Abstract
Techniques of comparative genomics are being used to identify candidate functional DNA sequences, and objective evaluations are needed to assess their effectiveness. Different analytical methods score distinctive features of whole-genome alignments among human, mouse, and rat to predict functional regions. We evaluated three of these methods for their ability to identify the positions of known regulatory regions in the well-studied HBB gene complex. Two methods, multispecies conserved sequences and phastCons, quantify levels of conservation to estimate a likelihood that aligned DNA sequences are under purifying selection. A third function, regulatory potential (RP), measures the similarity of patterns in the alignments to those in known regulatory regions. The methods can correctly identify 50%–60% of noncoding positions in the HBB gene complex as regulatory or nonregulatory, with RP performing better than do other methods. When evaluated by the ability to discriminate genomic intervals, RP reaches a sensitivity of 0.78 and a true discovery rate of ∼0.6. The performance is better on other reference sets; both phastCons and RP scores can capture almost all regulatory elements in those sets along with ∼7% of the human genome.
A major aim of genomics is to identify the functional segments of DNA (Collins et al. 2003; The ENCODE Project Consortium 2004), and comparative methods play a critical role in achieving this goal (Miller et al. 2004). Inclusion of comparative data has improved the accuracy of bioinformatic methods for predicting exons and gene structures (Brent and Guigó 2004), but full annotation of genes has not yet been achieved for complex genomes (Lander et al. 2001; Waterston et al. 2002; Gibbs et al. 2004; International Human Genome Sequencing Consortium 2004).
DNA segments needed to control the level, developmental timing, and spatial pattern of gene expression, termed cis-regulatory modules (CRMs), are even more difficult to identify accurately. Few constraints analogous to the rules of the genetic code, used for coding exons, can be universally applied to CRMs (Wasserman and Sandelin 2004), and thus bioinformatic predictions of CRMs commonly use one or more sources of information in addition to the DNA sequence. These can include the presumptive start site for transcription (Trinklein et al. 2003), matches to transcription factor binding sites (Wingender et al. 2001; Sandelin et al. 2004a), overrepresentation of motifs in coexpressed genes (Spellman et al. 1998), and noncoding sequence conservation (for review, see Pennacchio and Rubin 2001; Cooper and Sidow 2003; Frazer et al. 2003; Hardison 2003). However, CRMs are difficult to distinguish from neutral DNA in mammalian genomes by relatively simple human–mouse conservation scores (Waterston et al. 2002; Elnitski et al. 2003).
Interspecies comparisons can be used to infer function if aligned sequences are scored for the likelihood that they are under evolutionary constraint (purifying selection), which is often measured by an evolutionary rate slower than that observed in neutral DNA (Waterston et al. 2002; Cooper et al. 2004). Hidden Markov models and phylogenetic information have been used to estimate rates of evolution (Felsenstein and Churchill 1996) and to improve predictions of functional elements such as genes (Pedersen and Hein 2003; McAuliffe et al. 2004). Phylogenetic hidden Markov models (Siepel and Haussler 2003) have been applied to multiple sequence alignments to estimate a likelihood that a particular sequence is among the most highly conserved in a genome. A recent implementation of these models computes a score called phastCons (Siepel et al. 2005), which allows for rate variation in different lineages and assumes that adjacent bases score similarly. An earlier method finds multispecies conserved sequences (MCSs) (Margulies et al. 2003) as blocks of highly constrained aligned sequences. This algorithm weights the score by phylogenetic distance and adjusts estimates of significance by a neutral substitution rate.
In addition, aligned sequences can be analyzed for features other than degree of constraint in order to discriminate between alignments in distinct functional classes. Elnitski et al. (2003) introduced a regulatory potential (RP) score that evaluates the extent to which patterns in an alignment (strings of alignment columns) are more similar to patterns found in alignments of known regulatory elements than in alignments of ancestral repeats, which are a model for neutral DNA. This approach has been adapted to three-way alignments among human, mouse, and rat sequences (Kolbe et al. 2004). Alignments with positive scores have patterns similar to those found in the regulatory region training set, whereas those with negative scores have patterns more similar to those in aligned ancestral repeats. Although this method examines patterns in alignments, it does not use information about known factor binding sites.
In this article, we derive a set of all the known regulatory elements in the intensively studied β-globin gene complex of mammals (HBB gene complex) and use it as a reference set to evaluate the sensitivity (Sn) and specificity (Sp) of the constraint-based alignment scores (phastCons and MCSs) and the pattern-matching RP score. This calibration of the scores allows their effectiveness to be evaluated genome-wide, using alignments of the human, mouse, and rat genomes.
Results
Reference set of known regulatory elements from the HBB complex
DNA sequences needed to regulate the set of developmentally controlled, erythroid-specific genes encoding β-globin and its relatives have been studied intensively, and in this gene complex, the fraction of human sequences aligning with mouse and rat (35%) is very close to the genome average (Gibbs et al. 2004). Thus, regulatory elements in this gene complex comprise a good (but not perfect) data set with which to assess false-positive and false-negative rates for bioinformatic predictions of CRMs. This cluster of genes at human chromosome 11p15.5, referred to here as the HBB complex, includes the embryonically expressed HBE1, the fetally expressed HBG1 and HBG2, and HBD and HBB, which are expressed in adult life, along with a pseudogene HBBP1. A reference set of all known CRMs was assembled from the literature describing this gene complex, including promoters for the genes, upstream sequences (adjacent to the promoter) implicated in regulation, and five DNase hypersensitive sites in the distal strong enhancer called the locus control region (for review, see Hardison et al. 1997; Forget 2001; Hardison 2001; Li et al. 2002). The reference set includes 23 CRMs (Table 1).
Table 1.
Experimentally determined cis-regulatory modules in the HBB gene cluster.
| Chr11 Start | Chr11 Stop | CRM name | Function in regulation | Conserved in mousea | Conserved in rata | Conserved in chickena | References |
|---|---|---|---|---|---|---|---|
| 5276843 | 5277003 | HS5 | LCR, enhancer blocker | Yes | Yes | No | (Dhar et al. 1990; Yu et al. 1994; Bender et al. 1998; Li et al. 2002; Tanimoto et al. 2003; Wai et al. 2003) |
| 5273728 | 5274016 | HS4 | LCR, modulate enhancer | Yes | Yes | No | (Pruzina et al. 1991; Fraser et al. 1993; Stamatoyannopoulos et al. 1995; Molete et al. 2001) |
| 5271123 | 5271701 | HS3.2 | LCR, modulate enhancer | Yes | Yes | No | (Slightom et al. 1997; Molete et al. 2002) |
| 5270665 | 5270727 | HS3.1 | LCR | Yes | Yes | No | (Philipsen et al. 1990; Shelton et al. 1997) |
| 5270191 | 5270478 | HS3 | LCR, modulate enhancer | Yes | Yes | No | (Fraser et al. 1990, 1993; Philipsen et al. 1990, 1993; Jackson et al. 1996) |
| 5266399 | 5266483 | HS2_neg | LCR, negative | Yes | Yes | No | (Cavallesco and Tuan 1997; Elnitski et al. 2001) |
| 5266104 | 5266398 | HS2_pos | LCR, enhancer | Yes | Yes | No | (Ryan et al. 1989; Tuan et al. 1989; Fraser et al. 1990, 1993; Talbot et al. 1990) |
| 5261203 | 5261826 | HS1 | LCR | Yes | Yes | No | (Tuan et al. 1985; Forrester et al. 1986; Fraser et al. 1990, 1993) |
| 5258391 | 5258617 | HBE1_NRA | Negative | No | No | No | (Li et al. 1998) |
| 5258291 | 5258390 | HBE1_PRA | Positive | Yes | No | No | (Li et al. 1998) |
| 5257195 | 5257237 | HBE1_NRB | Negative | Yes | Yes | No | (Li et al. 1998) |
| 5256999 | 5257195 | HBE1_PRB | Positive | Yes | Yes | No | (Li et al. 1998) |
| 5255653 | 5255919 | HBE1_up | Negative | Yes | Yes | No | (Cao et al. 1989; Trepicchio et al. 1993; Watt et al. 1993) |
| 5255484 | 5255652 | HBE1_prom | Promoter | Yes | Yes | Yes | (Allan et al. 1983; Gong et al. 1991; Yu et al. 1991) |
| 5240524 | 5241054 | HBG2_up | Positive, negative | Yes | Yes | No | (Perez-Stable and Costantini 1990; Stamatoyannopoulos et al. 1993) |
| 5240320 | 5240523 | HBG2_prom | Promoter | Yes | Yes | Yes | (Gimble et al. 1988; McDonagh et al. 1991; Stamatoyannopoulos et al. 1993) |
| 5235600 | 5236122 | HBG1_up | Positive, negative | Yes | Yes | No | (Perez-Stable and Costantini 1990; Stamatoyannopoulos et al. 1993) |
| 5235395 | 5235599 | HBG1_prom | Promoter | Yes | Yes | Yes | (Gimble et al. 1988; McDonagh et al. 1991; Stamatoyannopoulos et al. 1993) |
| 5232674 | 5233423 | HBG1_3′enh | Enhancer | No | No | No | (Bodine and Ley 1987; Lloyd et al. 1994) |
| 5220022 | 5220469 | HBD_prom | Promoter | Yes | Yes | No | (Tang et al. 1997) |
| 5212610 | 5212865 | HBB-prom | Promoter | Yes | Yes | Yes | (Chao et al. 1983; Wright et al. 1984; Myers et al. 1986; Antoniou et al. 1988; Cowie and Myers 1988) |
| 5210185 | 5210449 | HBB_3′enh | Enhancer | Yes | Yes | No | (Behringer et al. 1987; Trudel and Costantini 1987; Antoniou et al. 1988) |
| 5190322 | 5190802 | 3′HS1 | Enhancer blocker | Nob | Nob | No | (Fleenor and Kaufman 1993; Farrell et al. 2002; Bulger et al. 2003) |
Coordinates are for hg16 (July 2003 freeze) of the human genome.
Conserved in mouse, rat, or chicken means that this segment of human chromosome 11 aligns in a level 1 or level 2 chain in the nets of BLASTZ alignments with human sequences (Kent et al. 2003; Schwartz et al. 2003), which is an indication of aligning to an orthologous region. Alignments to nonorthologous regions are not included.
About 30 bp aligns with a nonorthologous region of mouse chromosome 7.
One limitation to using interspecies conservation to predict CRMs is that some bona fide regulatory elements do not align between the species being examined. Of the 23 CRMs in the human HBB complex, 20 are conserved in mouse, 19 are conserved in rat, and only four are conserved in chicken (Table 1), based on BLASTZ pairwise alignments (Schwartz et al. 2003b). Fortunately, a substantial majority (19 of 23) is conserved among human, mouse, and rat, and 18 are in the genome-wide multiple alignments considered here (see Methods). The four CRMs conserved between human and chicken are in the promoters of the genes (Table 1); none of the upstream or distal CRMs, including enhancers, align at the stringencies used for the whole-genome human–chicken alignments (Hillier et al. 2004). It is possible that more sensitive alignment methods will discover more distantly related sequences in future studies.
It is important to realize that knowledge of CRMs is still incomplete, even in a rigorously studied region such as the HBB complex. DNA intervals identified by comparative genomics methods but not in our reference set are considered false positives (FPs), but in reality, they could be regulatory elements not yet tested for function.
Calibration of discriminatory thresholds
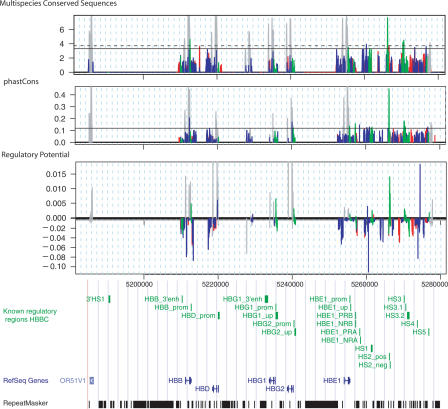
Since the conservation and RP scores are computed on human–mouse–rat three-way alignments, there is an associated score for the 18 CRMs that are in the alignments (green peaks in Fig. 1). Thus at a sufficiently low score, each of the methods can find all the conserved CRMs. The challenge is to distinguish these from other sequences. Some of the known CRMs, such as the HBB promoter and HS2 and HS3 of the LCR, score higher than do the adjacent noncoding DNA segments for MCS, phastCons, and RP (Fig. 1). In contrast, scores for other CRMs are similar to scores obtained in most of the aligning DNA, and some, such as the HBB enhancer, have negative RP scores, indicating a greater similarity to patterns in alignments of neutral DNA. Exons give high scores with all three methods (gray peaks in Fig. 1), and thus the locations of exons were masked and not considered in the performance evaluation for detection of regulatory regions. In general, the positive RP scores seem to distinguish many CRMs from other noncoding DNA (Fig. 1). To examine this quantitatively, each position in the noncoding DNA in the human HBB gene complex that aligns in all three species was designated as either within a regulatory region or not. The distribution of scores for all three functions shows considerable overlap between the regulatory (positive) and nonregulatory (negative) positions (Fig. 2, left panel), showing that discrimination of these two sets is challenging. The distribution of RP scores presents two peaks, one of which is higher than the distribution of RP scores in nonregulatory regions, indicating that in that score interval RP may provide good discrimination.
Figure 1.
Conservation and RP scores for human–mouse–rat alignments in the HBB complex. The scores for MCS, phastCons, and regulatory potential (RP) are plotted in sliding windows along the HBB gene complex. Gray peaks show the scores for exons, which give the most pronounced signals in this region for all scoring methods. Scores overlapping known regulatory regions are shown by the green peaks, those overlapping repeats found by RepeatMasker (A.F.A. Smit and P. Green, unpub., http://ftp.genome.washington.edu/RM/RepeatMasker.html) are red, and scores in uncharacterized regions are shown as blue. A horizontal solid line represents the threshold with optimal performance (interval evaluation) for each score. The dashed line in the MCS graph is the threshold calculated by WebMCS, according to the 95th percentile of conserved sites. Below the graphs is a panel from the UCSC Genome Browser with the known CRMs in the HBB complex as a custom track (green), the RefSeq genes in blue, and repeats in black. The interval chr11:5227344–5229500 contains hemoglobinβ pseudogene 1 (HBBP1, RefSeq: NR_001589), which is also masked from further analyses.
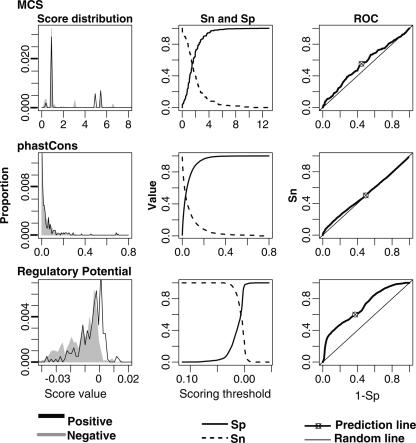
Figure 2.
Ability to identify positions in CRMs in the HBB gene complex for three scoring methods based on human–mouse–rat alignments. The graphs in the left column display the lowess-smoothed distribution of scores at positions in noncoding alignments for the regulatory regions (positives, black line) and the nonregulatory regions (negatives, gray area). The graphs in the center column display the sensitivity (Sn, dashed line) and specificity (Sp, solid line) of each method, determined by the fraction of each distribution in the left columns that is above and below the scoring thresholds. The receiver-operator characteristic (ROC) graphs plot Sn versus 1 – Sp for each scoring threshold (thick line). The values at the optimal threshold are plotted as the circle with the cross hairs. The expectation for a random signal follows the diagonal thin line.
Our goal is to find the threshold for each score that optimizes the ability to find the CRMs (high Sn) while minimizing the amount of other DNA that also passes the threshold (high Sp; see Methods). As expected, Sn decreases and Sp increases with increasing score thresholds for each method (Fig. 2, center panel). Optimal performance occurs at the crossover point between the Sn and Sp curves. The Sn for RP at this point is higher than that for phastCons or MCS (Table 2), but it is only ∼60%. The superior ability of RP scores to discriminate known regulatory elements from other aligning sequences is illustrated in the receiver-operator characteristic (ROC) curves (Fig. 2, right panel). Good discriminatory functions show a pronounced upward deviation from the diagonal (which is the line for “noise” or a random signal) in these curves, indicating both high Sn and high Sp. None of the three functions is an ideal discriminator, but RP scores show the greatest upward deflection, indicating better discrimination.
Table 2.
Thresholds and effectiveness of scores derived from human–mouse–rat genome alignments for finding CRMs in the reference set for the HBB complex
| Binary discrimination by position
|
Discrimination of intervalsb
|
||||
|---|---|---|---|---|---|
| Method | Threshold | Sn, Spa | Threshold | Snint | True discovery rate |
| MCS | 1.55 | 0.55 | 3.3 | 0.44 | 0.54 |
| PhastCons | 0.031 | 0.50 | 0.13 | 0.50 | 0.54 |
| Regulatory potential | -0.006 | 0.61 | 0 | 0.78 | 0.63 |
| Regulatory potential, no HBBc | -0.007 | 0.60 | 0 | 0.78 | 0.59 |
Sn indicates sensitivity; Sp, specificity. Optimal Sn and Sp are determined at the crossover point and thus are equal.
The intervals are evaluated by the maximum score within each interval.
RP scores computed using a training set that excludes CRMs from the HBB complex.
In this binary discrimination analysis, the optimal threshold for RP scores is –0.006 (Table 2). The fact that it is a negative number is initially surprising, because negative values mean that the patterns of the alignments are more like those in the negative training set (aligned ancestral repeats) than those in the known regulatory elements. However, it is important to realize that in this binary discrimination analysis, the methods are evaluated by how much of all the regulatory regions are found.
Another important feature to evaluate is whether any part of a regulatory element passes a given threshold. Thus, we conducted a second analysis, in which the regulatory regions are considered as intervals (not individual positions) and the relevant score is the maximum within the interval. The intervals containing nonregulatory regions are continuous runs of positions whose RP scores meet or exceed the threshold; thus their size and number varies with the threshold. They also were evaluated by the maximum score within the interval. We computed the fraction of regulatory region intervals that exceed a threshold score, called the interval Sn, or Snint, and the fraction of intervals exceeding a threshold that are regulatory regions, called the “true discovery rate.” With this approach, an RP threshold of zero achieves a Snint of 0.78 and a true discovery rate of ∼0.6 (Table 2). Thus an RP of zero is a useful operational threshold for the human–mouse–rat RP scores. The interval-based evaluation did not reveal an improved performance for MCS and phastCons (Table 2).
The RP scores were trained on a set of 93 known regulatory regions, which included four of the CRMs in the reference set from the HBB gene complex, namely, HS2 of the LCR and promoters for the HBE1, HBG2, and HBB genes. To remove bias introduced by this overlap in training and testing sets, we repeated the training of the RP model excluding the CRMs from the HBB gene complex. The threshold, Sn, and Sp of RP scores generated in this way are similar to the ones generated by including the CRMs from the HBB gene complex (Table 2).
Genome-wide evaluation of alignment scores for regulatory elements
The HBB complex provides a continuous region of intensively tested regulatory regions for evaluation, but it is important to know how well the results on this locus apply to known regulatory elements in the rest of the genome. Additional sets of functional elements distributed across the human genome were collected, and the maximum RP and phastCons scores within each interval were determined. (The MCS score was not included because it has not been computed across the entire human genome.) Because of the sparseness of known regulatory elements across the entire human genome, it is not possible to assess a false-positive or true-negative rate for these data sets. Hence, Sp cannot be determined, but it is useful to compare the distribution of scores for each functional set with the genome-wide scores.
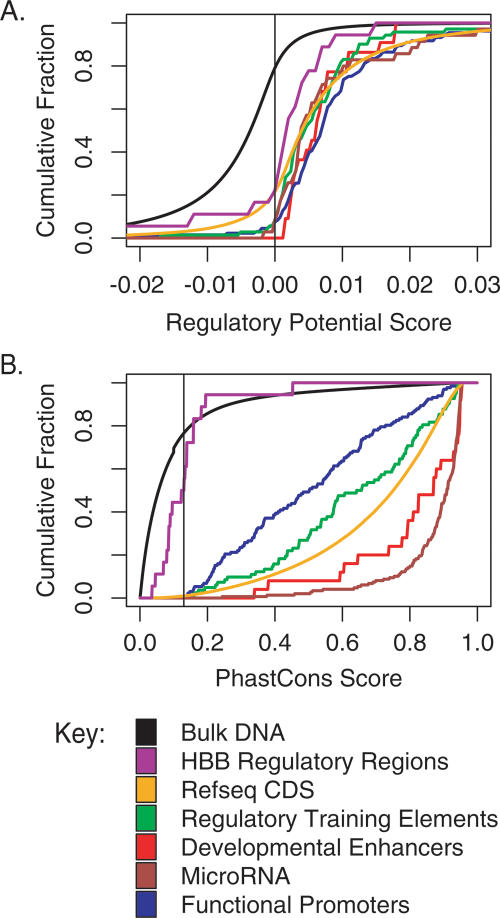
The distribution of RP scores for positions in the human genome (including coding regions) that align with mouse and rat shows that ∼20% has RP scores above zero (Fig. 3A). RP scores can be computed only for the ∼35% of the human genome that aligns with mouse and rat (Gibbs et al. 2004). Thus, ∼7% of the human genome has RP scores in the range that is effective in finding CRMs in the HBB complex. Almost all of the known regulatory regions and miRNA from dispersed loci have positive RP scores (Fig. 3A). Thus, the RP threshold of zero should capture most of the functional regions whose genomic alignments have properties similar to those in these data sets.
Figure 3.
Cumulative distributions of RP and phastCons scores in functional regions compared to the total aligned genomic DNA. The cumulative fraction with a maximal score below a scoring threshold for RP (A) and phastCons (B) is shown for each of six sets of functional sequences (colored lines). The purple line is for the CRMs in the HBB gene complex, gold is for the RefSeq coding exons (Pruitt and Maglott 2001), green is for the regulatory element training set (Elnitski et al. 2003), red is for a set of developmental enhancers (Plessy et al. 2005), brown is for miRNAs (Griffiths-Jones 2004), and blue is for functional promoters (Trinklein et al. 2003). The evaluation is based on the highest score within each interval for the functional elements. The cumulative distributions of scores for all the human–mouse–rat aligned positions are the black lines in each graph. For RP, every fifth base pair in alignments was scored (as the center of a 100-bp window), and for phastCons, all base pairs in alignments were scored. A vertical line is drawn at the optimal threshold for discriminating intervals (Table 2).
The distribution of phastCons scores for the sequences in the human genome that align with mouse and rat are dramatically skewed toward low values (Fig. 3B). About 22% of the aligning human positions have phastCons scores that exceed 0.13, the threshold determined from CRMs in the HBB gene complex. PhastCons scores in all the sets of functional DNA examined exceed this threshold, with the scores for miRNAs and developmental enhancers being particularly high (Fig. 3B). These results show that phastCons is a strong discriminatory function genome-wide, with a considerable dynamic range between scores for the known functional elements and the bulk genome scores.
One of the intriguing results for both methods is that their diagnostic effectiveness for the HBB complex reference set is less than that observed for the other sets of regulatory elements. The cumulative distributions for both scores in the HBB complex CRMs are shifted considerably to the left of the scores for other sets of CRMs, coding sequences, and miRNAs (Fig. 3). This illustrates the difficulty of finding all the CRMs in this gene complex.
Discussion
We organized the extensive experimental results on the DNA segments regulating expression of the HBB gene complex into a reference data set, and then used this data set to evaluate three different approaches for analyzing multispecies alignments to find CRMs. Two of the methods, MCSs and phastCons, are based exclusively on conservation, whereas a third method, RP, uses a pattern-matching discriminatory function within the conserved regions. All three methods had some success in detecting the CRMs in the reference set, with sensitivities and specificities ranging from 50%–60% when evaluated on all nucleotide positions. The RP function performed better than did the conservation measures on the reference set of CRMs in the HBB gene complex. This is expected, given that high conservation should reflect the effects of purifying selection for any function, whereas the RP function was trained to find patterns in alignments similar to those in transcriptional regulatory regions. When the performance was evaluated on the maximum score in each interval, RP scores reached a Sn of 78% with a true discovery rate of ∼60%. Strikingly, both conservation-based scores and RP scores perform much better against other sets of CRMs.
The RP and phastCons scores are deposited in databases such as the Genome Browser (Kent et al. 2002; Karolchik et al. 2004) and GALA (Giardine et al. 2003; Elnitski et al. 2005), and MCS scores can be computed on a Web server. Investigators can apply the thresholds determined in the current study to search the databases and predict CRMs with reasonable but not perfect reliability. Given that the fraction of the human genome with positive RP scores (∼7%) exceeds the fraction estimated to be under selection between primates and rodents (∼5%) (Waterston et al. 2002; Chiaromonte et al. 2003), one should expect some FPs in the RP-based predictions. Recent studies show that Sp of predictions is improved by combining bioinformatic features associated with regulatory elements, such as conservation with overrepresented motifs (Blanchette et al. 2002; Liu et al. 2004) or conservation with clusters of transcription factor binding sites (Berman et al. 2004). We find that when RP scores above the discriminatory threshold are combined with a conserved transcription factor binding site, the predicted CRMs are validated at a high rate (Hardison et al. 2003a; Welch et al. 2004).
Although the prospects for application of the current measures to experimental investigation of gene regulation are promising, our study also illustrates some important limitations in using multispecies alignments to predict CRMs. First, RP scores fail to distinguish at least 20% of the conserved CRMs in the HBB complex, and other methods have less Sn. Fortunately, for other reference data sets, the performance is better, but it is important to realize that some conserved CRMs will be missed using current methods.
Second, some human CRMs have no reliable matches with mouse and sequence, as is the case for four of the 23 CRMs known in the human HBB complex. Obviously, CRMs such as these are invisible to predictive algorithms based on primate–rodent alignments, but they may be detectable over shorter phylogenetic distances using techniques such as phylogenetic shadowing (Boffelli et al. 2003). The failure of these CRMs to align could be explained in at least three different ways. One is that homologs to some of the “nonconserved” human CRMs could be present in rodent or avian species, but current alignment algorithms are not sufficiently sensitive to detect them. For example, the HS2 enhancer in the LCR does not match in whole-genome human–chicken alignments generated with BLASTZ, but it does align between mammals and chicken when TBA (Blanchette et al. 2004) is used to align homologs to this ENCODE region (The ENCODE Project Consortium 2004). However, in this case the functional protein-binding sites are not preserved in the aligned chicken sequence (data not shown), which raises the issue of whether the alignment is biologically meaningful. It is also possible that some of the nonconserved human CRMs function only in one lineage and are not preserved in other lineages. Still other nonconserved CRMs possibly could be explained by turnover in the factor binding sites (Ludwig et al. 2000; Dermitzakis and Clark 2002; Berman et al. 2004). The hypersensitive site 3′HS1 may be an example of turnover. A hypersensitive site is located at a similar location downstream of the β-globin gene in both mouse and human (Fleenor and Kaufman 1993; Bulger and Groudine 1999), and this region has been implicated in boundary function (Bulger et al. 2003). However, the sequences of these HSs between mouse and human are not very similar, perhaps because the critical factor binding sites have changed order or location between the species.
A third limitation to the use of comparative genomics approaches for finding potential cis-regulatory elements is the incomplete knowledge of protein-coding regions. All the methods examined here, including RP, give high scores to exons. Exons that have not been annotated will not be excluded from the analysis of “noncoding” regions, and thus they can contribute to FPs in the predictions.
The reasons for Sn differing among data sets are of considerable interest. Recent studies show that genes encoding proteins involved in developmental and transcriptional regulation tend to have highly constrained CRMs (Sandelin et al. 2004b; Plessy et al. 2005; Woolfe et al. 2005). In contrast, the extensive studies in the HBB gene complex, many of which were not driven by sequence conservation, may have revealed some types of regulatory elements that do not have as strong a conservation signal as do those in developmental regulatory genes. Detailed analyses of the evolutionary features of different types of regulatory elements are an important area for future research.
Improvements are expected in the predictive power of all the scores being computed on multispecies alignments. The discriminatory power of alignments increases as more sequences are added, both for a particular locus (Thomas et al. 2003) and genome-wide (Gibbs et al. 2004). Indeed, all three of the methods evaluated here perform better on three-way human–mouse–rat alignments than on pairwise human–mouse alignment (data not shown). Including the sequences of other species, such as dog and opossum, should improve the discriminatory power. Other studies that address statistical challenges in developing discriminatory models (Kolbe et al. 2004) should also lead to improved performance.
Methods
Reference sets of transcriptional regulatory regions
The β-globin gene (HBB) complex contains several regulatory regions that have been well studied experimentally. A set of 23 experimentally determined CRMs was compiled from a literature survey and mapped within a 95-kb interval (chr11:5185001–5280000 in hg16), which encompasses the HBB complex and terminates at the surrounding olfactory receptor genes (Bulger et al. 2000). Several types of experimental data were used in establishing that a CRM is functional, including naturally occurring thalassemia mutations in humans, analysis of large DNA constructs in transgenic mice, effects on expression of reporter genes in either transient transfections or stably transformed cultured cells, DNase hypersensitive sites in chromatin, and in vivo footprints (see references in Table 1). Regions identified solely by electrophoretic mobility shift assays were not included.
Of the 23 CRMs in this reference set, 19 can be found in multiple alignments of the human, mouse, and rat sequences. However, only 18 were available for the evaluation of the scores computed on the multiple alignment of hg16, mm3, and rn3 (see below) because much of the sequence of hypersensitive site HS4 (Stamatoyannopoulos et al. 1995) was masked as a repeat. Specifically, it is within an ERV1 transposable element, a member of a family that was active around the time of the primate-rodent divergence. This history makes it difficult to accurately determine whether to include the repeats in alignments (soft-masking) or to exclude them entirely (hard-masking) (Schwartz et al. 2003b). For the whole-genome multiple alignment set used in this study, the ERV1 family was hard-masked, and consequently, we could not include HS4 in the evaluations. However, it is well-known that the sequence of HS4 aligns among mammals, including humans and rodents (Stamatoyannopoulos et al. 1995; Hardison et al. 1997), and hence it is listed as conserved in rat and mouse in Table 1.
A set of 40,000 predicted promoters were compiled by Trinklein et al. (2003). Of these, 152 were tested for promoter activity in transient transfection assays, with 138 verified (termed functional promoters). The 93 known regulatory regions were compiled from the literature and comprise the training set of RP (Elnitski et al. 2003). The developmental enhancers are the human homologs of a collection of 26 enhancers for mouse genes whose products regulate early development (Plessy et al. 2005). Other sets of functional sequences were the 176 miRNAs obtained from the miRNA Registry (Griffiths-Jones 2004; http://www.sanger.ac.uk/Software/Rfam/mirna/index.shtml) and the ∼200,000 coding exons from RefSeq (Pruitt and Maglott 2001).
Alignments
Three-way human–mouse–rat alignments were computed on the July 2003 human genome assembly (hg16, NCBI build 34), the February 2003 mouse genome assembly (mm3), and the June 2003 rat assembly (rn3), using MULTIZ (Blanchette et al. 2004) on the relevant pairwise BLASTZ alignments (Schwartz et al. 2003b). Of the 95,000 bp in the HBB gene complex, 33,642 bp (35%) are in the whole-genome human–mouse–rat alignments, similar to the fraction obtained genome-wide (Gibbs et al. 2004).
Scores based on alignments
Regulatory potential
The regulatory model was trained by using the frequencies of patterns in the alignment of a set of known regulatory regions (Elnitski et al. 2003). The coordinates of the known regulatory regions are available from GALA (Giardine et al. 2003; Elnitski et al. 2005) at http://www.bx.psu.edu/. The neutral model was trained by using frequencies of patterns in the alignment of a set of ancestral repeats, a model for neutral DNA (Hardison et al. 2003b). Patterns in the human–mouse–rat alignment were described by a 10-symbol alphabet using an order 2 Markov model (Kolbe et al. 2004). The RP score assigned to each position is the RP score computed for a 100-bp window centered on the position. Details on the computation are provided at http://www.bx.psu.edu/projects/rp/.
MCS
MCSs were calculated on MultiPipMaker alignments (Schwartz et al. 2003a) of human, mouse, and/or rat sequences of the HBB complex downloaded from University of California, Santa Cruz (UCSC) Genome Browser (Kent et al. 2002). The MCSs were computed by using WebMCS at http://research.nhgri.nih.gov/MCS/ (Margulies et al. 2003). The data were averaged into 25-bp windows with 1-bp slides. The scoring threshold representing the top 5% fraction was 3.7 for three-way (human–mouse–rat) alignment scores.
phastCons
The phastCons scores (Siepel et al. 2005) were calculated by using the software package Woody, obtained from Adam Siepel (Center for Biomolecular Science and Engineering, UCSC). The parameters used were λ = 0.9, k = 10 rate categories, Rev model; these were the same as those used to generate the data available on the UCSC Genome Browser when the calibration study was done. The probability that each alignment column is observed in the first of the k rate categories, hence the most conserved category, constituted the raw data. These data were then taken as 100-bp averages, in increments of 1-bp slides.
Evaluation of alignment scores for detecting known CRMs
For binary classification, all noncoding aligned positions in the HBB gene complex were assigned to be positive (regulatory) or negative (not regulatory), based on the positions of the known CRMs. Each aligned position was also assigned a score by each method, as described in the previous section. The distributions of scores for the “regulatory” and “nonregulatory” positions were evaluated to determine Sn and Sp at each score threshold. Positions in regulatory regions were classified as true positives (TPs) if they scored at or above a threshold and as false negatives (FNs) if they scored below it. The Sn was calculated as TP/(TP + FN). The positions in nonregulatory regions were classified as true negatives (TNs) if they scored below the threshold and as FPs if they scored above it. These results gave the Sp, which is TN/(TN + FP). The threshold at which Sn and Sp were most similar (crossover point) was taken as optimal for the binary classification by position.
A separate analysis was used to evaluate the ability of each score to discriminate the regulatory intervals from nonregulatory DNA, based on the highest score in each interval. In this analysis, the average value for each of the three scores was computed in 100-bp windows (with a 1-bp slide) for all the aligned positions in the HBB complex (and genome-wide for phastCons and RP). Windows whose average score met or exceeded a given threshold comprised the predicted set for that threshold. Overlapping windows were combined to make a single contiguous interval that passed the threshold. Regulatory regions that overlapped an interval that passed the threshold were counted as TPs, and those that did not were FNs. The intervals that passed the threshold but did not overlap with a regulatory region were counted as FPs. Note that the size of each regulatory interval is determined experimentally as the region required for regulation. The sizes vary among CRMs but are not affected by the score threshold. The sizes also vary among the FP intervals, being determined by the scores of overlapping windows; in addition, the sizes can differ for each threshold. Defining a TN interval is difficult, and thus the evaluation was based on interval-based Sn (Snint = TP/[TP + FN]) and the true discovery rate (TP/[TP + FP]). The optimal threshold is approximately the crossover between Snint and the true discovery rate. This evaluation is similar to procedures used to evaluate gene prediction programs (Burset and Guigó 1996).
A similar discrimination based on the maximum RP or phastCons score in each interval was performed for several sets of functional elements in the human genome, and cumulative distributions are shown in Figure 3. These were compared with the cumulative distributions of the phastCons scores in all aligned positions in the human genome (“bulk DNA”) and of every fifth aligned position for RP scores.
Availability
The whole-genome alignments, RP scores, and phastCons scores can be downloaded from or queried upon at the UCSC Genome Browser (Kent et al. 2002) and Table Browser (Karolchik et al. 2004; http://www.genome.ucsc.edu/) and GALA (Giardine et al. 2003) (http://www.bx.psu.edu/). The promoter, known regulatory region, and miRNA data sets are available from GALA or the original investigators.
The reference set of regulatory regions for the HBB gene complex is available at http://www.bx.psu.edu/~ross/dataset/DatasetHome.html, in both hg16 and hg17 coordinates. More information, along with references to the supporting literature, is recorded in dbERGE II (Elnitski et al. 2005), which allows users to obtain the data in a variety of formats (graphical or textual) and in various depths of detail. This resource can be accessed from the home page for Penn State Center for Comparative Genomics and Bioinformatics (http://www.bx.psu.edu/). After selecting the link to dbERGE II, on the Start page select dbERGE II v.2 (for hg16), on the Quick Links page select “Other” for Type of Data and ENCODE region “ENm009β Globin” for Restrict to Region, place no additional filters on the Detailed Experiments page, and select the desired output from the Display Options page. If viewed in the UCSC Genome Browser, links are provided back to dbERGE II for detailed information. A table of the intervals is available upon request.
The operations for the evaluations reported here can be performed in a UNIX environment using command-line pipes and wrapper scripts for software that is available on request.
Acknowledgments
This work was supported by NIH grants DK65806 (R.H.), HG02238 (W.M.), and HG02325 (L.E.). We thank Adam Siepel and David Haussler for access to the phastCons scores and programs prior to publication.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3642605. Article published online before print in July 2005.
References
- Allan, M., Lanyon, G., and Paul, J. 1983. Multiple origins of transcription in the 4.5 kb upstream of the ε-globin gene. Cell 35: 187–197. [DOI] [PubMed] [Google Scholar]
- Antoniou, M., deBoer, E., Habets, G., and Grosveld, F. 1988. The human β-globin gene contains multiple regulatory regions: Identification of one promoter and two downstream enhancers. EMBO J. 7: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer, R.R., Hammer, R.E., Brinster, R.L., Palmiter, R.D., and Townes, T.M. 1987. Two 3′ sequences direct adult erythroid-specific expression of human β-globin genes in transgenic mice. Proc. Natl. Acad. Sci. 84: 7056–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, M., Reik, A., Close, J., Telling, A., Epner, E., Fiering, S., Hardison, R., and Groudine, M. 1998. Description and targeted deletion of 5′ HS5 and 6 of the mouse β-globin locus control region. Blood 92: 4394–4403. [PubMed] [Google Scholar]
- Berman, B.P., Pfeiffer, B.D., Laverty, T.R., Salzberg, S.L., Rubin, G.M., Eisen, M.B., and Celniker, S.E. 2004. Computational identification of developmental enhancers: Conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 5: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, M., Schwikowski, B., and Tompa, M. 2002. Algorithms for phylogenetic footprinting. J. Comput. Biol. 9: 211–223. [DOI] [PubMed] [Google Scholar]
- Blanchette, M., Kent, W.J., Riemer, C., Elnitski, L., Smit, A.F.A., Roskin, K.M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E.D., et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14: 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine, D. and Ley, T. 1987. An enhancer element lies 3′ to the human A γ globin gene. EMBO J. 6: 2997–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K.D., Ovcharenko, I., Pachter, L., and Rubin, E.M. 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391–1394. [DOI] [PubMed] [Google Scholar]
- Brent, M.R. and Guigó, R. 2004. Recent advances in gene structure prediction. Curr. Opin. Struct. Biol. 14: 264–272. [DOI] [PubMed] [Google Scholar]
- Bulger, M. and Groudine, M. 1999. Looping versus linking: Toward a model for long-distance gene activation. Genes & Dev. 13: 2465–2477. [DOI] [PubMed] [Google Scholar]
- Bulger, M., Bender, M.A., von Doorninck, J.H., Wertman, B., Farrell, C., Felsenfeld, G., Groudine, M., and Hardison, R. 2000. Comparative structural and functional analysis of the olfactory receptor genes flanking the human and mouse β-globin gene clusters. Proc. Natl. Acad. Sci. 97: 14560–14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger, M., Schubeler, D., Bender, M.A., Hamilton, J., Farrell, C.M., Hardison, R.C., and Groudine, M. 2003. A complex chromatin “landscape” revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 23: 5234–5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset, M.R. and Guigó, R. 1996. Evaluation of gene structure prediction programs. Genomics 34: 353–367. [DOI] [PubMed] [Google Scholar]
- Cao, S.X., Gutman, P.D., Dave, H.P.G., and Schechter, A.N. 1989. Identification of a transcriptional silencer in the 5′-flanking region of the human ε-globin gene. Proc. Natl. Acad. Sci. 86: 5306–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallesco, R. and Tuan, D. 1997. Modulatory subdomains of the HS2 enhancer differentially regulate enhancer activity in erythroid cells at different developmental stages. Blood Cells Mol. Dis. 23: 8–26. [DOI] [PubMed] [Google Scholar]
- Chao, M.V., Mellon, P., Charnay, P., Maniatis, T., and Axel, R. 1983. The regulated expression of β-globin genes introduced into mouse erythroleukemia cells. Cell 32: 483–493. [DOI] [PubMed] [Google Scholar]
- Chiaromonte, F., Weber, R.J., Roskin, K.M., Diekhans, M., Kent, W.J., and Haussler, D. 2003. The share of human genomic DNA under selection estimated from human–mouse genomic alignments. In The genome of Homo sapiens, pp. 245–254. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Collins, F.S., Green, E.D., Guttmacher, A.E., and Guyer, M.S. 2003. A vision for the future of genomics research. Nature 422: 835–847. [DOI] [PubMed] [Google Scholar]
- Cooper, G.M. and Sidow, A. 2003. Genomic regulatory regions: Insights from comparative sequence analysis. Curr. Opin. Genet. Dev. 13: 604–610. [DOI] [PubMed] [Google Scholar]
- Cooper, G.M., Brudno, M., Stone, E.A., Dubchak, I., Batzoglou, S., and Sidow, A. 2004. Characterization of evolutionary rates and constraints in three mammalian genomes. Genome Res. 14: 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowie, A. and Myers, R.M. 1988. DNA sequences involved in transcriptional regulation of the mouse β-globin promoter in murine erythroleukemia cells. Mol. Cell. Biol. 8: 3122–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E.T. and Clark, A.G. 2002. Evolution of transcription factor binding sites in mammalian gene regulatory regions: Conservation and turnover. Mol. Biol. Evol. 19: 1114–1121. [DOI] [PubMed] [Google Scholar]
- Dhar, V., Nandi, A., Schildkraut, C.L., and Skoultchi, A.I. 1990. Erythroid-specific nuclease-hypersensitive sites flanking the human β-globin gene cluster. Mol. Cell. Biol. 10: 4324–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski, L., Li, J., Noguchi, C.T., Miller, W., and Hardison, R. 2001. A negative cis-element regulates the level of enhancement of hypersensitive site 2 of the β-globin locus control region. J. Biol. Chem. 276: 6289–6298. [DOI] [PubMed] [Google Scholar]
- Elnitski, L., Hardison, R.C., Li, J., Yang, S., Kolbe, D., Eswara, P., O'Connor, M.J., Schwartz, S., Miller, W., and Chiaromonte, F. 2003. Distinguishing regulatory DNA from neutral sites. Genome Res. 13: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski, L., Giardine, B., Shah, P., Zhang, Y., Riemer, C., Weirauch, M., Burhans, R., Miller, W., and Hardison, R.C. 2005. Improvements to GALA and dbERGEII: Databases featuring genomic sequence alignment, annotation and experimental results. Nucl. Acids Res. 33: D466–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636–640. [DOI] [PubMed] [Google Scholar]
- Farrell, C.M., West, A.G., and Felsenfeld, G. 2002. Conserved CTCF insulator elements flank the mouse and human β-globin loci. Mol. Cell. Biol. 22: 3820–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. and Churchill, G.A. 1996. A hidden Markov model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 13: 93–104. [DOI] [PubMed] [Google Scholar]
- Fleenor, D.E. and Kaufman, R.E. 1993. Characterization of the DNaseI hypersensitive site 3′ of the human β-globin gene domain. Blood 81: 2781–2790. [PubMed] [Google Scholar]
- Forget, B.G. 2001. Molecular genetics of the human globin genes. In Disorders of hemoglobin: Genetics, pathophysiology, and clinical management (eds. M.H. Steinberg et al.), pp. 117–130. Cambridge University Press, Cambridge, UK.
- Forrester, W.C., Thompson, C., Elder, J.T., and Groudine, M. 1986. A developmentally stable chromatin structure in the human β-globin gene cluster. Proc. Natl. Acad. Sci. 83: 1359–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, P., Hurst, J., Collis, P., and Grosveld, F. 1990. DNase I hypersensitive sites 1, 2 and 3 of the human β-globin dominant control region direct position-independent expression. Nucleic Acids Res. 18: 3503–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, P., Pruzina, S., Antoniou, M., and Grosveld, F. 1993. Each hypersensitive site of the human β-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes & Dev. 7: 106–113. [DOI] [PubMed] [Google Scholar]
- Frazer, K.A., Elnitski, L., Church, D., Dubchak, I., and Hardison, R.C. 2003. Cross-species sequence comparisons: A review of methods and available resources. Genome Res. 13: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine, B.M., Elnitski, L., Riemer, C., Makalowska, I., Schwartz, S., Miller, W., and Hardison, R.C. 2003. GALA, a database for genomic sequence alignments and annotations. Genome Res. 13: 732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs, R.A., Weinstock, G.M., Metzker, M.L., Muzny, D.M., Sodergren, E.J., Scherer, S., Scott, G., Steffen, D., Worley, K.C., Burch, P.E., et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521. [DOI] [PubMed] [Google Scholar]
- Gimble, J.M., Max, E.E., and Ley, T.J. 1988. High-resolution analysis of the human γ-globin gene promoter in K562 erythroleukemia cell chromatin. Blood 72: 606–612. [PubMed] [Google Scholar]
- Gong, Q.-H., Stern, J., and Dean, A. 1991. Transcriptional role of a conserved GATA-1 site in the human ε-globin gene promoter. Mol. Cell. Biol. 11: 2558–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S. 2004. The microRNA Registry. Nucleic Acids Res. 32: D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison, R. 2001. Organization, evolution and regulation of the globin genes. In Disorders of hemoglobin: Genetics, pathophysiology, and clinical management (eds. M.H. Steinberg et al.), pp. 95–116. Cambridge University Press, Cambridge, UK.
- Hardison, R.C. 2003. Comparative genomics. PLoS Biol. 1: 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison, R., Slightom, J.L., Gumucio, D.L., Goodman, M., Stojanovic, N., and Miller, W. 1997. Locus control regions of mammalian β-globin gene clusters: Combining phylogenetic analyses and experimental results to gain functional insights. Gene 205: 73–94. [DOI] [PubMed] [Google Scholar]
- Hardison, R.C., Chiaromonte, F., Kolbe, D., Wang, H., Petrykowska, H., Elnitski, L., Yang, S., Giardine, B., Zhang, Y., Riemer, C., et al. 2003a. Global predictions and tests of erythroid regulatory regions. In The genome of Homo sapiens, pp. 335–344. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Hardison, R.C., Roskin, K.M., Yang, S., Diekhans, M., Kent, W.J., Weber, R., Elnitski, L., Li, J., O'Connor, M., Kolbe, D., et al. 2003b. Covariation in frequencies of substitution, deletion, transposition and recombination during eutherian evolution. Genome Res. 13: 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier, L.W., Miller, W., Birney, E., Warren, W., Hardison, R.C., Ponting, C.P., Bork, P., Burt, D.W., Groenen, M.A.M., Delany, M.E., et al. 2004. Sequencing and comparative analysis of the chicken genome. Nature 432: 695–716. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945. [DOI] [PubMed] [Google Scholar]
- Jackson, J.D., Petrykowska, H., Philipsen, S., Miller, W., and Hardison, R. 1996. Role of DNA sequences outside the cores of DNase hypersensitive sites (HSs) in functions of the β-globin locus control region: Domain opening and synergism between HS2 and HS3. J. Biol. Chem. 271: 11871–11878. [DOI] [PubMed] [Google Scholar]
- Karolchik, D., Hinrichs, A.S., Furey, T.S., Roskin, K.M., Sugnet, C.W., Haussler, D., and Kent, W.J. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe, D., Taylor, J., Elnitski, L., Eswara, P., Li, J., Miller, W., Hardison, R., and Chiaromonte, F. 2004. Regulatory potential scores from genome-wide three-way alignments of human, mouse and rat. Genome Res. 14: 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Li, J., Noguchi, C., Miller, W., Hardison, R., and Schechter, A. 1998. Multiple regulatory elements in the 5′-flanking sequence of the human ε-globin gene. J. Biol. Chem. 273: 10202–10209. [DOI] [PubMed] [Google Scholar]
- Li, Q., Peterson, K., Fang, X., and Stamatoyannopoulos, G. 2002. Locus control regions. Blood 100: 3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Liu, X.S., Wei, L., Altman, R.B., and Batzoglou, S. 2004. Eukaryotic regulatory element conservation analysis and identification using comparative genomics. Genome Res. 14: 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, J.A., Case, S.S., Ponce, E., and Lingrel, J.B. 1994. Positive transcriptional regulation of the human γ-globin gene: gPE is a novel nuclear factor with multiple binding sites near the gene. J. Biol. Chem. 269: 26–34. [PubMed] [Google Scholar]
- Ludwig, M.Z., Bergman, C., Patel, N.H., and Kreitman, M. 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403: 564–567. [DOI] [PubMed] [Google Scholar]
- Margulies, E.H., Blanchette, M., NISC Comparative Sequencing Program, Haussler, D., and Green, E.D. 2003. Identification and characterization of multi-species conserved sequences. Genome Res. 13: 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe, J.D., Pachter, L., and Jordan, M.I. 2004. Multiple-sequence functional annotation and the generalized hidden Markov phylogeny. Bioinformatics 20: 1850–1860. [DOI] [PubMed] [Google Scholar]
- McDonagh, K.T., Lin, H.J., Lowrey, C.H., Bodine, D.M., and Nienhuis, A.W. 1991. The upstream region of the human γ-globin gene promoter: Identification and functional analysis of nuclear protein binding sites. J. Biol. Chem. 266: 11965–11974. [PubMed] [Google Scholar]
- Miller, W., Makova, K.D., Nekrutenko, A., and Hardison, R.C. 2004. Comparative genomics. Annu. Rev. Genomics Hum. Genet. 5: 15–56. [DOI] [PubMed] [Google Scholar]
- Molete, J.M., Petrykowska, H., Bouhassira, E.E., Feng, Y.Q., Miller, W., and Hardison, R.C. 2001. Sequences flanking hypersensitive sites of the β-globin locus control region are required for synergistic enhancement. Mol. Cell. Biol. 21: 2969–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molete, J.M., Petrykowska, H., Sigg, M., Miller, W., and Hardison, R. 2002. Functional and binding studies of HS3.2 of the β-globin locus control region. Gene 283: 185–197. [DOI] [PubMed] [Google Scholar]
- Myers, R.M., Tilly, K., and Maniatis, T. 1986. Fine structure genetic analysis of a β-globin promoter. Science 232: 613–618. [DOI] [PubMed] [Google Scholar]
- Pedersen, J.S. and Hein, J. 2003. Gene finding with a hidden Markov model of genome structure and evolution. Bioinformatics 19: 219–227. [DOI] [PubMed] [Google Scholar]
- Pennacchio, L.A. and Rubin, E.M. 2001. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2: 100–109. [DOI] [PubMed] [Google Scholar]
- Perez-Stable, C. and Costantini, F. 1990. Role of fetal G γ-globin promoter elements and the adult β-globin 3′ enhancer in the stage-specific expression of globin genes. Mol. Cell. Biol. 10: 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen, S., Talbot, D., Fraser, P., and Grosveld, F. 1990. The β-globin dominant control region: hypersensitive site 2. EMBO J. 9: 2159–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen, S., Pruzina, S., and Grosveld, F. 1993. The minimal requirements for activity in transgenic mice of hypersensitive site 3 of the β-globin locus control region. EMBO J. 12: 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessy, C., Dickmeis, T., Chalmel, F., and Strahle, U. 2005. Enhancer sequence conservation between vertebrates is favoured in developmental regulator genes. Trends Genet. 21: 207–210. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzina, S., Hanscombe, O., Whyatt, D., Grosveld, F., and Philipsen, S. 1991. Hypersensitive site 4 of the human β-globin locus control region. Nucleic Acids. Res. 19: 1413–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, T.M., Behringer, R.R., Martin, N.C., Townes, T.M., Palmiter, R.D., and Brinster, R.L. 1989. A single erythroid-specific DNase I super-hypersensitive site activates high levels of human β-globin gene expression in transgenic mice. Genes & Dev. 3: 314–323. [DOI] [PubMed] [Google Scholar]
- Sandelin, A., Alkema, W., Engstrom, P., Wasserman, W.W., and Lenhard, B. 2004a. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32: D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin, A., Bailey, P., Bruce, S., Engstrom, P.G., Klos, J.M., Wasserman, W.W., Ericson, J., and Lenhard, B. 2004b. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Elnitski, L., Li, M., Weirauch, M., Riemer, C., Smit, A., NISC Comparative Sequencing Program, Green, E.D., Hardison, R.C., and Miller, W. 2003a. MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 31: 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003b. Human–mouse alignments with BLASTZ. Genome Res. 13: 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton, D.A., Stegman, L., Hardison, R., Miller, W., Slightom, J.L., Goodman, M., and Gumucio, D.L. 1997. Phylogenetic footprinting of hypersensitive site 3 of the β-globin locus control region. Blood 89: 3457–3469. [PubMed] [Google Scholar]
- Siepel, A. and Haussler, D. 2003. Combining phylogenetic and hidden Markov models in biosequence analysis. In: Proceedings of the Seventh Annual International Conference on Computational Molecular Biology (RECOMB 2003), pp. 277–286. ACM Press, New York. [DOI] [PubMed]
- Siepel, A., Bejerano, G., Pedersen, J.S., Hinrichs, A., Hou, M., Rosenbloom, K., Clawson, H., Kent, W.J., Miller, W., and Haussler, D. 2005. Evolutionarily conserved elements in vertebrate, fly, worm and yeast genomes. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Slightom, J., Bock, J., Tagle, D., Gumucio, D., Goodman, M., Stojanovic, N., Jackson, J., Miller, W., and Hardison, R. 1997. The complete sequences of the galago and rabbit β-globin locus control regions: Extended sequence and functional conservation outside the cores of DNase hypersensitive sites. Genomics 39: 90–94. [DOI] [PubMed] [Google Scholar]
- Spellman, P.T., Sherlock, G., Zhang, M.Q., Iyer, V.R., Anders, K., Eisen, M.B., Brown, P.O., Botstein, D., and Futcher, B. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos, G., Josephson, B., Zhang, J.U., and Li, Q. 1993. Developmental regulation of human γ-globin gene in transgenic mice. Mol. Cell. Biol. 13: 7636–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos, J.A., Goodwin, A., Joyce, T., and Lowrey, C.H. 1995. NFE2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human β-globin locus control region. EMBO J. 14: 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, D., Philipsen, S., Fraser, P., and Grosveld, F. 1990. Detailed analysis of the site 3 region of the human β-globin dominant control region. EMBO J. 9: 2169–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, D.C., Ebb, D., Hardison, R.C., and Rodgers, G.P. 1997. Restoration of the CCAAT box and insertion of the CACCC motif activate d-globin gene expression. Blood 90: 421–427. [PubMed] [Google Scholar]
- Tanimoto, K., Sugiura, A., Omori, A., Felsenfeld, G., Engel, J.D., and Fukamizu, A. 2003. Human β-globin locus control region HS5 contains CTCF- and developmental stage–dependent enhancer-blocking activity in erythroid cells. Mol. Cell. Biol. 23: 8946–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.W., Touchman, J.W., Blakesley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788–793. [DOI] [PubMed] [Google Scholar]
- Trepicchio, W., Dyer, M., and Baron, M. 1993. Developmental regulation of the human embryonic β-like globin gene is mediated by synergistic interactions among multiple tissue- and stage-specific elements. Mol. Cell. Biol. 13: 7457–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinklein, N.D., Aldred, S.J., Saldanha, A.J., and Myers, R.M. 2003. Identification and functional analysis of human transcriptional promoters. Genome Res. 13: 308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudel, M. and Costantini, F. 1987. A 3′ enhancer contributes to the stage-specific expression of the human β-globin gene. Genes & Dev. 1: 954–961. [DOI] [PubMed] [Google Scholar]
- Tuan, D., Solomon, W., Li, Q., and London, I.M. 1985. The β-like globin gene domain in human erythroid cells. Proc. Natl. Acad. Sci. 82: 6384–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan, D., Solomon, W., London, I., and Lee, D. 1989. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human “β-like globin” genes. Proc. Natl. Acad. Sci. 86: 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai, A.W., Gillemans, N., Raguz-Bolognesi, S., Pruzina, S., Zafarana, G., Meijer, D., Philipsen, S., and Grosveld, F. 2003. HS5 of the human β-globin locus control region: A developmental stage-specific border in erythroid cells. EMBO J. 22: 4489–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman, W.W. and Sandelin, A. 2004. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 5: 276–287. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Watt, P., Lamb, P., and Proudfoot, N.J. 1993. Distinct negative regulation of the human embryonic globin genes ζ and ε. Gene Exp. 3: 61–75. [PMC free article] [PubMed] [Google Scholar]
- Welch, J.J., Watts, J.A., Vakoc, C.R., Yao, Y., Wang, H., Hardison, R.C., Blobel, G.A., Chodosh, L.A., and Weiss, M.J. 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104: 3136–3147. [DOI] [PubMed] [Google Scholar]
- Wingender, E., Chen, X., Fricke, E., Geffers, R., Hehl, R., Liebich, I., Krull, M., Matys, V., Michael, H., Ohnhauser, R., et al. 2001. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 29: 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe, A., Goodson, M., Goode, D.K., Snell, P., McEwen, G.K., Vavouri, T., Smith, S.F., North, P., Callaway, H., Kelly, K., et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., Rosenthal, A., Flavell, R., and Grosveld, F. 1984. DNA sequences required for regulated expression of β-globin genes in murine erythroleukemia cells. Cell 38: 265–273. [DOI] [PubMed] [Google Scholar]
- Yu, C.-Y., Motamed, K., Chen, J., Bailey, A.D., and Shen, C.K.J. 1991. The CACC box upstream of human embryonic ε globin gene binds Sp1 and is a functional promoter element in vitro and in vivo. J. Biol. Chem. 266: 8907–8915. [PubMed] [Google Scholar]
- Yu, Z., Bock, J., Slightom, J., and Villeponteau, B. 1994. A 5′ β-globin matrix-attachment region and the polyoma enhancer together confer position-independent transcription. Gene 139: 139–145. [DOI] [PubMed] [Google Scholar]
Web site references
- http://www.bx.psu.edu/; GALA and dbERGEII databases
- http://www.bx.psu.edu/~ross/dataset/DatasetHome.html; reference set of CRMs in HBB gene complex
- http://genome.ucsc.edu/; Genome Browser at UCSC
- http://research.nhgri.nih.gov/MCS/; WebMCS for computing multispecies conserved sequences
- http://www.sanger.ac.uk/Software/Rfam/mirna/index.shtml miRNA Registry