Abstract
Diagnostic peptides previously isolated from phage-displayed libraries by affinity selection with serum antibodies from patients with Lyme disease were found to give reproducible serum reactivity patterns when tested in two different enzyme-linked immunosorbent assay formats. In addition, the hypothetical possibility that peptides selected by this type of “epitope discovery” technique might identify the original antigens eliciting antibody responses was tested by searching for sequence similarities in bacterial protein databases. In support of this hypothesis, our search uncovered similarities between peptides representing two different sequence motifs and sequences in the VlsE and BBA61 antigens of Borrelia burgdorferi. Utilizing synthetic peptides, we verified that the sequence KAASKETPPALNK, located at the C terminus of the VlsE antigen, had the same reactivity pattern to sera from patients with extracutaneous Lyme disease as the diagnostic peptide SKEKPPSLNWPA, with which it shared a 7-amino-acid-residue match (consensus residues are underlined). A peptide with conservative mutations of five of the consensus residues was nonreactive, strongly suggesting that the VlsE sequence represents the epitope that originally elicited antibody responses in these patients. The diagnostic sensitivity of this new VlsE epitope was relatively low (30%) compared to that (100%) of the well-documented C6 diagnostic peptide of VlsE when tested in our small cohort of 10 patients with Lyme disease. Nonetheless, the identification of this previously unknown epitope serves as a proof of the principle of the hypothetical ability of “epitope discovery” techniques to detect specific microbial antigens with diagnostic relevance in infectious diseases.
Identifying serological markers for the diagnosis of acute infectious diseases with standard immunological approaches is laborious and requires, at a minimum, prior knowledge of the etiologic agents and the ability either to culture them as sources of antigens or to clone the appropriate antigens and produce recombinant proteins. The advent of phage-displayed peptide technology, in which large, complex libraries of filamentous bacteriophage bearing random peptide sequences on their coat proteins can be generated and screened with antibodies, has provided a new approach called “epitope discovery” that can circumvent these problems. In this approach, phage-displayed peptide libraries are screened with sera from patients who have suffered a particular disease or pathological condition to discover peptide epitopes that are specifically recognized by sera from patients with the same disease or condition (1, 3, 4, 6, 8, 9, 14, 16). The sequence of peptides from phage clones bearing diagnostic epitopes can be readily determined, and the peptide epitopes can be synthesized relatively cheaply to provide inexpensive diagnostic tests (18).
Perhaps of equal importance, “epitope discovery” has the potential to identify the antigen(s) responsible for eliciting antibody responses in patients experiencing a particular infection or condition. In theory, “mimotopes” identified by antibody screening are likely to have some sequence similarity to epitopes on the original antigen that elicited the antibody response. Thus, by performing similarity searches of protein databases, one may be able to identify the original antigenic stimulus to the patient's immune system. This reverse-discovery approach has the potential to identify previously unknown antigens and/or new epitopes on already known antigens. Moreover, epitope discovery has the potential to identify antigens in diseases of unknown etiology that stimulate an antibody response. However, one obvious limitation of applying “epitope discovery” to the identification of antigens with diagnostic relevance is that databases of microbial protein sequences are by no means complete, and hence, similarity searches will miss any antigens that are not represented in the available databases. A second problem arises in that phage libraries displaying peptides with the potential for forming “constrained epitopes” due to sulfhydryl bonds between cystine residues in the peptide sequence may mimic the conformation but not the sequence of an eliciting antigenic epitope. This problem can be minimized by screening libraries displaying linear, unconstrained peptides but carries the risk of missing conformational epitopes that might be valuable as diagnostic reagents.
The value of “epitope discovery” in developing diagnostics for infectious diseases is strongly supported by the report of Kouzmitcheva et al. (10), who identified a set of diagnostic peptides in patients with Lyme disease. They devised a protocol in which 12 different phage-displayed peptide libraries were first absorbed with immunoglobulin G (IgG) from pooled sera of healthy donors to remove epitopes recognized by normal human sera. The depleted libraries were then panned over all possible pairwise combinations of IgG purified from the sera of eight different Lyme disease patients. From the 336 sublibraries that were created by this panning procedure, they identified a set of 12 peptides comprising five different sequence motifs that could specifically identify Lyme disease patients. While the diagnostic sensitivity of any individual peptide was not more than 50%, the combined set of peptides was 100% sensitive and highly specific for the small cohorts (10 patients each) of Lyme disease and control patients that were tested.
We wished to evaluate the diagnostic utility of these peptides and to determine whether similarities existed between the peptide motifs and proteins in sequence databases that might reveal the bacterial antigens responsible for eliciting antibodies in patients with Lyme disease. The 12 peptide libraries employed by Kouzmitcheva et al. (10) included random 8-mer and 15-mer linear peptides and 15-mer peptides containing two cystine residues spaced between 0 and 6 residues apart that could potentially introduce conformational constraints on the epitopes. Although searches conducted in the original report did not reveal any convincing sequence similarities with Borrelia burgdorferi proteins or other bacterial proteins in the TIGR protein database, we found similarities between several of their peptide motifs (designated A, B, C, F, and H) and bacterial antigens in the NCBI database by utilizing the phi-BLAST search algorithm. The most notable similarity was between the motif A sequence and a nine-peptide C-terminal sequence in the VlsE antigen of B. burgdorferi. We present evidence which indicates that this VlsE sequence represents a previously undiscovered epitope recognized by antibodies in Lyme disease patient sera.
MATERIALS AND METHODS
Reagents received for evaluation.
George P. Smith of the Division of Biological Sciences, University of Missouri—Columbia, provided human serum samples, purified phage preparations, and synthetic peptides for evaluation in this study. The serum samples were obtained from Lyme disease patients and from patients with rheumatoid and psoriatic arthritis as previously described (10). Phages from 12 clones bearing different peptides and the wild-type f88-4 phage vector were supplied as cesium chloride-purified preparations. The number, amino acid sequence, and sequence motif of each peptide, as designated by Kouzmitcheva and coworkers (10), are presented in Table 1. Peptides no. 7 (KPRDTLPPPLNRPPC) and 24 (GNNSVSKEKPPSLNWPAC), representing motif A sequences, and peptide no. 12 (VPVDAPHAGTKPHSAC), representing a unique motif B sequence, were synthesized at the University of Virginia Biomolecular Research Facility, UVA Health Science Center, Charlottesville, and were also supplied courtesy of George P. Smith.
TABLE 1.
Sequence motifs of peptides displayed on purified phage clonesa
| Peptide motif | Peptide no. | Amino acid sequence |
|---|---|---|
| A | 24 | GNNSVSKEKPPSLNWPP |
| A | 5 | TELKLAPPVLNAPPL |
| A | 11 | YPKESPPRLNAPWYQ |
| A | 1 | ESKLTPPPLNPIRVV |
| A | 7 | KPRDTLPPPLNRPPP |
| A | 4 | KAHPPLLNSPRDVPL |
| B | 12 | VPVDAPHAGTKPHSA |
| C | 23 | PKSSCTQNPILCAILS |
| F | 14 | QKDFACKHCKLPSP |
| F | 17 | EKRFACKPLCNTPA |
| H | 19 | QRSECSTSKCFVRK |
| H | 20 | YREACTNGKCFVLK |
Motif and peptide numbers are as assigned by Kouzmitcheva et al. (10). Consensus residues within a motif are underlined.
Patient serum samples.
Samples of sera from a convenience sample of 10 patients from New York State with extracutaneous Lyme disease (6 patients had Lyme arthritis, 2 patients had early neuroborreliosis, and 2 patients [one of whom also had seventh-nerve palsy] had cardiac manifestations) were studied.
Synthetic peptides.
The C6 peptide (CMKKDDQIAAAMVLRGMAKDGKFALK) of the B. burgdorferi VlsE antigen that is used commercially for Lyme disease serodiagnosis (11, 15); peptide no. 2 (SKEKPPSLNWPAC), representing the motif A consensus sequence; peptide no. 3 (KAASKETPPALNKC), representing the C-terminal sequence of the B. burgdorferi VlsE antigen; and peptide no. 4 (KAASREKGGAVQKC), representing a mutated motif A and VlsE C-terminal consensus sequence, were synthesized and supplied as products that were at least 75% pure by high-performance liquid chromatography-mass spectrometry (Sigma-Genosys, The Woodlands, TX). C-terminal cystines were incorporated into all sequences to permit covalent linkage via sulfhydryl groups to enzyme-linked immunosorbent assay (ELISA) plates. All the peptides dissolved readily in deionized water and were stored at a 10 mM concentration at −20°C.
Purified-phage ELISA.
Purified phage diluted to a concentration of 4 × 1011 virions/ml in Tris-buffered saline (TBS) (50 mM Tris, 137 mM NaCl, pH 7.4) was added in 50-μl/well volumes to 96-well Maxisorb plates (Nalge Nunc International, Rochester, NY) and allowed to adsorb overnight at 4°C. Plates were washed twice with TTDBA (10 mM Tris, 137 mM NaCl, pH 7.4, containing 0.5% Tween 20 [Sigma, St. Louis, MO] and 0.1% dialyzed bovine serum albumin [Sigma; product no. A7906]). Sera were diluted 1:100 in TTDBA, added to plates in a volume of 50 μl/well, and incubated for a minimum of 2 h at room temperature or at 4°C overnight. Plates were washed 10 times with TBS containing 0.5% Tween 20 (TBS-Tween) and incubated for 1 h at room temperature with either anti-human γ-chain-specific (IgG-specific) or anti-human μ-chain-specific (IgM-specific) alkaline phosphatase-conjugated goat antibodies (Jackson ImmunoResearch, West Grove, PA) diluted 1:20,000 in TBS-Tween. Plates were washed 10 times with TBS-Tween and developed with 1 M diethanolamine, pH 9.8, containing 0.5 mg/ml p-nitrophenyl phosphate substrate and 1 mM MgCl2.
Synthetic-peptide ELISA.
Synthetic peptides were covalently linked to Covalink 96-well flat-bottomed plates (Nalge Nunc International) with an EMCS (N-[ɛ-maleimidocaproyloxy]succinimide ester) amine-sulfhydryl-reactive linker (Pierce Biotechnology, Inc., Rockford, IL) containing a 9.4-Å spacer arm. Plates were first reacted via amine groups with 1 mM EMCS dissolved in dimethyl sulfoxide containing 1 mM N-ethyldiisopropylamine (Sigma, St. Louis, MO). After a 3-h incubation at room temperature, unreacted EMCS linkers were removed by filling plates twice with dimethyl sulfoxide and discarding the unbound EMCS. Plates were washed once with deionized water, and peptides containing terminal cystine residues diluted to concentrations ranging between 10 and 1,000 μM in 7 M guanidine-HCl (buffered to a pH of 7 with 1 M Na phosphate) were added in volumes of 100 μl/well to plates for binding via SH groups of the cystine residues. Peptides were reacted in EMCS-derivatized plates overnight at room temperature in a humidified chamber. Unbound peptides were removed by washing three times with TBS-Tween, and plates were then refrigerated with TBS-Tween added until ELISAs were performed.
The ELISA for human antibody reactivity was performed by adding 100 μl/well of sera diluted 1:100 in TTDBA to duplicate wells of peptide-linked plates, incubating for a minimum of 90 min, and washing 10 times with TBS-Tween. Antibody binding was quantitated by adding 100 μl/well of 1:20,000 dilutions of alkaline phosphate-conjugated goat anti-human γ-chain- and μ-chain-specific antibodies to detect IgG and IgM, respectively. After a 90-min incubation, plates were washed 10 times with TBS-Tween and developed by adding 100 μl/well of p-nitrophenyl phosphate substrate solution. Absorbance readings were taken at an optical density at 405 nm (OD405) at 3-min intervals for 30 min on a Benchmark (Bio-Rad, Hercules, CA) 96-well microplate reader, and the maximum kinetic velocity (mOD/min) of the reaction in each well was calculated by Microplate Manager 4.0 (Bio-Rad) software. Serum IgG reactivity was expressed as the average mOD/min of duplicate sample wells.
Statistical analysis.
A general linear-model analysis of variance was applied to evaluate the reproducibility of the synthetic-peptide ELISA. Cutoff values for comparing the diagnostic sensitivities of different peptides were established by generating receiver operating curves and selecting the lowest cutoff value that was 100% specific for all peptides.
RESULTS AND DISCUSSION
Phage-displayed peptide and synthetic-peptide ELISAs are reproducible and give equivalent results.
We examined the reproducibility of the ELISA protocol published by Kouzmitcheva et al. (10) by utilizing the same cesium chloride-purified phage clones, patient sera, and control sera that were used in their report. We obtained the same IgG reactivity patterns as they reported, although there were not sufficient amounts of some serum samples to completely replicate their experiments (Table 2). We also tested sera for IgM reactivity and observed the same unique IgM reactivity of the serum from Lyme disease patient 9 for motif A and C peptides and the absence of IgM reactivity of sera from any other Lyme disease or control patient for the different peptide motifs as reported by Kouzmitcheva et al. (10).
TABLE 2.
Reproducibility of serum IgG reactivity patterns determined by ELISAs with purified phage-displayed peptides
| Peptide motif | IgG reactivitya
|
||||||
|---|---|---|---|---|---|---|---|
| LD1 | LD5 | LD6 | LD8 | LD9 | C11 | C12 | |
| A | −/− | −/− | +/+ | −/− | +/NT | −/− | −/− |
| B | +/+ | −/− | −/− | −/NT | −/− | −/− | −/− |
| C | −/− | −/− | −/− | −/NT | −/NT | −/NT | −/NT |
| F | +/+ | −/− | −/− | +/NT | −/NT | −/NT | −/NT |
| H | +/+ | −/− | −/− | +/NT | −/NT | −/NT | −/NT |
Serum reactivity reported by Kouzmitcheva et al. (10)/serum reactivity we observed. LD, Lyme disease serum; C, control serum; NT, not tested. Numbers correspond to patient numbers.
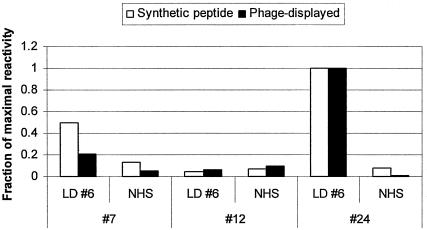
We next determined whether ELISA results obtained with purified phage preparations were equivalent to those obtained with synthetic peptides covalently linked to ELISA plates. Synthetic peptides no. 7 and 24, representing motif A sequences, and synthetic peptide no. 12, representing a motif B sequence, were reacted in 1 mM concentrations in EMCS-derivatized Covalink plates (Nalge Nunc International) and washed with TBS prior to the performance of ELISAs. Purified preparations of phage bearing the corresponding peptides were diluted in TBS and adsorbed to 96-well Maxisorb plates, as described in Materials and Methods. Patient and control sera (provided by George P. Smith) were tested at the same dilution of 1:1,600 as was previously reported for ELISAs detecting IgG reactivity with purified phage preparations (10). We found that the IgG reactivity patterns of serum from a Lyme disease patient and control sera from healthy humans were the same for synthetic peptides as for the corresponding phage-displayed peptides (Fig. 1). The reproducibility of the synthetic-peptide ELISA was evaluated by preparing duplicate plates containing each of the three synthetic peptides (no. 7, 12, and 24) and performing ELISAs on two different days with Lyme disease patient no. 6 and control arthritis patient no. 12 sera at 1:800 and 1:1,600 dilutions. The IgG reactivities in each assay were normalized by dividing the average kinetic velocity of each serum sample for each peptide with the highest kinetic velocity observed in the assay, to yield the fraction of maximal IgG reactivity to each peptide. These data were analyzed by a general linear-model analysis of variance to generate F ratios which relate the variance ascribable to any particular variable to the random error of the total experiment. In this experiment, the between-assay variability (F ratio, 0.04; probability level, 0.8514) was low, indicating that the assay is reproducible, and also the variability due to serum dilution (F ratio, 0.31; probability level, 0.5854) was low, indicating that twofold differences in serum dilutions give equivalent results. The difference between Lyme disease patient no. 6 and control patient no. 12 serum reactivities for the entire experiment was highly significant (F ratio, 13.81; probability level, 0.0017), indicating that the assay is highly sensitive for detecting differences in serum reactivities to the peptides being tested.
FIG. 1.
Synthetic peptides and phage-displayed peptides give similar patterns of reactivity in ELISAs. Synthetic peptides no. 7 and 24, representing motif A sequences, and synthetic peptide no. 12, representing a motif B sequence, were reacted in 1 mM concentrations in EMCS-derivatized Covalink plates (Nalge Nunc International) and washed with TBS prior to the performance of ELISAs. Purified preparations of phage bearing the corresponding peptides were diluted to a concentration of 4 × 1011 phage particles/ml in TBS and added in 50-μl/well volumes to 96-well Maxisorb plates for overnight absorption. Sera from Lyme disease patients (LD) and control sera from healthy (normal) human subjects (NHS) were tested at a dilution of 1:1,600 in TTDBA buffer for IgG reactivity, as described in Materials and Methods. Serum reactivities observed with the two ELISA methods were normalized by expressing the IgG reactivity of each serum sample with each peptide as a fraction of the maximal IgG reactivity observed in each set of experiments.
Sequence motifs have homologies with B. burgdorferi and other bacterial proteins.
To test the hypothesis that “epitope discovery” is useful for identifying antigens that elicit antibodies over the course of an infection, we conducted phi-BLAST searches of the nonredundant NCBI database of bacterial proteins for similarities with peptide sequences representing five different motifs reported by Kouzmitcheva et al. (10). Sequence similarities with proteins from a number of bacteria that are either normal flora or potential pathogens of human hosts were found in four out of the five motifs (Table 3). Sequence similarities with proteins from bacteria that do not infect or colonize human hosts were also found but are not considered here, since the patients in this study are unlikely to have had immunologic exposures to such antigens. The most notable findings from the phi-BLAST search were the strong similarities of two sequence motifs with B. burgdorferi proteins. The motif A sequence had a 7 out of 9 amino acid residue match with the 9 C-terminal residues of the VlsE antigen, for which the high-resolution crystal structure (5) can be accessed online through the GenBank protein database (gi|21730691|pdb|1L8W|A). The motif C sequence had complete identity with a 5-residue stretch spanning positions 19 to 24 in the N-terminal portion of the BBA61 protein (7). Both the A and C motifs also had sequence similarities with other bacterial proteins. In the case of motif A, similarities were found with hypothetical proteins encoded by Escherichia coli and Clostridium tetani. It seems unlikely that the E. coli protein is responsible for eliciting an antibody response, since if it were responsible, one would expect pooled normal sera and control sera to have these reactivities, and they did not (Table 1 and Fig. 1). The hypothetical C. tetani protein CTC0157 can probably be eliminated as the eliciting antigen, since it is not a constituent of the Tetanus toxoid vaccine and since these Lyme disease patients are unlikely to have experienced a C. tetani infection. In the case of motif C, similarities were found with hypothetical proteins encoded by Listeria monocytogenes, Helicobacter pylori, Haemophilus influenzae, and Bacteroides thetaiotaomicron. The close similarity of this motif with protein sequences from bacteria that frequently colonize or infect humans could represent a case where antibodies that cross-react with the BBA61 epitope were elicited. However, these sequence similarities may be only coincidental, and serologic reactivity may be truly related to B. burgdorferi infection, since 3 of 10 serum samples from Lyme disease patients and 0 of 10 serum samples from control patients reacted with B. burgdorferi in the report of Kouzmitcheva et al. (10).
TABLE 3.
Peptide motif similarities with sequences in bacterial protein databases
| Peptide motif | Query sequencea | Protein match | Organism |
|---|---|---|---|
| A | SKEKPPSLNWPA | ||
| SKETPPALN | VlsE protein | Borrelia burgdorferi | |
| SKEKPPTFN | Hypothetical protein CTC0157 | Clostridium tetani | |
| PSLNWP | Hypothetical protein | Escherichia coli | |
| B | VPVDAPHAGTKPSA | ||
| ADQAGTKP | Type 1 M protein | Streptococcus pyogenes | |
| VEVDAPHAGT | Dihydrolipoamide S-acetyltransferase | Brucella melitensis | |
| C | PKSSCTQNPILCAILS | ||
| NPILCA | Hypothetical protein BBA61 | Borrelia burgdorferi | |
| ILCAIL | Hypothetical protein lmo0990 | Listeria monocytogenes | |
| ILCAILS | Hypothetical protein HP1015 | Helicobacter pylori | |
| TQNPIL | Hypothetical protein HI0493 | Haemophilus influenzae | |
| NPILGAILS | Integral membrane protein | Bacteroides thetaiotaomicron | |
| F | EKRFACKPLCNTPA | ||
| KGFACKVL | Hypothetical protein VC1354 | Vibrio cholerae | |
| H | RSECSTSKCVRK | No matches found |
Underlined residues match residues in query sequences, also shown by vertical alignment.
Motif B had a high degree of similarity to a sequence in the type 1 M protein of Streptococcus pyogenes and to a sequence in the dihydrolipoamide S-acetyltransferase of Brucella melitensis. The similarity with the type 1 M protein of S. pyogenes is intriguing and suggests that perhaps this peptide was selected by sera from Lyme disease patients who coincidentally had high antistreptococcal antibody titers due to prior group A streptococcal infections. If this is indeed the case, this motif will obviously have little diagnostic utility for Lyme disease. Motif F was similar to a sequence in a hypothetical protein encoded by Vibrio cholerae. We believe that the similarity of motif B with the dihydrolipoamide S-acetyltransferase of B. melitensis and the similarity of motif F with a hypothetical protein of V. cholerae are likely to represent chance sequence similarities unrelated to antigenic exposures, since infections with either of these organisms are extremely rare in the United States. No similarities were found between the motif H sequence and any proteins contained in the bacterial database by using the default parameters of the phi-BLAST search algorithm. It should be noted that both the motif F and H peptides were selected from libraries containing two cystine residues and hence may be conformationally “constrained.” Such peptides may represent conformational epitopes whose linear sequence may or may not have any similarity with that of the eliciting antigenic epitope.
Verification of a new VlsE epitope identified by similarity searches.
Of the five motifs we examined, the similarity between the motif A sequence and a 9-residue sequence at the C terminus of the B. burgdorferi VlsE antigen appears to represent the most likely instance where “epitope discovery” identified the antigen responsible for eliciting the antibody response. In fact, prior work with a 51-mer synthetic peptide of the C terminus of the VlsE antigen has shown that this region contains an immunodominant epitope(s) that is apparently associated with the B31 strain of B. burgdorferi (13). The similarity of motif C with the BBA61 protein of B. burgdorferi may also represent another case where “epitope discovery” identified an eliciting antigen. However, due to the possible presence of cross-reacting antibodies to the BBA61 protein, we chose to focus our attention on verifying the immunologic reactivity of the putative VlsE epitope.
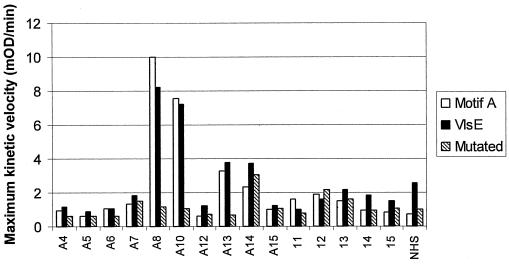
To verify that the reactivity of Lyme disease patient sera to the motif A peptide-represented antibodies elicited by the VlsE1 protein epitope, we synthesized peptides of similar lengths (13 to 14 amino acids) derived from the motif A sequence (SKEKPPSLNWPAC) and the actual VlsE sequence (KAASKETPPALNKC) with C-terminal cystines added (consensus sequences are underlined). In addition, to verify that the consensus residues were required for antibody reactivity, a peptide containing conservative amino acid substitutions at five of the consensus sequence residues was synthesized (KAASREKGGAVQKC). These peptides were tested in parallel with our own set of sera from patients with extracutaneous Lyme disease. The ELISA results demonstrated that the motif A sequence and VlsE sequence had the same pattern and level of reactivity to the Lyme disease patient sera and that there was little or no reactivity to the mutated consensus sequence (Fig. 2).
FIG. 2.
Similar serum reactivity patterns of the motif A consensus peptide and the VlsE peptide. Peptides representing the motif A consensus sequence, the VlsE sequence, and the mutated motif A consensus sequence were adjusted to 10 μM concentrations in 7 M guanidine-HCl and linked via free sulfhydryl groups in C-terminal cystine residues to EMCS-derivatized 96-well CovaLink plates, as described in Materials and Methods. Sera were diluted 1:100 in TTDBA and incubated in duplicate wells (100 μl/well) of 96-well plates for 2 h at room temperature. Human IgG binding was detected by ELISA as described in Materials and Methods. Sera A4 to A15 were from patients with extracutaneous Lyme disease, and sera 11 to 15 were from control patients with rheumatoid arthritis without Lyme disease. Serum samples pooled from healthy (normal) human subjects (NHS) were obtained commercially (Sigma) and were also tested as a control.
Diagnostic sensitivity of the VlsE C-terminal epitope relative to the immunodominant VlsE C6 epitope.
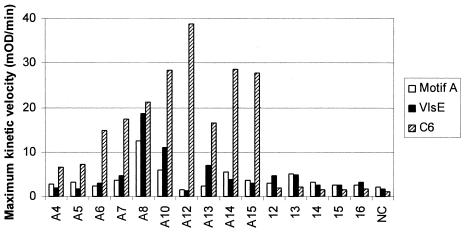
Once we had determined that the C-terminal peptide sequence, SKETPPALNK, of the VlsE antigen was likely to represent an epitope eliciting antibody responses in patients infected with B. burgdorferi, we wished to determine its diagnostic sensitivity relative to the well-characterized immunodominant C6 diagnostic peptide contained in the sixth invariant region of the VlsE antigen (11, 12, 19). Synthetic peptides comprising the C6 peptide sequence (2), the VlsE C-terminal sequence, and the motif A consensus sequence were diluted to 10 μM concentrations in 7 M guanidine-HCl and linked to EMCS-derivatized Covalink plates, as described in Materials and Methods. The IgG reactivities of patients with extracutaneous Lyme disease and control patients with rheumatoid arthritis were detected by ELISA as described above and expressed as the maximal kinetic velocity of substrate conversion. Receiver operating curves were constructed from the data in order to establish a cutoff value for evaluating the diagnostic sensitivity of each peptide. A cutoff value of 6 mOD/min was chosen to evaluate the sensitivity of each peptide based on the criteria of selecting the lowest value for all three peptides that was 100% specific. At this cutoff value, the C6 peptide was 100% sensitive for the serodiagnosis of Lyme disease, the newly defined VlsE epitope was 30% sensitive, and the motif A peptide was 20% sensitive (Fig. 3). These results are consistent with reports that identify the C6 peptide as an immunodominant epitope (11) and with the report of Liang et al. (13) that identifies the C-terminal region as containing strain-dependent epitopes with less diagnostic sensitivity in human populations than the C6 region. It appears likely that we have identified a strain-specific epitope that would obviously be of interest to examine for sequence variation among different sensu lato strains of B. burgdorferi (13).
FIG. 3.
Reactivities of serum IgG from Lyme disease and rheumatoid arthritis patients to the motif A peptide, VlsE C-terminal peptide, and VlsE C6 peptide. Peptides representing the motif A consensus sequence, the C-terminal VlsE sequence, and the VlsE C6 peptide were adjusted to 10 μM concentrations in 7 M guanidine-HCl and linked via free sulfhydryl groups in C- or N-terminal cystine residues to EMCS-derivatized 96-well CovaLink plates, as described in Materials and Methods. Sera were diluted 1:100 in TTDBA and incubated in duplicate wells (100 μl/well) of 96-well plates for 2 h at room temperature. Human IgG binding was detected by ELISA as described in Materials and Methods. The designations for the patient sera are the same as those described in the legend for Fig. 2. NC, serum samples from healthy (normal) human subjects that were tested as a control.
Summary and conclusions.
The identification of a new linear epitope in the VlsE antigen of B. burgdorferi described in this investigation serves as a proof of the principle of the utility of “epitope discovery” in detecting antigens responsible for serologic responses to infectious agents. Our results provide some insight into the promise and potential pitfalls of this approach, as applied to discovering diagnostic reagents. To be most effective in identifying diagnostically relevant antigens, “epitope discovery” requires a source of disease-specific antibodies, diverse phage-displayed peptide libraries for epitope selection, an efficient procedure to deplete phage libraries of irrelevant specificities found in healthy-human serum, and a complete DNA or protein database of the etiologic agent in question. We were aided in this investigation by having access to peptides that were selected under protocols designed to satisfy the first three conditions and by the availability of the complete genomic sequence of the infectious entity under study, B. burgdorferi. The promise of “epitope discovery” was demonstrated by finding sequence similarities between two B. burgdorferi antigens, VlsE and BBA61, and two of the five sequence motifs we searched. In the case of the VlsE antigen, we verified that its C-terminal sequence and the motif A sequence had the same patterns of reactivity with patient sera and that the consensus sequence was required for antibody binding. Pitfalls of this technique were also evident in three different respects. (i) In the case of the motif C match with the BBA61 antigen, there were matches with several other bacterial proteins that could potentially elicit cross-reacting antibodies. Hence, it is unclear which antigen actually elicited the original antibody response. (ii) Matches were found between B and H motifs and bacterial proteins that the human subjects whose sera were used for selection were unlikely to have encountered. This result highlights the inherent risk of finding matches with irrelevant proteins due to chance sequence similarities when conducting searches of protein databases. (iii) Finally, it is somewhat surprising that other known immunodominant epitopes of B. burgdorferi, such as the C6 peptide on the VlsE antigen and epitopes on antigens such as DbpA, OspC, and BmpA, were not discovered. The failure to detect more of such epitopes may be related to several theoretical limitations of “epitope discovery” utilizing phage-displayed peptides that have been treated in depth by Smith and Petrenko (17) in their excellent review of phage display technology. The limitations that they discuss basically fall into three categories: (i) insufficient peptide diversity, (ii) insufficient peptide structural stability (i.e., “floppiness”), and (iii) selection of conformational “mimotopes” that have little or no primary sequence similarity to actual antigenic epitopes. Taken together, these limitations could account for the limited number of epitopes that were found in this investigation. However, our results illustrate the promise of this technique, in that by identifying specific epitopes, new information can be gained about the basis for heterogeneity in serologic reactivities observed for known antigens. Moreover, the identification of even one epitope can be extremely valuable when attempting to develop diagnostic reagents and/or identify the causative agent for diseases of unknown etiology. Taken together, we believe these results validate “epitope discovery” with phage-displayed peptide libraries as a valuable approach for specifically identifying epitopes with diagnostic utility on new or previously known antigens. Moreover, as sequence information continues to accumulate in protein databases, this approach will have an increasing likelihood of success when applied to identifying the etiologies of various diseases and pathologies whose causes are currently unknown.
Acknowledgments
We thank George P. Smith for his generous donations of synthetic diagnostic peptides, purified phage clones, and patient sera for this study. We also thank him for providing detailed technical advice that was critical for the successful implementation of synthetic-peptide ELISA protocols.
REFERENCES
- 1.Adda, C. G., R. F. Anders, L. Tilley, and M. Foley. 2002. Random sequence libraries displayed on phage: identification of biologically important molecules. Comb. Chem. High Throughput Screen. 5:1-14. [DOI] [PubMed] [Google Scholar]
- 2.Bacon, R. M., B. J. Biggerstaff, M. E. Schriefer, R. D. Gilmore, Jr., M. T. Philipp, A. C. Steere, G. P. Wormser, A. R. Marques, and B. J. B. Johnson. 2003. Serodiagnosis of Lyme disease by kinetic enzyme-linked immunosorbent assay using recombinant VlsE1 or peptide antigens of Borrelia burgdorferi compared with 2-tiered testing using whole-cell lysates. J. Infect. Dis. 187:1187-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortese, I., R. Tafi, L. M. Grimaldi, G. Martino, A. Nicosia, and R. Cortese. 1996. Identification of peptides specific for cerebrospinal fluid antibodies in multiple sclerosis by using phage libraries. Proc. Natl. Acad. Sci. USA 93:11063-11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cortese, R., F. Felici, G. Galfre, A. Luzzago, P. Monaci, and A. Nicosia. 1994. Epitope discovery using peptide libraries displayed on phage. Trends Biotechnol. 12:262-267. [DOI] [PubMed] [Google Scholar]
- 5.Eicken, C., V. Sharma, T. Klabunde, M. B. Lawrenz, J. M. Hardham, S. J. Norris, and J. C. Sacchettini. 2002. Crystal structure of Lyme disease variable surface antigen VlsE of Borrelia burgdorferi. J. Biol. Chem. 277:21691-21696. [DOI] [PubMed] [Google Scholar]
- 6.Folgori, A., R. Tafi, A. Meola, F. Felici, G. Galfre, R. Cortese, P. Monaci, and A. Nicosia. 1994. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 13:2236-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilmore, R. D., Jr., K. J. Kappel, and B. J. B. Johnson. 1997. Molecular characterization of a 35-kilodalton protein of Borrelia burgdorferi, an antigen of diagnostic importance in early Lyme disease. J. Clin. Microbiol. 35:86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iniguez, P., S. Zientara, M. Marault, I. B. Machin, D. Hannant, and C. Cruciere. 1998. Screening of horse polyclonal antibodies with a random peptide library displayed on phage: identification of ligands used as antigens in an ELISA test to detect the presence of antibodies to equine arteritis virus. J. Virol. Methods 73:175-183. [DOI] [PubMed] [Google Scholar]
- 9.Irving, M. B., O. Pan, and J. K. Scott. 2001. Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr. Opin. Chem. Biol. 5:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kouzmitcheva, G. A., V. A. Petrenko, and G. P. Smith. 2001. Identifying diagnostic peptides for Lyme disease through epitope discovery. Clin. Diagn. Lab. Immunol. 8:150-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang, F. T., A. C. Steere, A. R. Marques, B. J. B. Johnson, J. N. Miller, and M. T. Philipp. 1999. Sensitive and specific serodiagnosis of Lyme disease by enzyme-linked immunosorbent assay with a peptide based on an immunodominant conserved region of Borrelia burgdorferi VlsE. J. Clin. Microbiol. 37:3990-3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang, F. T., A. L. Alvarez, Y. Gu, J. M. Nowling, R. Ramamoorthy, and M. T. Philipp. 1999. An immunodominant conserved region within the variable domain of VlsE, the variable surface antigen of Borrelia burgdorferi. J. Immunol. 163:5566-5573. [PubMed] [Google Scholar]
- 13.Liang, F. T., L. C. Bowers, and M. T. Philipp. 2001. C-terminal invariable domain of VlsE is immunodominant but its antigenicity is scarcely conserved among strains of Lyme disease spirochetes. Infect. Immun. 69:3224-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motti, C., M. Nuzzo, A. Meola, G. Galfre, F. Felici, R. Cortese, A. Nicosia, and P. Monaci. 1994. Recognition by human sera and immunogenicity of HBsAg mimotopes selected from an M13 phage display library. Gene 146:191-198. [DOI] [PubMed] [Google Scholar]
- 15.Peltomaa, M., G. McHugh, and A. C. Steere. 2003. Persistence of the antibody response to the VlsE sixth invariant region (IR6) peptide of Borrelia burgdorferi after successful antibiotic treatment of Lyme disease. J. Infect. Dis. 187:1178-1186. [DOI] [PubMed] [Google Scholar]
- 16.Santamaria, H., K. Manoutcharian, L. Rocha, E. Gonzalez, G. Acero, T. Govezensky, L. I. Uribe, A. Olguin, J. Paniagua, and G. Gevorkian. 2001. Identification of peptide sequences specific for serum antibodies from human papillomavirus-infected patients using phage display libraries. Clin. Immunol. 101:296-302. [DOI] [PubMed] [Google Scholar]
- 17.Smith, G. P., and V. A. Petrenko. 1997. Phage display. Chem. Rev. 97:391-410. [DOI] [PubMed] [Google Scholar]
- 18.Yu, Z., J. M. Carter, S. Y. Huang, H. Lackland, L. H. Sigal, and S. Stein. 1996. Presentation of peptide antigens as albumin conjugates for use in detection of serum antibodies by enzyme-linked immunosorbent assay. Bioconjug. Chem. 7:338-342. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]