Abstract
Group A streptococci cause a wide spectrum of clinical illness. One of several strategies for vaccine prevention of these infections is based on the type-specific M protein epitopes. A multivalent M protein-based vaccine containing type-specific determinants from 26 different M serotypes is now in clinical trials. Recent epidemiologic studies have shown that, within some serotypes, the amino-terminal M protein sequence may show natural variation, giving rise to subtypes. This raises the possibility that vaccine-induced antibodies against the parent type may not be as effective in promoting bactericidal killing of variant subtypes. In the present study we used rabbit antisera against the 26-valent M protein-based vaccine in bactericidal tests against M1, M3, and M5 streptococci, which were represented by multiple subtypes. We show that the vaccine antibodies effectively promoted in vitro bactericidal activity despite the fact that the M proteins contained naturally occurring variant sequences in the regions corresponding to the vaccine sequence. Our results show that the variant M proteins generally do not result in significant differences in opsonization promoted by rabbit antisera raised against the 26-valent vaccine, suggesting that a multivalent M protein vaccine may not permit variant subtypes of group A streptococci to escape in a highly immunized population.
The M protein of group A streptococci is one of the major virulence determinants of these organisms and also functions as a major protective antigen (6). Immunization of animals with purified M protein or M protein fragments elicits opsonic antibodies that correlate with protection from lethal challenge infections by the same serotype. Serum bactericidal antibodies also appear following natural infection of humans and are associated with protection against subsequent infection with the same serotype (8). These observations have served as the basis for the development of multivalent vaccines containing type-specific fragments of M proteins designed to evoke opsonic antibodies against the prevalent serotypes of group A streptococci (11, 12). We have recently conducted studies of the safety and immunogenicity of a 26-valent vaccine in animals (5) and humans (10). This vaccine contains type-specific peptides of M proteins from group A streptococcal serotypes that account for approximately 85% of cases of uncomplicated pharyngitis and invasive infections in North America (9, 11, 12).
One of the issues related to this vaccine strategy is whether natural variations in N-terminal M protein sequences might affect the functional activity of vaccine-induced opsonic antibodies. Variations in emm gene sequences occur which result in amino acid substitutions or insertion of short sequences within the N-terminal, hypervariable segments of the M proteins (9). Ongoing epidemiologic studies in the United States and Canada (9, 12) have revealed that the vast majority of 5′ emm sequences within a given serotype are conserved and are identical to those included in the 26-valent vaccine. However, within some serotypes there are occasional isolates that express M proteins with amino acid sequences that differ from the prevalent “parent” strain. If the sequence variations within a serotype result in changes in functional activity of vaccine-induced antibodies, this raises the theoretical possibility that in a highly immunized population these relatively rare subtypes could eventually emerge as predominant pathogens.
In the present study, we assessed indirect bactericidal activity of rabbit antisera raised against the 26-valent M protein-based vaccine against a collection of serotype M1, M3, and M5 strains that express variant M proteins. The results show that the subtypes within each type were opsonized by the vaccine antisera, indicating that the variation in M protein sequences may not be immunologically significant.
MATERIALS AND METHODS
Rabbit antisera against the 26-valent vaccine.
The 26-valent vaccine consists of four different recombinant proteins, each containing six or seven N-terminal M protein fragments linked in tandem (5). The individual peptide components range in size from 30 to 80 amino acids and represent M proteins from 26 different serotypes of group A streptococci. New Zealand White rabbits were immunized with three doses consisting of 400 μg of the 26-valent vaccine adsorbed to 750 μg alum via the intramuscular route, as previously described (5). The antisera used in these studies were obtained from rabbits that were immunized at 0, 4, and 8 weeks or 0, 4, and 16 weeks. All sera were obtained at least 2 weeks following the last booster injection of vaccine.
Group A streptococcal strains.
The bacterial strains used in this study were obtained from the ongoing North America Group A Streptococcal Pharyngitis Serotype Surveillance Study (12) and from the Active Bacterial Core surveillance of invasive group A streptococcal infections at the Centers for Disease Control and Prevention (9). The M1, M3, and M5 serotypes were selected because each was represented by a substantial number of different subtypes.
Bactericidal assays.
Indirect bactericidal assays were performed essentially as described by Lancefield (7), with minor modifications. Test organisms were grown overnight at 25°C in Todd-Hewitt broth (THB) supplemented with 1% yeast extract and 20% normal rabbit serum, subcultured in fresh broth the next day, and grown to an optical density of ∼0.05 at 530 nm. Tenfold dilutions in THB were prepared, 0.05 ml of diluted streptococci was added to 0.1 ml of test serum, and the mixture was incubated for 15 min at 37°C and 15 min on ice. The inoculum for each test was determined by making pour plates using melted sheep's blood agar. After preincubation with the test serum, 0.35 ml of nonimmune whole human blood was added and the mixture was rotated end over end at 12 rpm for 3 h at 37°C. At the end of the incubation, the number of surviving bacteria was quantitated using the pour plate method.
Multiple bactericidal tests were performed, ranging from 3 to 10 assays with each strain of group A streptococcus. Four different rabbit antisera against the 26-valent vaccine were used in these studies and were selected based on the levels of bactericidal antibodies observed using the “parent” vaccine strain of each serotype. Criteria for including the results of an assay in the analysis were the following: (i) growth of the test organism in control tubes (normal rabbit serum) increased during the 3-h rotation by a factor more than 32 times the inoculum (five generations) and (ii) growth of the test organism in the control tubes was quantifiable, meaning that the plate was not “laked,” i.e., that the colonies were not too numerous to count. Bactericidal activity was expressed as percent kill and was calculated using the formula [(CFU after 3 h of growth with normal rabbit serum − CFU after 3 h of growth with immune serum)/CFU after 3 h of growth with normal rabbit serum] ×100.
Statistical analyses.
Statistical comparisons were made using a one-way analysis of variance on GraphPad Prism software (GraphPad Software, Inc., San Diego, CA).
RESULTS AND DISCUSSION
The overall goal of these studies was to determine whether the bactericidal activity of vaccine-induced antibodies was affected by natural variation in sequences observed in the N-terminal regions of M proteins expressed by subtypes of group A streptococci. The 26-valent vaccine contains amino acid residues 1 to 50 of the mature M1.0 and M1.2 M proteins (Table 1). M1.4, whose sequence is significantly different from M1.0 and M1.2, is not included in the vaccine. M3.1 is the vaccine strain and is represented by amino acid residues 21 to 70. Deletion of the first 20 amino acids was based on experimental evidence showing that antisera evoked by a synthetic peptide copying the N-terminal region of M3.1 were much less opsonic than those raised against peptides from the 36 to 70 sequence (2). The vaccine contains the amino-terminal 30 amino acids of M5.0 (Table 1).
TABLE 1.
Amino acid sequences of the M subtypes and their frequencies of occurrence
| Subtype | No. of isolates recovereda | M sequence corresponding to vaccine subunitb |
|---|---|---|
| 11020304050 | ||
| M1.0 | 586 | NGDGNPREVIEDLAANNPAIQNIRLRHENKDLKARLENAMEVAGRDFKRA |
| M1.2 | 0 | NNDGRSRDVTEEIAANNTTVQNIRLRNENKNLKAKNEDLKARLENAMNVA |
| M1.4 | 1 | NENVGPRDVVKELVEKDPVLQNKRLRSENQKLKESLENARDVAGRDFKRA |
| M1.8 | 1 | NGDGNPREVIEDLAANNPAIQNIRLRHHENKDLKARLENAMEVAGRDFKRA |
| M1.9 | 1 | NGDGNPREVIEDLAANNPAIRNIRLRHENKDLKARLENAMEVAGRDFKRA |
| M1.10 | 1 | NGDGNPREVIEDLAANNPVIQNIRLRHENKDLKARLENAMEVAGRDFKRA |
| M1.11 | 1 | NGDGNPREVIEDLAANNPAIQNIRLRHENKVLKARLENAMEVAGRDFKRA |
| M1.13 | 1 | NDDGNPREVIEDLAANNPAIQNIRLRHENKDLKARLENAMEVAGRDFKRA |
| 213040506070 | ||
| M3.1 | 196 | NLLDQVTQLYTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.2 | 9 | NLLDQVTQLYTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLDVKK |
| M3.4 | 51 | NLLNQVTQLYTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.6 | 2 | NLLNQVTQLHTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.7 | 1 | NLLAQVTQLYTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.8 | 1 | NLLDQVTQLYTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.9 | 1 | NLLDQVTQLYTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.10 | 1 | NLLNQVTQLYAKHNSNHQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.11 | 1 | NLLDQVTQLYTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.12 | 8 | NLLNQVTQLYTRHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.13 | 1 | NLLDQVTQLYTKHNSNYQLYTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.14 | 1 | NLLDQVTQLYTKHNSNYQQYNAQAGTLDLRQKAEYLKGLNDWAERLLQEL |
| M3.15 | 2 | NLLDQVTQLYTKHNSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| M3.16 | 1 | NLLDQVTQLYTKHNSNYQQYNAQDSNYQQYNAQAGRLDLRQKAEYLKGLNDWAERLLQEL |
| 1102030 | ||
| M5.0 | 1 | AVTRGTINDPQRAKEALDKYELENHDLKTK |
| M5.6 | 6 | TVTRGTINDPQRAKEALDKYELENHDLKTK |
| M5.7 | 1 | TVTRGTINDPQRAKEALDKYELENHDLKTK |
| M5.8 | 1 | TVTRGTINDPQRAKEALDKYELENHDLKTE |
| M5.10 | 1 | TVTRSTINDPQRAKEALDKYELENHDLKTK |
| M5.13 | 2 | VVTRSTINDPQRAKEALDKYELENHDLKTE |
| M5.14 | 27 | TVTRGTINDPQRAKEALDKYELENHDLKTK |
| M5.18 | 2 | TVTRGTVNDPQRAKEILDKYELENHDLKTK |
| M5.19 | 1 | TVTGGTINDPQRAKEALDKYELENHDLKTK |
| M5.22 | 1 | TVTRGTINDPQRAKAALDKYELENHDLKTK |
The vast majority of M1 isolates recovered from patients with uncomplicated pharyngitis or invasive infections expressed the M1.0 sequence (586/592, 99%). All other M1 subtypes were recovered only once. Similarly, M3.1 accounted for 196/276 (71%) of the total M3 serotypes isolated. M3.2, M3.4, and M3.12 were recovered on multiple occasions. Serotype M5 was not a common serotype in the surveillance studies (9, 12), and the vaccine strain M5.0 was represented by a single isolate among 43 M5 strains. Interestingly, M5.14 was the most common subtype isolated (27/43, 63%).
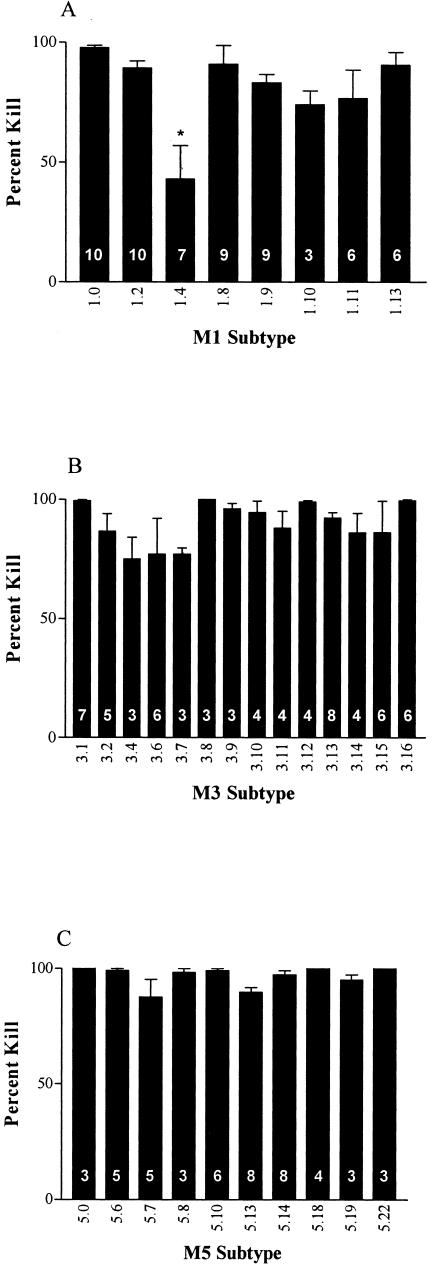
To determine the potential protective efficacy of the 26-valent vaccine against subtypes of group A streptococci, indirect bactericidal tests were performed (Fig. 1). The M1.0 and M1.2 strains of streptococci, which are both vaccine strains, were effectively killed in the presence of the 26-valent antiserum. The remaining subtypes of M1, with the exception of M1.4, were also opsonized and killed in the presence of vaccine-induced antiserum (Fig. 1A). Similar observations were made with the subtypes of M3 (Fig. 1B). All of the M3 strains were opsonized by the vaccine antiserum, despite the fact that some M3 subtypes (M3.13 and M3.16) contained insertion sequences within the region of the vaccine peptide (Table 1). Additionally, all of the M5 subtypes tested were killed in the presence of the 26-valent vaccine antiserum. Taken together, these results indicate that minor variations in M protein sequences generally do not significantly affect the ability of the vaccine-induced antibodies to opsonize subtypes of the same serotype of group A streptococci. However, major variations in N-terminal M protein sequence, such as in M1.4, result in significantly lower levels of bactericidal killing.
FIG. 1.
In vitro bactericidal tests performed with rabbit antisera against the 26-valent M protein-based group A streptococcal vaccine and subtypes of M1 (A), M3 (B), and M5 (C). Percent kill (±standard error of the mean) was calculated based on the formula provided in Materials and Methods. The number within the base of each bar indicates the number of assays performed for each strain. *, statistically significant difference (P < 0.001) compared to the result obtained with M1.0 based on a one-way analysis of variance.
Our results suggest that, in a highly vaccinated population, subtypes of group A streptococci would be unlikely to emerge as escape variants. Previous studies have shown that synonymous base substitutions are almost nonexistent within type-specific regions of M proteins and that significant variation of amino acid sequences within these type-specific regions is extremely rare (9, 12). In addition, during the past 60 years the most common type-specific sequences have remained constant in the vast majority of isolates. These observations suggest that, while M proteins are likely under intense immunologic selective pressure, there may be structural constraints on the hypervariable region of the M proteins that confer some advantage in the host-pathogen interaction and this may prevent an infinite number of sequences from arising through selective pressure.
In the present study, M1.4 was the only subtype that was not opsonized effectively by the 26-valent vaccine antisera. Based on current emm-typing criteria and nomenclature (4), neither M1.2 nor M1.4 would be considered M1 strains since the sequences of the N-terminal regions are considerably different from that of the parent M1.0. Since M1.2 is represented in the vaccine, it was effectively killed in the in vitro assays. However, the bactericidal activity against M1.4 was significantly lower than that against the vaccine strain M1.0. Previous studies with human sera indicated that opsonization of M1 streptococci may be strain specific (3, 13). Human sera variably opsonized genetically different strains of M1 (3), and, in some instances, strains with identical M1 protein sequences showed highly divergent levels of opsonization by the same human serum (13). Interestingly, not all convalescent-phase human sera showed variable levels of opsonic antibodies against different M1 strains, suggesting that differences in individual human immune responses together with subtle differences among M1 strains might account for these results. Whether M protein-based vaccines that contain a limited region of the M1 protein (50 amino acids) will also evoke a spectrum of functional human immune responses will best be determined during large-scale clinical trials.
A more recent study (1) of type 3 streptococci (M3.1 and M3.2) that caused two different peaks of infection in Canada implicated a 4-amino-acid duplication at the N-terminal end of the M3 protein as a contributing factor. The authors reasoned that the M3.2 variant became more prominent in the second wave of infection as a consequence of host immune pressure. In previous studies, we showed that antibodies evoked in rabbits against a synthetic peptide copying the first 35 amino acids of M3.1 had little to no opsonic activity (2). The antisera reacted strongly with the purified M3.1 protein but reacted poorly with the native protein on the surface of the organism, suggesting that the N terminus of the M3.1 protein may be masked or conformationally altered so as not to allow antibody recognition. Additional studies revealed that antibodies evoked by residues 36 to 70 of the M3.1 protein were strongly opsonic (2). For this reason, the 26-valent vaccine contains the amino acid sequence 21 to 70 of M3.1. Thus, our previous studies together with the data reported here showing that the 26-valent vaccine evoked bactericidal antibodies against both M3.1 and M3.2 suggest that the addition of 4 amino acids to the N terminus of M3.2 may not have a significant effect on opsonic activity. Studies using convalescent-phase sera from patients with M3.1 and M3.2 infections would be needed to provide definitive evidence of the precise role of the N-terminal antibodies in protection against infection by each subtype.
Taken together, the results of the present study and those of previous surveillance studies suggest that (i) the majority of clinical isolates of group A streptococci express M proteins that are identical to the Centers for Disease Control and Prevention reference strains and to the vaccine strains that are included in the 26-valent vaccine (9, 12), (ii) when sequence variation gives rise to subtypes, most vary by only one or two amino acid substitutions, and (iii) the variant M proteins generally do not result in significant differences in opsonization promoted by rabbit antisera raised against the 26-valent vaccine. These results suggest that a multivalent M protein vaccine is unlikely to evoke antibodies that would permit variant subtypes of group A streptococci to escape in a highly immunized population. Large-scale clinical trials will be required to provide definitive evidence for this conclusion.
Acknowledgments
This work was supported by National Institutes of Health grant R37 AI100085 to James B. Dale, research funds from the Department of Veterans Affairs to James B. Dale, and research funds from ID Biomedical Corporation to Stanford T. Shulman. James B. Dale and Stanford T. Shulman receive research support from ID Biomedical Corporation.
James B. Dale owns stock in ID Biomedical Corporation.
REFERENCES
- 1.Beres, S. B., G. L. Sylva, D. E. Sturdevant, C. N. Granville, M. Liu, S. M. Ricklefs, A. R. Whitney, L. D. Parkins, N. P. Hoe, G. J. Adams, D. E. Low, F. R. DeLeo, A. McGeer, and J. M. Musser. 2004. Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc. Natl. Acad. Sci. USA 101:11833-11838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dale, J. B., M. Simmons, E. C. Chiang, and E. Y. Chiang. 1996. Recombinant, octavalent group A streptococcal M protein vaccine. Vaccine 14:944-948. [DOI] [PubMed] [Google Scholar]
- 3.de Malmanche, S. A., and D. R. Martin. 1994. Protective immunity to the group A Streptococcus may be only strain specific. Med. Microbiol. Immunol. 183:299-306. [DOI] [PubMed] [Google Scholar]
- 4.Facklam, R., B. Beall, A. Efstratiou, V. Fischetti, D. Johnson, E. Kaplan, P. Kriz, M. Lovgren, D. Martin, B. Schwartz, A. Totolian, D. Bessen, S. Hollingshead, F. Rubin, J. Scott, and G. Tyrrell. 1999. emm typing and validation of provisional M types for group A streptococci. Emerg. Infect. Dis. 5:247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu, M., M. Walls, S. Stroop, M. Reddish, B. Beall, and J. Dale. 2002. Immunogenicity of a 26-valent group A streptococcal vaccine. Infect. Immun. 70:2171-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lancefield, R. C. 1962. Current knowledge of the type specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 7.Lancefield, R. C. 1957. Differentiation of group A streptococci with a common R antigen into three serologic types with special reference to the bactericidal test. J. Exp. Med. 106:525-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lancefield, R. C. 1959. Persistence of type-specific antibodies in man following infection with group A streptococci. J. Exp. Med. 110:271-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, Z., V. Sakota, D. Jackson, A. R. Franklin, and B. Beall. 2003. Array of M protein gene subtypes in 1064 recent invasive group A streptococcus isolates recovered from the active bacterial core surveillance. J. Infect. Dis. 188:1587-1592. [DOI] [PubMed] [Google Scholar]
- 10.McNeil, S. A., S. A. Halperin, J. Langley, B. Smith, A. Warren, D. Baxendale, M. A. Reddish, M. C. Hu, S. D. Stroop, M. A. Walls, J. Linden, G. H. Lowell, L. F. Fries, P. Vink, and J. B. Dale. Safety and immunogenicity of a 26-valent group A streptococcus vaccine in healthy adult volunteers. Clin. Infect. Dis., in press. [DOI] [PubMed]
- 11.O'Brien, K. L., B. Beall, N. L. Barrett, P. R. Cieslak, A. Reingold, M. M. Farley, R. Danila, E. R. Zell, R. Facklam, B. Schwartz, and A. Schuchat. 2002. Epidemiology of invasive group a streptococcus disease in the United States, 1995-1999. Clin. Infect. Dis. 35:268-276. [DOI] [PubMed] [Google Scholar]
- 12.Shulman, S. T., R. R. Tanz, W. Kabat, K. Kabat, E. Cederlund, D. Patel, Z. Li, V. Sakota, J. B. Dale, and B. Beall. 2004. Group A streptococcal pharyngitis serotype surveillance in North America, 2000-2002. Clin. Infect. Dis. 39:325-332. [DOI] [PubMed] [Google Scholar]
- 13.Villasenor-Sierra, A., W. M. McShan, D. Salmi, E. L. Kaplan, D. R. Johnson, and D. L. Stevens. 1999. Variable susceptibility to opsonophagocytosis of group A streptococcus M-1 strains by human immune sera. J. Infect. Dis. 180:1921-1928. [DOI] [PubMed] [Google Scholar]