 × 106. Assuming (i) a stable number of Borrelia in the EM, (ii) a generation time of Borrelia of 12 h (2) and (ii) a urine volume of 1.5 liters per day, 0.03% of these Borrelia passing into the urine would lead to a positive PCR.
Since it is questionable whether whole Borrelia can pass through an intact kidney barrier, it would be likely that Borrelia organisms which reach the urine only derive from a persistent infection of the urinary tract.
Assuming that the urine of LB patients contains Borrelia DNA, negative results could be due to a small amount of DNA or to the presence of nucleases, whereas the first point may result from the second. As in the case of DNA, the average yield by centrifugation of 10 ml of spiked urine was only about 26% (Fig. 7); 0.07% DNA equivalents reaching the urine of the assumed number of 4 × 106 Borrelia organisms in an EM would result in a positive PCR.
The kidney plays a role in the clearance of plasma DNA, and the human kidney barrier is permeable to DNA molecules (6, 33). In an experiment with laboratory mice, only 0.06% of subcutaneously injected DNA was excreted into the urine within 3 days in a polymeric form (6). Thus, it appears unlikely that sufficient Borrelia DNA for a positive PCR result would be contained in 10 ml of urine.
Addition of EDTA to the samples immediately after collection inhibits nuclease activity in human urine, but DNA destruction due to nuclease activity in the bladder could not be ruled out. The predominant fraction of DNA fragments found in human urine was 150 bp to 250 bp (6, 33). This is in line with our results, where the nuclease activity on plasmid DNA did not lead to complete degradation (Fig. 6). Since the amplicon of our standard PCR is about 350 bp in length, most of the DNA fragments would be too short for amplification. But it could be that Borrelia DNA is bound to proteins and may thus be protected from nucleases better than the naked DNA used in our experiments. If urine of LB patients did contain Borrelia DNA, improved detection by inhibition of nuclease activity already in the bladder might be attained by systematic adjustment of the pH in the urine to at least pH 8 by bicarbonate-mediated alkalization of urine in vivo.
Taken together, we have developed an optimized, well-controlled, and highly sensitive PCR protocol for the species-specific detection of Borrelia DNA. Extrapolations indicate that even with this protocol, it is unlikely that Borrelia DNA will be detected in patient urine. In line, we were only able to detect Borrelia DNA in the urine of 1 of 12 EM patients. Although only 12 patient samples were tested, the combined results of spiked and patient urine samples enforce doubts that urine is a suitable material for the diagnosis of LB.
Acknowledgments
We thank Jürgen Jürgensens (Frickingen, Germany) and Margot Grau (Konstanz, Germany) for collecting patient urines, Sonja von Aulock for revision of the manuscript, and Petra Eisele for technical assistance.
REFERENCES
- 1.Aberer, E., B. L. Schmidt, F. Breier, T. Kinaciyan, and A. Luger. 1999. Amplification of DNA of Borrelia burgdorferi in urine samples of patients with granuloma annulare and lichen sclerosus et atrophicus. Arch. Dermatol. 135:210-212. [DOI] [PubMed] [Google Scholar]
- 2.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 3.Bauerfeind, R., U. Kreis, R. Weiss, L. H. Wieler, and G. Baljer. 1998. Detection of Borrelia burgdorferi in urine specimens from dogs by a nested polymerase chain reaction. Zentralbl. Bakteriol. 287:347-361. [DOI] [PubMed] [Google Scholar]
- 4.Behzadbehbahani, A., P. E. Klapper, P. J. Vallely, and G. M. Cleator. 1997. Detection of BK virus in urine by polymerase chain reaction: a comparison of DNA extraction methods. J. Virol. Methods 67:161-166. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann, A. R., B. L. Schmidt, A. M. Derler, and E. Aberer. 2002. Importance of sample preparation for molecular diagnosis of Lyme borreliosis from urine. J. Clin. Microbiol. 40:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botezatu, I., O. Serdyuk, G. Potapova, V. Shelepov, R. Alechina, Y. Molyaka, V. Ananev, I. Bazin, A. Garin, M. Narimanov, V. Knysh, H. Melkonyan, S. Umansky, and A. Lichtenstein. 2000. Genetic analysis of DNA excreted in urine: a new approach for detecting specific genomic DNA sequences from cells dying in an organism. Clin. Chem. 46:1078-1084. [PubMed] [Google Scholar]
- 7.Brettschneider, S., H. Bruckbauer, N. Klugbauer, and H. Hofmann. 1998. Diagnostic value of PCR for detection of Borrelia burgdorferi in skin biopsy and urine samples from patients with skin borreliosis. J. Clin. Microbiol. 36:2658-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crameri, R., and M. Suter. 1993. Display of biologically active proteins on the surface of filamentous phages: a cDNA cloning system for selection of functional gene products linked to the genetic information responsible for their production. Gene 137:69-75. [DOI] [PubMed] [Google Scholar]
- 9.Demaerschalck, I., A. BenMessaoud, M. De Kesel, B. Hoyois, Y. Lobet, P. Hoet, G. Bigaignon, A. Bollen, and E. Godfroid. 1995. Simultaneous presence of different Borrelia burgdorferi genospecies in biological fluids of Lyme disease patients. J. Clin. Microbiol. 33:602-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diterich, I., L. Härter, D. Hassler, A. Wendel, and T. Hartung. 2001. Modulation of cytokine release in ex vivo-stimulated blood from borreliosis patients. Infect. Immun. 69:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumler, J. S. 2001. Molecular diagnosis of Lyme disease: review and meta-analysis. Mol. Diagn. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Dumler, J. S. 2003. Molecular methods for ehrlichiosis and Lyme disease. Clin. Lab. Med. 23:867-884. [DOI] [PubMed] [Google Scholar]
- 13.Goodman, J. L., P. Jurkovich, C. Kodner, and R. C. Johnson. 1991. Persistent cardiac and urinary tract infections with Borrelia burgdorferi in experimentally infected Syrian hamsters. J. Clin. Microbiol. 29:894-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman, J. L., P. Jurkovich, J. M. Kramber, and R. C. Johnson. 1991. Molecular detection of persistent Borrelia burgdorferi in the urine of patients with active Lyme disease. Infect. Immun. 59:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmeister, E. K., R. B. Markham, J. E. Childs, and R. R. Arthur. 1992. Comparison of polymerase chain reaction and culture for detection of Borrelia burgdorferi in naturally infected Peromyscus leucopus and experimentally infected C.B-17 scid/scid mice. J. Clin. Microbiol. 30:2625-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyde, F. W., R. C. Johnson, T. J. White, and C. E. Shelburne. 1989. Detection of antigens in urine of mice and humans infected with Borrelia burgdorferi, etiologic agent of Lyme disease. J. Clin. Microbiol. 27:58-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, K., N. Minamiura, and T. Yamamoto. 1984. Human urine DNase I: immunological identity with human pancreatic DNase I, and enzymic and proteochemical properties of the enzyme. J. Biochem. (Tokyo) 95:1399-1406. [DOI] [PubMed] [Google Scholar]
- 18.Khan, G., H. O. Kangro, P. J. Coates, and R. B. Heath. 1991. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J. Clin. Pathol. 44:360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebech, A. M. 2002. Polymerase chain reaction in diagnosis of Borrelia burgdorferi infections and studies on taxonomic classification. APMIS Suppl. 2002:1-40. [PubMed] [Google Scholar]
- 20.Lebech, A. M., K. Hansen, F. Brandrup, O. Clemmensen, and L. Halkier-Sorensen. 2000. Diagnostic value of PCR for detection of Borrelia burgdorferi DNA in clinical specimens from patients with erythema migrans and Lyme neuroborreliosis. Mol. Diagn. 5:139-150. [DOI] [PubMed] [Google Scholar]
- 21.Lebech, A. M., P. Hindersson, J. Vuust, and K. Hansen. 1991. Comparison of in vitro culture and polymerase chain reaction for detection of Borrelia burgdorferi in tissue from experimentally infected animals. J. Clin. Microbiol. 29:731-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liveris, D., G. Wang, G. Girao, D. W. Byrne, J. Nowakowski, D. McKenna, R. Nadelman, G. P. Wormser, and I. Schwartz. 2002. Quantitative detection of Borrelia burgdorferi in 2-millimeter skin samples of erythema migrans lesions: correlation of results with clinical and laboratory findings. J. Clin. Microbiol. 40:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lunemann, J. D., S. Zarmas, S. Priem, J. Franz, R. Zschenderlein, E. Aberer, R. Klein, L. Schouls, G. R. Burmester, and A. Krause. 2001. Rapid typing of Borrelia burgdorferi sensu lato species in specimens from patients with different manifestations of Lyme borreliosis. J. Clin. Microbiol. 39:1130-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiwald, M., C. Stockinger, D. Hassler, M. von Knebel Doeberitz, and H. G. Sonntag. 1995. Evaluation of the detection of Borrelia burgdorferi DNA in urine samples by polymerase chain reaction. Infection 23:173-179. [DOI] [PubMed] [Google Scholar]
- 25.Mercier, G., A. Burckel, and G. Lucotte. 1997. Detection of Borrelia burgdorferi DNA by polymerase chain reaction in urine specimens of patients with erythema migrans lesions. Mol. Cell. Probes 11:89-94. [DOI] [PubMed] [Google Scholar]
- 26.Mueller, M., S. Postius, J. G. Thimm, K. Gueinzius, I. Muehldorfer, and C. Hermann. 2004. Toll-like receptors 2 and 4 do not contribute to clearance of Chlamydophila pneumoniae in mice, but are necessary for the release of monokines. Immunobiology 209:599-608. [DOI] [PubMed] [Google Scholar]
- 27.Rauter, C., R. Oehme, I. Diterich, M. Engele, and T. Hartung. 2002. Distribution of clinically relevant Borrelia genospecies in ticks assessed by a novel, single-run, real-time PCR. J. Clin. Microbiol. 40:36-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt, B., R. R. Muellegger, C. Stockenhuber, H. P. Soyer, S. Hoedl, A. Luger, and H. Kerl. 1996. Detection of Borrelia burgdorferi-specific DNA in urine specimens from patients with erythema migrans before and after antibiotic therapy. J. Clin. Microbiol. 34:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt, B. L. 1997. PCR in laboratory diagnosis of human Borrelia burgdorferi infections. Clin. Microbiol. Rev. 10:185-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt, B. L., E. Aberer, C. Stockenhuber, H. Klade, F. Breier, and A. Luger. 1995. Detection of Borrelia burgdorferi DNA by polymerase chain reaction in the urine and breast milk of patients with Lyme borreliosis. Diagn. Microbiol. Infect. Dis. 21:121-128. [DOI] [PubMed] [Google Scholar]
- 31.Schwaiger, M., O. Peter, and P. Cassinotti. 2001. Routine diagnosis of Borrelia burgdorferi (sensu lato) infections using a real-time PCR assay. Clin. Microbiol. Infect. 7:461-469. [DOI] [PubMed] [Google Scholar]
- 32.Schwan, T. G., W. Burgdorfer, M. E. Schrumpf, and R. H. Karstens. 1988. The urinary bladder, a consistent source of Borrelia burgdorferi in experimentally infected white-footed mice (Peromyscus leucopus). J. Clin. Microbiol. 26:893-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su, Y. H., M. Wang, D. E. Brenner, A. Ng, H. Melkonyan, S. Umansky, S. Syngal, and T. M. Block. 2004. Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J. Mol. Diagn. 6:101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]