Abstract
The patterns of CD8 T-cell epitopes recognized within the E6 protein in women who had cleared their human papillomavirus 16 infection were examined. T-cell lines were established using autologous dendritic cells infected with a recombinant vaccinia virus. Evidence of potential antigenic epitopes was shown in 8 of 23 (34.8%) women.
The association between human papillomavirus (HPV) infections, HPV type 16 (HPV 16) in particular, and the development of invasive cervical cancer has been well described (8). However, the exact mechanisms leading from HPV infection to malignancy are not well understood. One of the critical steps in the progression to cervical cancer appears to be the establishment of persistent infection. Epidemiologic evidence suggests that more than 90% of women are able to clear their HPV infections, including HPV 16 infections. The remainder, whose HPV infections persisted, are at risk for the development of cervical cancer. In a previously published study (6), we examined the role of cytotoxic T lymphocytes (CTL) in clearing HPV 16 infection in women without evidence of precancer or squamous intraepithelial lesion. We found that none of the women with persistent HPV 16 infection had a detectable CTL response to HPV 16 E6 protein compared to more than half of those with evidence of clearance. Given the importance of CTL recognition of HPV 16 E6 proteins in controlling HPV, we set out to examine the pattern that CD8 T-cell epitopes recognized within the E6 protein in women who had evidence of HPV 16 clearance.
A total of 654 female subjects, ranging in age from 13 to 20 years at entry were participants in a longitudinal study of HPV infection initiated in 1991 (4). As part of this parent study, the subjects were monitored via cervical HPV DNA testing by PCR (7), cytology, and colposcopy every 4 months. Subjects who had negative HPV 16 cervical specimens for a minimum of two consecutive visits, after previously testing positive, were selected for the present study. The study protocol was approved by the University of California at San Francisco Committee on Human Research.
HPV 16 E6-specific T-cell lines were established by in vitro stimulation of CD8 cells using autologous dendritic cells infected with recombinant vaccinia viruses expressing the E6 protein (E6-vac [6]). CD8 cells were selected from peripheral blood mononuclear cells (PBMC) using a commercially available magnetic kit (CD8 Isolation Kit; Miltenyi Biotec, Auburn, CA). Autologous dendritic cells were established by isolating monocytes from PBMC using CD14 antibody coupled to magnetic beads (Miltenyi Biotec) and by growing the autologous dendritic cells in the presence of granulocyte-macrophage colony-stimulating factor (50 ng/ml) and recombinant interleukin-4 (100 U/ml) for 7 days. They were matured by culture in wells containing irradiated L cells expressing CD40 ligand for 48 h, and E6-vac was added during the last 24 h. After 7 days, in vitro stimulation was repeated for an additional 7 days.
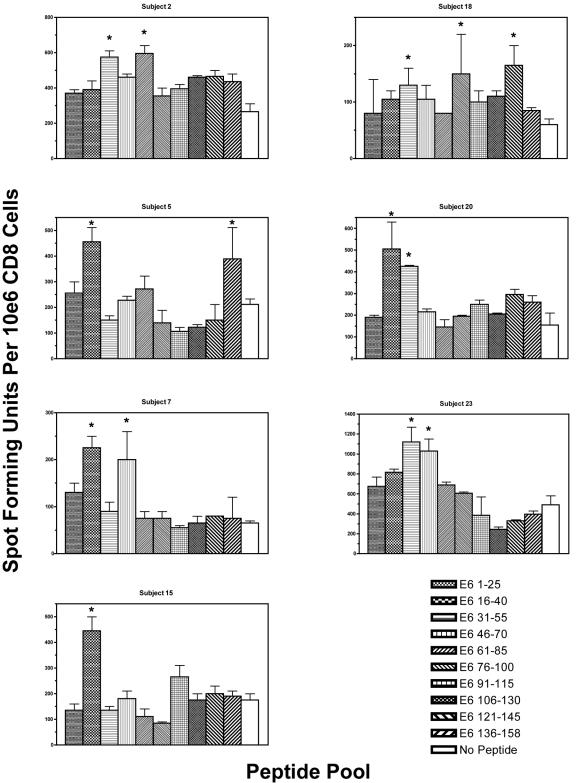
To examine the patterns of CD8 T-cell epitopes (Fig. 1), gamma interferon enzyme-linked immunospot (ELISPOT) assays were performed as described by Larsson et al. (3). Overlapping 15-mer peptides (overlapping by the 10 central amino acids) were pooled into 10 groups (each containing three peptides) and tested in duplicate (10 μM for each peptide), along with a no-peptide control. One hundred thousand cells of a CD8 T-cell line were plated to each well. Spot-forming units were counted using an automated ELISPOT analyzer (Cell Technology, Inc., Jessup, MD) and were normalized to the number of spot-forming units per 106 cells. The response was considered positive when the number of spot-forming units in wells with peptide pools was at least double that seen in the no-peptide control wells (2).
FIG. 1.
ELISPOT assays showing the pattern of CD8 T-cell immunodominance within the HPV 16 E6 protein in women who were able to clear their HPV 16 infection. The results of subjects who had at least one positive peptide pool are shown, except for subject 1, whose data have been described previously (5).  , Positive peptide pool.
, Positive peptide pool.
A total of 23 subjects enrolled in the parent study with a history of cleared HPV 16 infection were studied. The mean age at the time of blood sampling was 25.3 years. The mean time since the last positive HPV 16 result was 51.8 months. At least one positive peptide pool was identified in 8 of 23 subjects examined. The region within E6 that most often contained antigenic epitopes with the greatest magnitude of response was that of amino acids 16 to 40, which was observed in four subjects (17.4%). Two subjects had one positive region (subjects 1 and 15), five subjects had two positive regions (subjects 2, 5, 7, 20, and 23), and one subject had three positive regions (subject 18). Of 23 subjects studied, 8 (34.8%) showed the presence of potential antigenic epitopes in the N-terminal half (amino acids 1 to 85). In comparison, 2 of 23 subjects (8.7%) showed the presence of potential antigenic epitopes in the C-terminal half (amino acids 76 to 158), but this finding was not statistically significant (Fisher exact test, P = 0.07).
The observation that only one-third of the women tested showed the presence of potential CD8 T-cell epitopes raises the question of whether there is an alternate mechanism for viral clearance. This may be a reflection of weakened immune response simply because a number of years have passed for many subjects since the last HPV 16 detectable visit. In addition, the definition of a positive response as being a minimum of twice the background level is rather arbitrary and does not have a strong statistical basis. However, the specificity of the T-cell responses has been demonstrated by the fact that HPV-specific T-cell clones have been isolated from peaks that range from 2.5 to 5.4 in the ratio of spot-forming units with a peptide pool to spot-forming units with a no-peptide control. It is also possible that some peaks with a ratio of <2 may be weak but true T-cell responses. Alternatively, it may be possible that CD4 T cells and other immune cells may play an important role in viral clearance in a subset of individuals.
The minimal and optimal amino acid sequences of the CD8 T-cell epitopes and their restricting HLA molecules have been defined for two of the subjects: HPV 16 E6 52-61 (FAFRDLCIVY) restricted by HLA-B57 in subject 1 (5) and HPV 16 E6 29-37 (TIHDIILEC) restricted by HLA-B48 in subject 15 (unpublished data). Future studies will focus on evaluation of CD8 T-cell epitopes from other subjects. Recognition of HPV 16 E6 and E7 peptides by peripheral T cells in a woman whose grade 3 vulvar intraepithelial neoplasia regressed has also been described by others (1). These T-cell epitopes may be used as sources of antigens for dendritic cell immunotherapies to treat patients with HPV-related malignancies who express the particular HLA types. When sufficient numbers of these epitopes covering a large proportion of the population are described, they may be used in concert as vaccines.
Acknowledgments
This study was supported by grants from the National Institutes of Health (NCI CA51323, NCI K07 CA75974, and M01 RR01271) and from the Arkansas Biosciences Institute, the major component of the Tobacco Settlement Proceeds Act of 2000.
REFERENCES
- 1.Bourgault Villada, I., M. Moyal Barracco, M. Ziol, A. Chaboissier, N. Barget, S. Berville, B. Paniel, E. Jullian, T. Clerici, B. Maillere, and J. G. Guillet. 2004. Spontaneous regression of grade 3 vulvar intraepithelial neoplasia associated with human papillomavirus-16-specific CD4+ and CD8+ T-cell responses. Cancer Res. 64:8761-8766. [DOI] [PubMed] [Google Scholar]
- 2.Kaul, R., T. Dong, F. A. Plummer, J. Kimani, T. Rostron, P. Kiama, E. Njagi, E. Irungu, B. Farah, J. Oyugi, R. Chakraborty, K. S. MacDonald, J. J. Bwayo, A. McMichael, and S. L. Rowland-Jones. 2001. CD8+ lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J. Clin. Investig. 107:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larsson, M., D. T. Wilkens, J. F. Fonteneau, T. J. Beadle, M. J. Merritt, R. G. Kost, P. A. Haslett, S. Cu-Uvin, N. Bhardwaj, D. F. Nixon, and B. L. Shacklett. 2002. Amplification of low-frequency antiviral CD8 T-cell responses using autologous dendritic cells. AIDS 16:171-180. [DOI] [PubMed] [Google Scholar]
- 4.Moscicki, A. B., S. Shiboski, J. Broering, K. Powell, L. Clayton, N. Jay, T. M. Darragh, R. Brescia, S. Kanowitz, S. B. Miller, J. Stone, E. Hanson, and J. Palefsky. 1998. The natural history of human papillomavirus infection as measured by repeated DNA testing in adolescent and young women. J. Pediatr. 132:277-284. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa, M., K. Kim, and A.-B. Moscicki. 2004. Different methods of identifying new antigenic epitopes of human papillomavirus type 16 E6 and E7 proteins. Clin. Diagn. Lab. Immunol. 11:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagawa, M., D. P. Stites, S. Patel, S. Farhat, M. Scott, N. K. Hills, J. M. Palefsky, and A. B. Moscicki. 2000. Persistence of human papillomavirus type 16 infection is associated with lack of cytotoxic T lymphocyte response to the E6 antigens. J. Infect. Dis. 182:595-598. [DOI] [PubMed] [Google Scholar]
- 7.Ting, Y., and M. M. Manos. 1990. Detection and typing of genital human papillomaviruses. Academic Press, Inc., San Diego, Calif.
- 8.zur Hausen, H. 1996. Papillomavirus infections: a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]