Abstract
Recent studies of the genome architecture of vertebrates have uncovered two unforeseen aspects of its organization. First, large regions of the genome, called gene deserts, are devoid of protein-coding sequences and have no obvious biological role. Second, comparative genomics has highlighted the existence of an array of highly conserved non-coding regions (HCNRs) in all vertebrates. Most surprisingly, these structural features are strongly associated with genes that have essential functions during development. Among these, the vertebrate Iroquois (Irx) genes stand out on both fronts. Mammalian Irx genes are organized in two clusters (IrxA and IrxB) that span >1 Mb each with no other genes interspersed. Additionally, a large number of HCNRs exist within Irx clusters. We have systematically examined the enhancer activity of HCNRs from the IrxB cluster using transgenic Xenopus and zebrafish embryos. Most of these HCNRs are active in subdomains of endogenous Irx expression, and some are candidates to contain shared enhancers of neighboring genes, which could explain the evolutionary conservation of Irx clusters. Furthermore, HCNRs present in tetrapod IrxB but not in fish may be responsible for novel Irx expression domains that appeared after their divergence. Finally, we have performed a more detailed analysis on two IrxB ultraconserved non-coding regions (UCRs) duplicated in IrxA clusters in similar relative positions. These four regions share a core region highly conserved among all of them and drive expression in similar domains. However, inter-species conserved sequences surrounding the core, specific for each of these UCRs, are able to modulate their expression.
The next stage in the successful annotation of fully sequenced genomes involves the identification and characterization of functional elements responsible for the cis-regulation of gene expression. These elements represent the integration nodes in gene regulatory networks (GRNs), making their description of utmost importance. However, this effort will require far more complex approaches than those used in the prediction of the protein-coding portion of genes, where sequence-based ab initio tools have provided an adequate answer in most cases. The high variability and relaxed sequence constraint of transcription factor DNA binding sites makes computational prediction of these cis-regulatory elements difficult, necessitating functional assays to describe them. A possible shortcut to this problem comes from the use of comparative genomics to select those regions in the genome that have been conserved during evolution and are not related to protein-coding or transcribed regions. The logic of this approach runs that if these sequences have been conserved during evolution, they must fulfill some functional role, and one potential role is as transcriptional regulatory elements. However, in this way we have only reduced the portion of the genome to examine, and robust and reproducible functional assays are still needed to tackle the complexity of cis regulation.
Various whole-genome analyses of evolutionarily conserved regions have provided us with an extremely useful roadmap of the distribution and structural characteristics of these regions. Two unforeseen features of the organization of vertebrate genomes that have been uncovered by these studies are the existence of gene deserts, large regions of the genome devoid of protein-coding sequences (Nobrega et al. 2003; Ovcharenko et al. 2005), and the presence of extremely highly conserved elements between different vertebrate species (Bejerano et al. 2004; Sandelin et al. 2004; Woolfe et al. 2005). Most surprisingly, in both cases a strong correlation exists between the chromosomal location of these characters and that of developmentally controlled genes. Focusing on this group of genes and performing a detailed analysis of those cases where additional expression and functional information is available will surely provide us with a more thorough knowledge of how GRNs control early embryonic development.
During vertebrate development, a group of transcription factors present in broad and partially overlapping domains subdivides the neural plate in the anterior–posterior and dorsal–ventral axes. This gridded subdivision of the neural tube by the combinatorial expression of transcription factors, the so-called prepattern, is essential for proper regionalization of the nervous system and for the formation of specific neurons in precise locations (Lee and Jessell 1999; Briscoe and Ericson 2001; Gómez-Skarmeta et al. 2003).
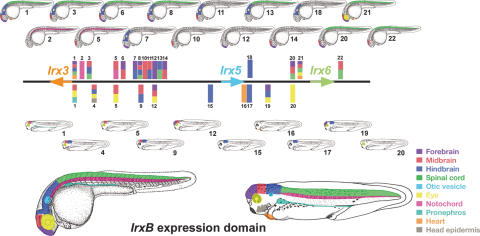
The Iroquois (Irx) genes are members of this prepattern that encode homeoproteins conserved throughout the animal kingdom (Gómez-Skarmeta and Modolell 2002). In mouse and humans, six Irx genes are grouped into two genomic clusters, IrxA, containing Irx1, Irx2, and Irx4, and IrxB, containing Irx3, Irx5, and Irx6 (Peters et al. 2000). These two mammalian clusters are the result of a segmental or chromosomal duplication of a single ancestral cluster, as the Irx1, Irx2, and Irx4 proteins are clear paralogs of Irx3, Irx5, and Irx6, respectively (Peters et al. 2000). The human IrxA and IrxB clusters span 1.8 and 1.3 Mb, respectively, and the mouse clusters are of comparable size (see Fig. 1A for a diagram showing the intergenic distances between the three IrxB genes in human chromosome 16). Both clusters contain large genomic regions that can be considered as gene deserts that lack any other transcribed or potential coding sequences (Nobrega et al. 2003).
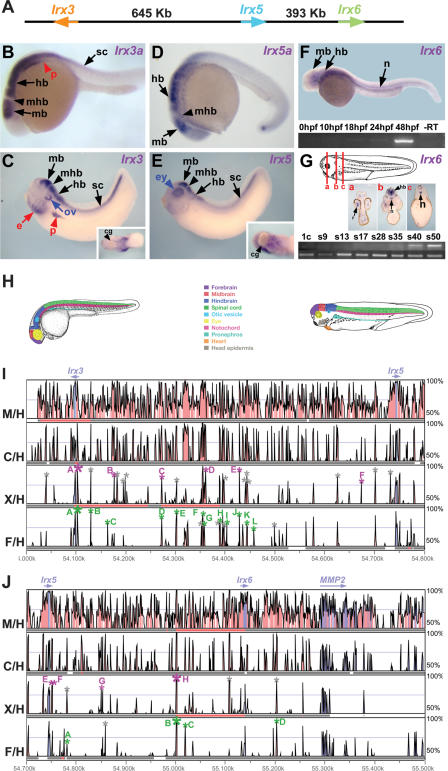
Figure 1.
Expression pattern of zebrafish and Xenopus IrxB genes and distribution of highly conserved non-coding regions within the IrxB locus. (A) Intergenic distances between the three IrxB genes in human chromosome 16. (B–G) Expression pattern of zebrafish and Xenopus IrxB genes. Lateral views of 24-hpf (B, D) or 48-hpf (F) zebrafish embryos, and stage 35 Xenopus embryos (C, E). (B) Zebrafish Irx3a is expressed in neural tissues and in the pronephros. (C) Xenopus Irx3 mRNA is detected in similar domains. In addition, it is detected in the ectodermal layer of the branchial arches and in the future heart region (inset). (D) Zebrafish Irx5a is present in neural tissues. (E) Xenopus Irx5 is expressed in the brain and in the neural tube in a pattern similar to that of Irx3. In addition it is expressed in the eye and in the future heart region (inset) but not in the otic vesicle, pronephros, or head epidermis. (F) In zebrafish, Irx6 is initially expressed at 48 hpf, as determined by RT-PCR (bottom). At this stage, in whole mounts Irx6 is detected in different neural domains and in the notochord (top). (G) Xenopus Irx6 mRNA onsets of expression occurs at stage 35 (bottom), as determined by RT-PCR. Compare Irx6 initial expression with the control Histone H4 mRNA shown below. (1c) One-cell stage; (s9–50) stages 9–50. Transverse sections of a stage 40 Xenopus laevis embryo at different levels, indicated by lines in drawing at top [(a) midbrain, (b) hindbrain, (c) spinal cord], show that Irx6 is expressed in the nervous system and in the notochord. (mb) Midbrain; (hb) hindbrain; (mhb) midbrain–hindbrain boundary; (sp) spinal cord; (ov) otic vesicle; (e) ectodermal layer of the branchial; (ey) eye; (p) pronephros; (cg) cement gland; (n) notochord. (H) Color-coded schematic representation of the zebrafish and Xenopus regions with IrxB gene expression. (I, J) VISTA view of the occurrence of conserved sequence domains in the gene deserts between the Irx3 and the Irx5 (I) or Irx5 and Irx6 (J) genes from vertebrate IrxB clusters. Shown from top to bottom are mouse vs. human (M/H), chick vs. human (C/H), Xenopus tropicalis vs. human (X/H), and Fugu vs. human (F/H) global alignments. Colored peaks (purple, coding; pink, non-coding) indicate regions of at least 100 bp and 75% similarity. Gray, green, and magenta asterisks mark the amplified genomic regions with no enhancer activity, with enhancer activity in zebrafish, and with enhancer activity in Xenopus, respectively. Letters in the colored peaks are referred to in the panels shown in Figure 2 (F/H row in I), Figure 3 (X/H row in I), and Figure 4 (F/H and X/H rows in J).
In non-mammalian vertebrates, five Irx genes have been identified in both Xenopus laevis and chick (Bellefroid et al. 1998; Gómez-Skarmeta et al. 1998; Garriock et al. 2001; Ogura et al. 2001) and zebrafish and pufferfish have up to eleven Irx genes (Dildrop and Ruther 2004; Feijóo et al. 2004). Several lines of evidence indicate that, in these species, Irx genes are organized in clusters similar to those of mammals (Tan et al. 1999; Ogura et al. 2001; Wang et al. 2001; Dildrop and Ruther 2004; Feijóo et al. 2004). The fact that fishes have more than six Irx genes is explained by an additional round of genomic duplication that likely occurred in the teleost lineage (Taylor et al. 2001). It is noteworthy that the overall extension of the fish and amphibian Irx clusters is comparable with that of mammals and that even in pufferfish the clusters are not reduced by the same ratio (1:8) that has been found for other genomic regions (Brenner et al. 1993; Aparicio et al. 2002). For instance, the pufferfish IrxBa cluster is only three times smaller than the mouse IrxB cluster (Dildrop and Ruther 2004), suggesting that there has been selective constraint to maintain these large gene deserts.
The regulation of the expression of the Irx genes remains largely unresolved. However, the conserved organization of vertebrate Irx genes in clusters could shed some light on the issue of transcriptional regulation of this gene family. Work carried out on vertebrate Hox genes has shown that the presence of shared and/or global cis-regulatory elements can be one of the reasons for the evolutionary conservation of their peculiar organization in gene clusters (Duboule 1998). Furthermore, such work has also highlighted the power of comparative genomics to identify cis-elements and the regulatory networks that act upon them (Manzanares et al. 1999). A similar scenario is emerging from the comparison of Irx gene expression patterns. In all organisms analyzed to date, the developmental expression patterns of Irx1 and Irx2 are almost identical, and this is also the case for Irx3 and Irx5 (Bellefroid et al. 1998; Gómez-Skarmeta et al. 1998; Garriock et al. 2001; Houweling et al. 2001). The expression of the third gene of each cluster, Irx4 or Irx6, is in general more divergent (Garriock et al. 2001; Houweling et al. 2001). Nonetheless, in certain tissues, all the genes of a cluster, or even of both clusters, are expressed identically. This suggests the existence of cis-regulatory elements (enhancers) that act on the entire set of the Irx genes in a cluster and that some of these enhancers are present in both clusters. Moreover, the expression of Irx orthologs is largely equivalent, which is indicative of cross-species conservation of regulatory elements (for review, see Gómez-Skarmeta and Modolell 2002).
The genomic organization of Irx genes, their similar expression domains in several tissues, and their equivalent patterns in different vertebrates indicate an evolutionary conservation of the underlying regulatory mechanisms. A powerful approach for the identification of regulatory elements has been the comparison of genomic sequences between species (phylogenetic footprinting) (Muller et al. 2002; Frazer et al. 2003). However, few studies have demonstrated the functional relevance of conserved non-coding sequences.
In this work, we examined, by means of functional expression assays in Xenopus and zebrafish, the transcriptional enhancer activity of highly conserved non-coding sequences present in the gene deserts that lie between the Irx genes of the vertebrate IrxB cluster. Our results suggest that several of these regions contain global enhancers, whereas others appear to modulate specific domains of expression of particular Irx genes. Several of these conserved regions are absent in fish, suggesting the appearance of novel regulatory mechanisms in tetrapods. Finally, we have analyzed the enhancer activity of four Irx ultraconserved regions (UCRs) present between the six Irx genes of all vertebrates. The evolutionary and structural genomic implications of these results are discussed.
Results
Expression of zebrafish and Xenopus IrxB genes
As a prerequisite to analyze the regulation of IrxB genes in early development, we determined the patterns of expression of these genes in zebrafish and Xenopus. In zebrafish, as a result of a genome duplication that occurred in teleosts, two IrxB clusters are present (Dildrop and Ruther 2004; Feijóo et al. 2004). Of these, the genes of the IrxBa cluster (Irx3a, Irx5a, and Irx6) are most similar to those from the IrxB cluster of higher vertebrates. Thus, in this study we have focused on this cluster.
Expression of the zebrafish Irx3a at 24 hours post-fertilization (hpf) has been described previously (Tan et al. 1999). Transcripts are detected at the anterior midbrain extending posteriorly towards the spinal cord but are absent in the midbrain–hindbrain boundary (mhb) (Fig.1B). Expression is also found in the otic vesicle (Tan et al. 1999 and not shown). Outside the nervous system, Irx3a expression is detected in the pronephros (Fig. 1B, red arrowhead). Similarly, Xenopus Irx3 expression is found in the midbrain, hindbrain, spinal cord, and otic vesicle (Fig. 1C; Bellefroid et al. 1998), and it is absent from the mhb (Fig. 1C, black arrowhead). Outside the neural domains, Xenopus Irx3 mRNA is present in the pronephros, in the ectodermal layer of the branchial arches, and in a ventral domain close to the cement gland (Fig. 1C; Bellefroid et al. 1998).
In the zebrafish nervous system, Irx5a expression (Fig. 1D) is detected in domains similar but not identical to those of Irx3a (Wang et al. 2001). In addition, expression is found in the otic vesicle but not in the pronephros. Xenopus Irx5, in the brain and spinal cord, is expressed in a pattern almost identical to that of Irx3 but with two differences: It is not present in the otic placode but it is expressed in the retina (Fig. 1E; Garriock et al. 2001). Outside the neuroectoderm, expression occurs in a ventral anterior territory close to the cement gland, a region from which the heart will form (Fig. 1E, inset).
The expression of Irx6 in zebrafish or Xenopus has not been described. RT-PCR experiments indicated that in both species this gene is first expressed at late neurula stages (48 hpf for zebrafish, Fig. 1F, and from stage 35 onwards in Xenopus, Fig. 1G; lower panels). In situ hybridizations showed that in zebrafish, Irx6 is expressed in the midbrain, hindbrain, and notochord (Fig. 1F). In Xenopus, its late onset of expression required examining the distribution of Irx6 mRNA in transverse sections of embryos at stages 38–40. Irx6 expression was found at higher levels in the retina, hindbrain, and notochord and at lower levels in the midbrain and in the spinal cord (Fig. 1Ga-c). A diagram summarizing the expression domains of zebrafish and Xenopus IrxB genes is shown in Figure 1H.
Highly conserved non-coding regions are present in vertebrate IrxB intergenic domains
Cross-species sequence comparison of the IrxB clusters reveals multiple highly conserved non-coding regions (HCNRs) (sequences longer than 100 bp with higher than 75% identity) in all vertebrate species examined (Fig. 1I,J). The number of these HCNRs varies when comparing different vertebrates. Therefore, when human and mouse IrxB clusters are compared, 358 HCNRs are identified. This extremely high number makes their systematic analysis via a functional transgenic assay for enhancer function unrealistic. However, this number is greatly reduced when the human cluster is compared with those from more distant vertebrates such as Xenopus (36 HCNRs) or Fugu (30 HCNRs). The different HCNRs were named by a numeral indicating their position in human chromosome 16 (NCBI build 33, assembly of July 2003) (Fig. 1I,J), preceded by a letter indicating the species. We have performed both local (using Pipmaker) and global (using Vista) alignments of these regions obtaining the same results.
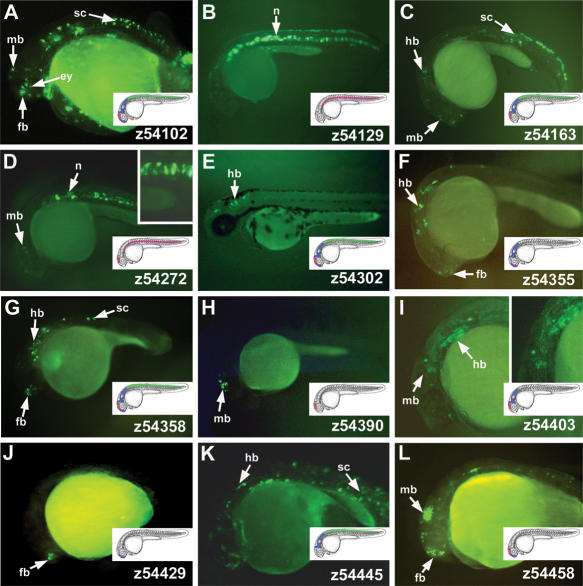
We next tested HCNRs for the presence of cis-regulatory elements. To that end, we attempted to amplify by PCR all of these HCNRs. Within the gene desert between Irx3 and Irx5 (GD 3–5), we succeeded in amplifying 16 HCNRs from the zebrafish genome (Fig. 1I, asterisks in fourth line). Of these sequences, 5 were not found in the Xenopus genome (Assembly 3.0). This could be explained because either they correspond to unsequenced or otherwise unavailable regions, or they correspond to sequences that have diverged beyond our ability to recognize them. A further possibility we cannot exclude at present is that these elements have been specifically lost in Xenopus. HCNRs were cloned upstream of a 0.7-kb proximal promoter of the zebrafish Irx3a gene driving the expression of the enhanced green fluorescent protein (EGFP) reporter gene. This promoter alone is silent during embryogenesis in zebrafish, assayed by transient expression, as well as in transgenic Xenopus. These constructs were used to generate transient transgenic zebrafish embryos. Twelve HCNRs (Fig. 1I, green asterisks) activated expression of EGFP in neural and non-neural domains (Fig. 2; Supplemental Table S2). Most HCNRs drove expression within subdomains of the territories where Irx3a and Irx5a are expressed at 24 hpf, such as the midbrain, hindbrain, and spinal cord (Fig. 2; Supplemental Table 2). In addition, some of them activated expression in domains that express Irx3a and Irx5a at earlier stages, such as the notochord (Fig. 2B,D; Supplemental Table 2). These HCNRs may contain enhancers shared by both Irx3a and Irx5a genes. Interestingly, many of the HCNRs promoted expression in partially overlapping domains, especially in the midbrain and hindbrain (Fig. 2; Supplemental Table 2). Thus, they appear to contain partially redundant enhancers. A few of the HCNRs promote expression in the forebrain, which is not part of the endogenous Irx3a or Irx5a expression domains (Fig. 2A,F,G,J,L). This suggests that silencer or repressor elements may be present within the IrxB cluster to suppress this forebrain expression.
Figure 2.
Highly conserved non-coding regions (HCNRs) located between Irx3 and Irx5 genes show enhancer activity in zebrafish. Lateral views of 24-hpf zebrafish showing enhanced green fluorescent protein localization promoted by different HCNRs (A–L). Insets show magnifications of some of the corresponding expression domains. The number in the lower righthand corner of each panel corresponds to the position of the human homologous region in chromosome 16 (NCBI build 33) as shown in Figure 1I. For better comparison, illustrations show the expression domains in which the HCNRs are active. Color codes are as those in Figure 1. Note that this is an oversimplification scheme, as many enhancers are active in subdomains of these territories. (fb) Forebrain; (mb) midbrain; (hb) hindbrain; (sp) spinal cord; (ey) eye; (n) notochord.
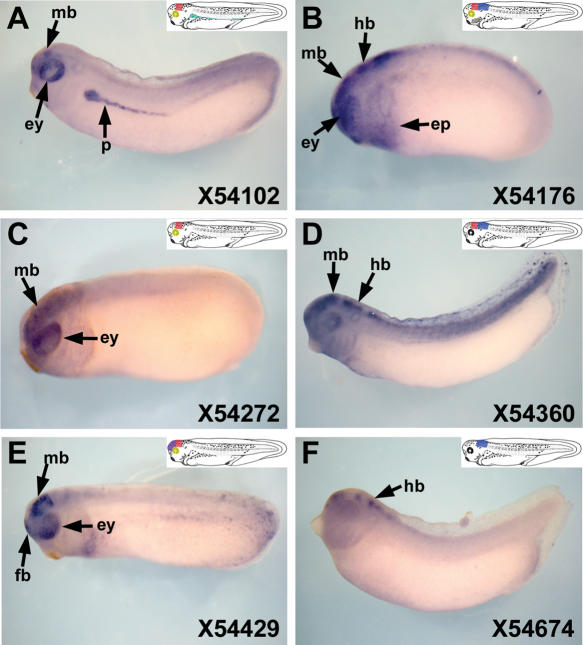
We next examined the enhancer activity of several HCNRs located in the GD3–5 region of Xenopus tropicalis. These sequences were used to drive expression of EGFP with either the zebrafish Irx3 or a similar 0.6-kb Irx3 promoter from X. tropicalis. These constructs were then assayed in X. laevis transgenic embryos. We PCR-amplified 18 Xenopus HCNRs (Fig. 1I, asterisks in third line). Eight of these are conserved between Xenopus and higher vertebrates and 10 are present in all vertebrates, including teleosts. Of the latter group, only three activate expression of the reporter; the cognate zebrafish elements were also active, as shown above (Fig. 3A,C,E, cf. Fig. 2A,D,J). The remaining seven HCNRs were inactive during early Xenopus development (up to stage 35), although three of the zebrafish homologous HCNRs activated expression in the zebrafish assays (Fig. 2B,E,K). The three HCNRs active in Xenopus drove expression, at least in part, in domains similar to those of their corresponding zebrafish homologs. Thus, both zebrafish and Xenopus element 54102 promoted expression in the eye, midbrain, and heart (Figs. 2A, 3A, and not shown). However, z54102 drove expression in the forebrain, hindbrain, and spinal cord (Fig. 2A; Supplemental Table 2), whereas X54102 did not promote expression in these territories but did so in the pronephros. These differences may be due to the transgenic methodology (transient in zebrafish versus stable in Xenopus) or to the sequence differences of the zebrafish and Xenopus HCNRs. Moreover, zebrafish but not Xenopus embryos are transparent, which allows the detection of expression in deeper tissues than can be observed in Xenopus embryos. Similarities as well as differences are also observed with the remaining two HCNRs tested in both systems. For example, z54272 and X54272 are both active in the midbrain. In contrast, z54272 drives EGFP in the notochord, whereas X54272 activates expression in the eye but not in the notochord (Figs. 2D, 3C). Finally, both z54429 and X54429 drive expression in the forebrain, but X54429 is also active in the midbrain and in the eye.
Figure 3.
Highly conserved non-coding regions (HCNRs) located between Irx3 and Irx5 genes show enhancer activity in Xenopus. Lateral views of late neurula Xenopus embryos showing enhanced green fluorescent protein mRNA localization promoted by different HCNRs (A–F). The number in the lower righthand corner of each panel corresponds to the position of the human homologous region in chromosome 16 (NCBI build 33) as shown in Figure 1I. Illustrations show a schematic representation of the expression domains in which the HCNRs are active. (fb) Forebrain; (mb) midbrain; (hb) hindbrain; (ep) epidermis; (ey) eye; (p) pronephros.
Of the eight Xenopus HCNRs that are not conserved in zebrafish, three were active during early Xenopus development. They promoted expression in different regions of the brain, in the eye, and in the epidermis of the head (Fig. 3B,D,F). In general, the territories where the Xenopus HCNRs are active correspond to subregions of Irx3 and/or Irx5 domains, although, as with the zebrafish assays, we observed that some HCNRs activate EGFP in the forebrain (Fig. 3B,E), an area devoid of Irx3 and Irx5 expression. This again suggests the presence of silencer elements within this gene desert. Like the zebrafish HCNRs, most of the Xenopus HCNRs were active in overlapping domains of the nervous system, further supporting the idea that this gene desert contains redundant enhancers. Interestingly, at least one of the enhancers present in all vertebrates but not in teleost fishes (zebrafish, tetraodon, or Fugu) promoted expression in the head epidermis (Fig. 3B), a territory that seems to lack expression of Irx3 and Irx5 in fish.
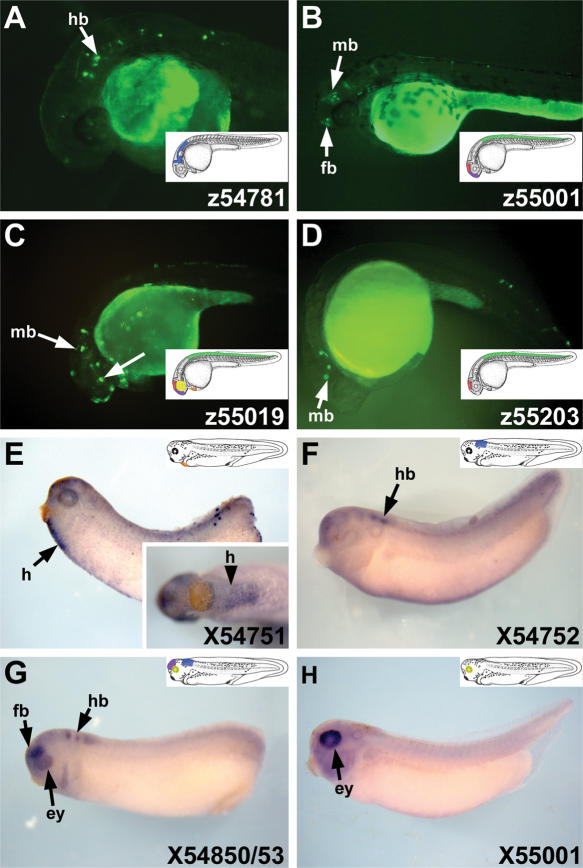
Next, we searched for functionally active HCNRs in the gene desert located between the Irx5 and Irx6 genes (GD5–6). This region is about 39% smaller than the previous one (393 kb versus 645 kb in the human genome NCBI build 33; Fig. 1A,I,J). In addition, GD5–6 contains a lower number of HCNRs (3 vs. 15; Fig. 1I,J). The number of HCNRs increases when teleosts are excluded from the genomic comparison (11 in GD5–6 and 25 in GD3–5; Fig. 1I,J). Finally, in GD5–6, we detected three HCNRs that are present in all other vertebrates but not in Xenopus. We assayed 4 HCNRs from zebrafish. Three activated expression within Irx5a and/or Irx6a domains, such as the eye, midbrain, and hindbrain (Fig. 4A,B,C; Supplemental Table 2). Of these four zebrafish HCNRs, only one (z55001) is conserved in Xenopus (X55001) and it was also active in Xenopus transgenic experiments. However, whereas z55001 drives EGFP expression in the forebrain, midbrain, and spinal cord, X55001 is active only in the eye (Fig. 4H).
Figure 4.
Highly conserved non-coding regions (HCNRs) located between Irx5 and Irx6 genes show enhancer activity in zebrafish and Xenopus.(A–D) Lateral views of 24-hpf zebrafish embryos showing distribution of enhanced green fluorescent protein (EGFP) promoted by different HCNRs. (E–H) Lateral views of late neurula Xenopus embryos showing EGFP mRNA localization. Inset in E shows a higher magnification of the embryo from a ventral view. The number in the lower righthand corner of each panel corresponds to the position of the human homologous region in chromosome 16 (NCBI build 33) as shown in Figure 1J. Illustrations show a schematic representation of the expression domains in which the HCNRs are active. (fb) Forebrain; (mb) midbrain; (hb) hindbrain; (ey) eye; (h) heart territory.
We also tested another five Xenopus HCNRs, three of which showed transcriptional activity in the prospective heart, forebrain, midbrain, and eye (Fig. 4E,F,G). As in GD3–5 HCNRs, the GD5–6 HCNRs also promoted expression in partially redundant territories that in most cases corresponded to Irx5 and Irx6 domains. However, some zebrafish and Xenopus GD5–6 HCNRs can also activate EGFP expression in the forebrain, which is devoid of Irx5a and Irx6a expression. Interestingly, one HCNR conserved in all vertebrates except fishes (zebrafish, tetraodon or Fugu) promotes expression in the heart primordium (Fig. 4E, inset). This territory lacks Irx5 or Irx6 expression in zebrafish but does show expression of at least Irx5 in higher vertebrates (Houweling et al. 2001; Wang et al. 2001; this paper).
In addition, we also examined an HCNR 3′ to the Irx6a gene isolated from both zebrafish and Xenopus and another HCNR located 3′ of Xenopus Irx3. Whereas both Xenopus HCNRs were inactive, the zebrafish sequence activated expression in the midbrain and in the spinal cord (Fig. 4D).
Finally, we examined whether non-conserved regions can promote any expression using similar constructs. To that end, we amplified three regions between 700 and 900 bp long from the IrxB Xenopus genome that were placed 5′ of the Xenopus Irx3 promoter driving EGFP expression. These regions are located between conserved regions X54129–X54176, X54578–X54627, and X55204–X55256. As expected, these regions did not activate expression at specific and reproducible domains.
We conclude that the gene deserts located between Irx3 and Irx5 or Irx5 and Irx6 harbor multiple HCNRs that contain enhancer elements and, probably, some silencer elements. A diagram summarizing the different zebrafish and Xenopus enhancers identified in this study is shown in Figure 7, below. Some enhancers may act on both neighboring genes, whereas others appear to be acting specifically on a single gene. Moreover, many of the enhancers appear to be redundant in terms of the territories in which they are active. Finally, some enhancers are conserved in all vertebrates, whereas others appear to have evolved after the separation of the teleosts and tetrapod lineages.
Figure 7.
Summary of our results. Diagram showing the distribution of the different enhancers detected in this study within the IrxB cluster. For simplicity, we have numbered these enhancers from 1–22. These numbers reflect their linear position in the genome from Irx3 to Irx6. Enhancers tested in both systems have the same number. The correspondence of these numbers with the enhancer position in human genome can be found in Supplemental Table 2. The color coding represents the different expression domains in which these enhancers are active in zebrafish (top) and Xenopus (middle). These domains are listed in the same color code. Note that this is an oversimplification scheme, as many enhancers are active in subdomains of these territories. At bottom, an illustration shows the expression domains of zebrafish and Xenopus IrxB genes.
Some HCNRs are interchangeable between different vertebrates
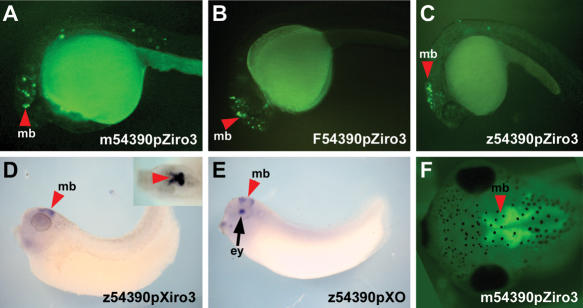
The HCNRs in the Irx complexes of different vertebrates suggest that these regions may contain enhancers capable of driving expression in equivalent territories of different species. However, in some cases, only the zebrafish HCNR and not its Xenopus homolog was active. Moreover, in those cases in which both HCNRs were active, we observed expression in both equivalent and non-equivalent territories in these two species. To test whether these differences can be due to variations in the DNA sequence, we isolated HCNR 54390 from zebrafish, Fugu, and mouse and examined its enhancer activity in both zebrafish and Xenopus. HCNR z54390 produces a very reproducible pattern in zebrafish transient expression assays. As shown in zebrafish assays (Fig. 5A,B,C), the mouse (A), Fugu (B), and zebrafish (C) versions of this HCNR activate expression in the same brain territory, the midbrain, in 24-hpf embryos.
Figure 5.
Highly conserved non-coding regions (HCNRs) are functionally active in different vertebrates. (A–C) Lateral views of 24-hpf zebrafish showing of enhanced green fluorescent protein (EGFP) fluorescence in the midbrain (mb) promoted by mouse (A), Fugu (B), or zebrafish (C) HCNR 54390. (D–E) Zebrafish 54390 placed 5′ of Xenopus Irx3 (D) or opsin (E) promoters activates EGFP expression in the same midbrain (mb) domain. Inset in D shows a higher magnification of a dorsal view of the embryo. The opsin promoter also activates EGFP expression in the eye (ey) (arrow in E). (F) Dorsal view of a stage 45 Xenopus embryo transgenic for the mouse 54390 region. EGFP expression is detected in the midbrain (mb).
We examined the activity of the zebrafish and mouse HCNRs in Xenopus transgenic assays. Using the Xenopus Irx3 promoter, the zebrafish HCNR drove expression, as in zebrafish, in the midbrain of Xenopus embryos (Fig. 5D), even though Xenopus lacks a homologous HCNR. We switched this enhancer from the Xenopus promoter to the zebrafish Irx3 or the Xenopus opsin promoters. In both cases, the HCNR still drove expression in the same territory (not shown and Fig. 5E). This was also the case when the mouse HCNR was assayed in Xenopus, although expression was delayed to later developmental stages (Fig. 5F).
We conclude that, at least in some cases, differences in the DNA sequence of homologous HCNRs from different vertebrates do not significantly alter the territorial specificity of the corresponding enhancer. However, they can affect the level or the timing of transcription. Thus, there may be species specificity associated with minor DNA differences. These differences would be optimized over evolutionary time for interactions with the proteins that activate transcription in the appropriate species. In addition, we have shown that HCNRs absent from Xenopus can be active in this organism.
The core domains of the ultraconserved Irx regions promote similar expression patterns that are modulated by adjacent conserved DNA
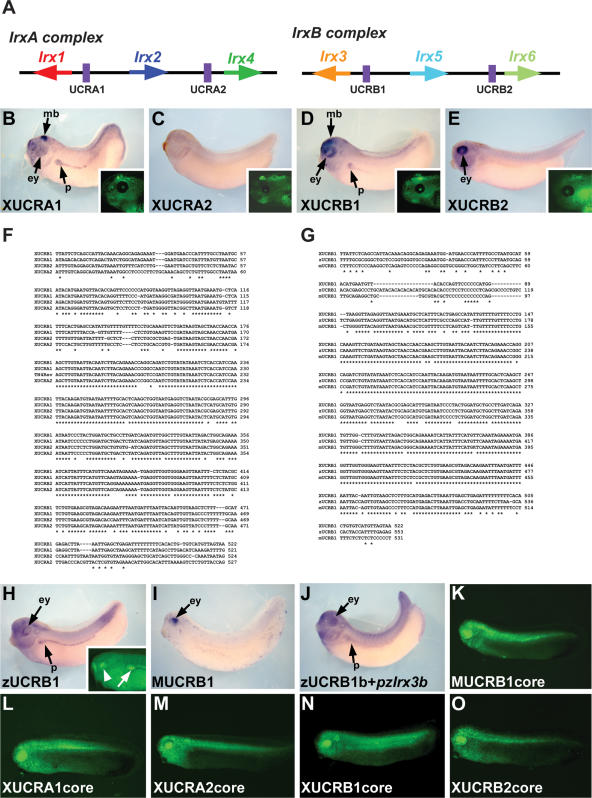
Within the IrxB cluster, two conserved non-coding regions are of particular interest. These belong to the group of so-called UCRs, which are longer than 200 bp and are 100% identical in humans and rodents (Bejerano et al. 2004). Interestingly, there is an extremely high degree of conservation between these two UCRs (UCRB1 and UCRB2), and they are highly similar to two other UCRs in the IrxA complex (UCRA1 and UCRA2), which are located in relatively similar genomic domains. Thus, in each Irx cluster, there is an UCR in each gene desert located in a similar relative position (Fig. 6A). We found that the four UCRs from Xenopus had enhancer activity and promoted similar, but not identical, patterns of expression (Fig. 6B–E). Thus, UCRA1 (Fig. 6B) and UCRB1 (Fig. 6D; see also X54102 in Fig. 3A) drove expression at stage 35 in the midbrain, eye, and pronephros. However, UCRB2 (Fig. 6E; see also X55001 in Fig. 4H) was active only in the eye, whereas the UCRA2 (Fig. 6E) was inactive at this stage. At later stages, the four UCRs promoted expression in the brain (Fig. 6B–E, insets). Thus, despite the striking identity of these four UCRs (Fig. 6F), they promote expression in slightly different domains. These differences may result from the small variations in the respective DNA sequences.
Figure 6.
Functional comparison of Irx ultraconserved non-coding regions (UCRs). (A) Schematic representation of the vertebrate Irx clusters showing the position of the UCRs (purple boxes) between the different Irx genes. (B–E) Lateral views of late neurula Xenopus embryos showing enhanced green fluorescent protein (EGFP) mRNA localization promoted by the different Xenopus UCRs. Insets show EGFP fluorescence in stage 45 embryos transgenic for the Xenopus UCRs. Note similar expression in the brain. (mb) Midbrain; (ey) eye; (p) pronephros. (F, G) Sequence alignment of the four Xenopus UCRs (F) or Xenopus, zebrafish, and mouse UCRB1s (G). Asterisks mark identical bases. (H, I) Lateral views of stage 35 Xenopus embryos showing EGFP expression directed by zebrafish (H) or mouse (I) UCRB1s. Inset in H is a higher magnification of a living embryo transgenic for zUCRB1 showing EGFP fluorescence in the eye (arrowhead) and in the pronephros (arrow). (J) Xenopus embryo transgenic for a construct in which EGFP is directed by a 3-kb Irx3b promoter that harbors a duplicated UCRB1b. (K–O) EGFP fluorescence in Xenopus promoted by the core domain conserved between different vertebrate Irx UCRs.
We next examined the activity of the UCRB1 from different vertebrates in Xenopus. As shown in Figure 6G, mouse, Xenopus and zebrafish UCRB1 have higher sequence identities than the four Xenopus UCRs compared with themselves. Both zebrafish and Xenopus UCRB1 promoted similar expression in the eye and pronephros (Fig. 6D,H). However, mouse UCRB1 only drove expression in the eye (Fig. 6I). Interestingly, the duplicate zebrafish IrxB cluster, IrxBb (Feijóo et al. 2004), contains a second UCR close to the Irx3b transcriptional unit (UCRB1b). We amplified a 3-kb region that contained this UCRB1 and the Irx3b promoter. When assayed in Xenopus, this enhancer plus promoter region, similar to Irx3a UCRB1b, activated expression in the eye, brain, and pronephros (Fig. 6J).
The regions tested above are of considerable length (1–3 kb) and include, for each UCR, the whole conserved domains between different vertebrates. The length of these regions varies for each UCR. Thus, IrxA UCRs are longer (up to 2 kb) than the IrxB ones (about 500 bp), the common conserved domain for all of them being about 400 bp (Fig. 6F,G). It is therefore possible that the different activities of these regions could be due to the size variations in our constructs. To test this, we prepared similar size constructs for all Xenopus UCRs and mouse UCRB1 that contain only the core-conserved region showed in Figure 6, F and G. When they were assayed in Xenopus with the Xenopus Irx3 promoter, these equivalent regions promoted expression in very similar domains (Fig. 6K–O). These domains were broader than those observed with the respective longer regions and included the eye, most of the nervous system, and the mesoderm. In addition, Xenopus UCRB1 and in some cases UCRA1 core regions could promote expression in the pronephros (Fig. 6N and not shown). Taken together, these data suggest that adjacent highly conserved DNA modulates the core-promoted expression pattern.
Discussion
The strong evolutionary conservation of the IrxB genomic organization and the similar expression patterns of IrxB genes suggest the presence of shared regulatory elements within the cluster. Here we provide evidence that many HCNRs located in the gene deserts between IrxB genes contain regulatory elements responsible for the activation of Irx genes within specific territories. Some of these elements may be shared among IrxB genes, whereas others are gene specific.
Irx6 expression in zebrafish and Xenopus
Here we report for the first time the expression pattern of Irx6 in zebrafish and Xenopus. The expression of Irx6 begins at late neurula stages, an indication that this gene does not participate in early neural patterning, as do the other members of the family (for review, see Gómez-Skarmeta and Modolell 2002). A similar situation is found for mouse Irx6 (Houweling et al. 2001). We determined that, like their mouse paralogs, zebrafish and Xenopus Irx6 share expression domains with the other IrxB genes, Irx3 and Irx5, mainly in neural tissues. Despite these similarities, in all vertebrates examined, the expression domains of Irx3 and Irx5 are more similar between themselves than either is to that of Irx6 (Bellefroid et al. 1998; Tan et al. 1999; Garriock et al. 2001; Houweling et al. 2001; Wang et al. 2001; this paper).
Enhancer activity in HCNRs within IrxB cluster
Whole-genome comparisons between different vertebrates have demonstrated the presence of many HCNRs in the genomes from fish to human (Bejerano et al. 2004; Sandelin et al. 2004; Woolfe et al. 2005). The number of these conserved regions varies from 250 to 3500, depending on the stringency of the selection method (Bejerano et al. 2004; Sandelin et al. 2004; Woolfe et al. 2005). Remarkably, these studies conclude that HCNRs tend to cluster in gene deserts surrounding key developmental regulators, suggesting strong evolutionary constraints against changes in these GRNs. Functional analyses of a small fraction of these HCNRs indicate that some of them contain enhancer elements (Gottgens et al. 2000; Bagheri-Fam et al. 2001; Ghanem et al. 2003; Nobrega et al. 2003; Santini et al. 2003; Spitz et al. 2003; Sumiyama et al. 2003; Uchikawa et al. 2003; Woolfe et al. 2005). The Irx genes are developmentally important genes organized in large genomic complexes that show patterns of expression highly conserved during evolution. We performed a detailed search for conserved sequences present in the two gene deserts spanning the intergenic regions between the Irx3–Irx5 (GD3–5) and Irx5–Irx6 (GD5–6) genes in the IrxB cluster. Our identification of multiple HCNRs present in these large intergenic regions is coincident with two recent reports that have highlighted the striking enrichment of HCNRs surrounding the Irx genes (Sandelin et al. 2004; Woolfe et al. 2005). These comparisons showed that Irx genomic regions contain between 2% and 5% of the total number of HCNRs present in all vertebrate genomes (Sandelin et al. 2004; Woolfe et al. 2005).
A recent classification of gene deserts divided them into two types. In the stable gene deserts, the density of conserved regions between human and chick is higher than 2%, whereas in the variable gene deserts this percentage is below 2% (Ovcharenko et al. 2005). Stable gene deserts also have about a 120-fold higher density of conserved non-coding regions between human and fish than variable gene deserts. According to this classification, IrxB gene deserts belong to the stable type. Within the IrxB complex, GD3–5 spans 60% of the total intergenic regions and harbors 83% of the HCNRs. The genes flanking GD3–5, Irx3 and Irx5, are expressed in more complex patterns than Irx6, indicating that perhaps the regulation of Irx3 and Irx5 requires a larger number of enhancers than Irx6. We speculate that these enhancers lie within the HCNRs.
Indeed, our functional analysis of HCNRs by means of zebrafish and Xenopus transgenesis clearly demonstrates the presence of enhancer activity within them. Most of these enhancers promote expression in a subset of the territories in which the flanking Irx genes are expressed. This indicates that the complex Irx patterns of expression are generated by the combinatorial action of multiple regulatory elements. Some of these enhancers may have redundant functions as they are expressed in partially overlapping territories, especially within the brain. In correspondence with their genomic location, an important fraction of these enhancers seems to activate expression in regions in which both flanking genes are expressed, suggesting that they may function bidirectionally. The existence of these shared enhancers may be responsible for keeping the Irx genes associated in genomic cluster, as it has been suggested for Hox genes (Duboule 1998). In contrast, some of the enhancers we have isolated drive expression in territories in which only one of the flanking genes is expressed, suggesting that they are gene-specific regulatory elements. We have also found that some elements promote expression in domains devoid of IrxB expression, such as the forebrain. This may indicate that there are also silencer elements within these intergenic regions, which may downregulate IrxB expression in this territory. Alternatively, we cannot rule out that this ectopic expression is a consequence of the disruption of the genomic architecture of these elements in our assays. This may also explain why some enhancers do not precisely recapitulate the expression pattern of Irx genes in specific territories or why we find that some elements lack transcriptional regulatory activity.
In our in silico analysis using genomic sequence from three vertebrate classes, fish, frogs, and mammals, we have found that most HCNRs are present in all species examined but that some are absent from either fish or Xenopus. We cannot rule out that these regions are as yet unsequenced, that they have been duplicated in the respective genome and subsequently lost, or that the corresponding sequence has diverged beyond our ability to recognize them. However, it is an attractive hypothesis that some of the tetrapod-specific HCNRs could be responsible for expression domains that evolved after the divergence of fish from the higher vertebrate lineages. Indeed, in each IrxB gene desert, we found at least one HCNR that was not present in zebrafish, Fugu, or tetraodon and that drives expression in territories in which the zebrafish orthologous IrxB genes are not expressed. The addition of new regulatory elements to key developmental regulators is a mechanism used by evolution for the generation of new forms without increasing the number of genes.
We conclude that many vertebrate HCNRs within the IrxB cluster harbor enhancer elements (for a summary, see Fig. 7). However, the ectopic expression promoted by some enhancers, as well as the modulation of the activity of some other by surrounding DNA (see below), indicates that the genomic context is likely playing an important function for the proper function of these elements. We note that this type of screening is not adequate for detection of inhibitory elements, which may lie within some of the HCNRs that showed no enhancer activity. In order to do so, it would be necessary to assay each of the candidate inhibitory elements in a condition in which GFP is broadly expressed, such as under a promoter driving generalized expression. Nevertheless, it is still possible that some of these inhibitory elements may be specific for individual enhancers; therefore, they would not be detected unless these two elements interact in the proper genomic context.
We would also like to remark on two important benefits of zebrafish and Xenopus over the mouse model systems in this kind of analysis. First, we were able to determine the activity of many enhancers in a very cost and time-efficient manner. Second, and in contrast to mouse studies, we were able to examine the enhancer activity in the same embryo during all development.
Are ortholog HCNRs active in similar domains?
Our results show that at least one of the HCNRs examined activates expression in the same territory regardless of the model used to test it or the source of the HCNR. This indicates that the HCNR probably binds the same transcription factors in different vertebrates. Moreover, some HCNRs are active independently of the promoter used or the orientation of the HCNR with respect to the promoter, behaving as classical enhancers. Other HCNRs promote expression in both identical and different territories when their activities are comparatively examined in zebrafish and in Xenopus. These differences may be due to small variations in the DNA sequences of orthologous HCNRs. Alternatively, some of the HCNRs may have evolved in specific vertebrate lineage to acquire distinct functional properties. Finally, it is possible that these dissimilar activities are a consequence of the different techniques used for the generation of transgenic zebrafish or Xenopus. Also, the transparency of zebrafish embryos may help to detect expression in more domains in zebrafish than is possible in Xenopus. Accordingly, we found many more active elements in zebrafish than in Xenopus.
Of the different IrxB HCNRs examined, two are of special interest. They are longer that 200 bp and display 100% conservation between humans and rodents, and are by this definition termed UCRs (Bejerano et al. 2004). Approximately 256 UCRs with no evidence of being transcribed are found in the human genome and they are prevalent in gene deserts near key developmental genes (Bejerano et al. 2004). Of these, four are within the Irx clusters (Bejerano et al. 2004; Sandelin et al. 2004; this paper). Surprisingly, they are not only located in equivalent regions of both clusters, with respect to the Irx genes, but they are extremely similar in sequence in a core region of about 400 bp. This is a unique situation in the vertebrate genomes (Sandelin et al. 2004). The functional significance of UCRs remains largely unknown, although some of them harbor enhancer elements (Nobrega et al. 2003). Here, we have performed a detailed comparative analysis of the four Irx UCRs isolated from Xenopus and of the UCR closest to Irx3 from different vertebrates. To our knowledge, this is the first detailed functional analysis of these intriguing genomic elements. Given the largely overlapping expression patterns of Irx genes, we expected to find enhancers responsible for the shared patterns lying within the Irx UCRs. Indeed, the conserved core of these UCRs, that is, the region conserved in all vertebrate UCRs from the two Irx clusters, activates expression in very similar territories. This pattern was modified when each UCR was analyzed as fragments of different length containing the entire conserved DNA in different vertebrates. These longer UCRs did not promote expression in exactly the same pattern in transgenic Xenopus. Indeed, compared with core-promoted expression, the pattern produced by the complete UCR becomes restricted. Hence, the highly conserved DNA that surrounds the core of all Irx UCRs seems to modulate the pattern promoted by the core negatively. Interestingly, in embryos transgenic for the different core Irx UCRs, we still observed some small differences that affected both the domains and the level of expression of the reported gene. Small DNA differences could account for these variations. Mutation of specific conserved or nonconserved bases or the swapping of domains between different UCRs may help determine how the small differences in DNA sequence affect the enhancer activity of vertebrate UCRs.
Although our study indicates that the Irx UCRs contain functional enhancers and that these may function by interacting with transcription factors, we cannot exclude the possibility that they operate in alternative and less conventional ways. It seems unlikely that this extreme conservation of DNA sequence is due solely to multiple overlapping of transcription factor binding sites. Indeed, UCRs are more highly conserved than sequences coding for identical proteins and are far more conserved than any other known cis-regulatory elements. Thus, it is likely that they have additional roles, such as the regulation of chromatin structure. This is further supported by the fact that the identity of each of the two IrxA UCRs in different vertebrates extends to almost 2 kb, compared with the 400 bp conserved when comparing the four Irx UCRs of any vertebrate. This possibility is especially attractive considering that the four Irx UCRs are situated in similar regions within the Irx clusters and that the genome organization of these clusters has been conserved throughout vertebrate evolution.
Interestingly, in Drosophila, there are three Iroquois (Iro) genes that are also organized in a genomic complex. These Drosophila Iro genes share at least some regulatory elements (for review, see Gómez-Skarmeta and Modolell 2002). However, the vertebrate Irx proteins are more similar among themselves than to the Drosophila Iro proteins, which are closely related to one another (Peters et al. 2000). Therefore, despite the similar genomic organization of vertebrate and invertebrate Irx/Iro genes and the presence of shared enhancers in vertebrate and invertebrate clusters, it is likely that independent duplication events gave rise to three-gene clusters in ancestors of the insect and vertebrate lineages. Supporting this, we have not found regions in the Drosophila genome homologous to the vertebrate Irx HCNRs. Moreover, Drosophila or vertebrate Irx/Iro enhancers are not active in vertebrates or Drosophila, respectively (A. Leticia, unpubl.).
Methods
Zebrafish and Xenopus whole-mount in situ hybridization
Antisense RNA probes were prepared from cDNAs using digoxigenin (Roche) as label. Xenopus and zebrafish specimens were prepared, hybridized, and stained as in Harland (1991) and Jowett and Lettice (1994), respectively. Transverse sections of 50 μm were generated with a Leica vibratome. Embryos were embedded in 5% agarose, post-fixed for 30 min, and then processed for in situ hybridization. The Xenopus Irx6 partial coding region fragment used for in situ hybridization was obtained by PCR from X. tropicalis genomic DNA with primers corresponding to the fifth exon: 5′-CAACATGCAAAAATGGGAAG-3′ and 5′-GAAA TGATTCTCCGAGTACGG-3′. The amplification product was subcloned in pGEMT-Easy (Promega) and sequenced. A publicly available cDNA clone (IMAGE: 7073836) was the source of a partial zebrafish Irx6 cDNA used for probe generation.
RT-PCR
Total RNA was extracted from different developmental stages of zebrafish or X. tropicalis embryos, treated with DNAse, and processed for RT-PCR as described (Gawantka et al. 1995). RT-PCRs were carried out using the zebrafish primers 5′-CGTGTCGGCA ACCCCCAGTAC-3′ and 5′-GGCCAGCATGATCTTTCACCC-3′ or the X. tropicalis primers 5′-CCTCAGTTTGGATATCCG-3′ and 5′-TACTGCCCCAAGGAGTG-3′. These primers amplify a 517-bp or a 296-bp fragment of the zebrafish or X. tropicalis irx6 cDNA, respectively.
Zebrafish transgenesis
Plasmid DNAs were linearized, dissolved in water at 50 μg/mL, and injected into the yolk at the one-cell stage. Injected embryos were raised to the desired stage and examined under fluorescence in a Leica MZ12 dissecting scope. Between 300 and 400 embryos were injected for each enhancer in at least two different experiments. In our study, 30%–50% of the injected embryos survived to gastrulation. From these, 10%–20% showed some GFP expression. Therefore, we considered an enhancer active when at least 10–15 embryos show similar GFP distribution.
Xenopus transgenesis
Transgenic embryos were generated with linearized constructs according to Kroll and Amaya (1996) with modifications (Sparrow et al. 2000). This method allowed us to obtain a transgenic efficiency of about 20% of the embryos that survived to gastrulation. For each construct, about 100 embryos were scored, from at least two independent experiments. Embryos expressing GFP were first analyzed by observation of fluorescence and, after fixation, by whole-mount in situ hybridization to detect GFP mRNA. An enhancer was considered active when it promotes the same pattern in at least five different embryos. It should be noted that each embryo corresponds to a different integration event.
Vector construction
Primers 5′-gggaatTCAGAGGACCTCGACTGGC-3′ and 5′-gggt cgACAAAAAGGCGGCGAATCCG-3′ were used to amplify the zebrafish Irx3a promoter. Lowercase letters correspond to sequences not present in the genome. The 0.7-kb PCR fragment was subcloned into pGEMT-Easy and sequenced. Using EcoRI and SalI sites (in boldface type in the primer sequences), this promoter was placed 5′ of EGFP in pGreen Lantern (pGL, GIBCO). The promoter, EGFP and SV40 polydenylation signal were excised with PstI and ClaI from pGL and placed in pBlue-script. The primers 5′-gcctgCAGATACACACCCGAACTCC-3′ and 5′-gcggATCCCCTTCTATGACCTGAG-3′ were used to amplify the X. tropicalis Irx3 promoter. The 0.6-kb PCR fragment was subcloned in pGEMT-Easy and sequenced. We replaced the Xenopus Opsin promoter from the XOP vector (in which this promoter is 5′ to EGFP) with the X. tropicalis Irx3 promoter using the PstI and BamHI sites. Genomic HCNRs were amplified with the corresponding primers (Supplemental Table 1) and subcloned into pGEMT-Easy. In most cases, they were excised with EcoRI and placed 5′ of the zebrafish Irx3a or the Xenopus Irx3 promoters driving the expression of EGFP disregarding the orientation of the inserted regions. When the genomic regions contained internal EcoRI sites, SacII and PstI were used to transfer the region 5′ to the zebrafish promoter construct (Supplemental Table 1). The short versions of the four Xenopus UCRs and the mouse UCRB1 regions were cloned in the same orientation. This orientation was the same that the endogenous UCRB1 has with respect to the Irx3 promoter.
DNA sequencing
DNA sequencing was performed with ABI chemistry in an automatic DNA sequencer. Custom synthesized oligonucleotides were from Sigma.
Genomic sequence analysis
Genomic sequences spanning the Irx clusters were downloaded from the Ensembl server (www.ensembl.org), annotated, and multiple alignments were performed using both the Vista package (Frazer et al. 2004) and PipMaker (Schwartz et al. 2000).
Acknowledgments
We are especially grateful to I. Rollán for her help with some of the zebrafish experiments, E. Amaya for advice with the Xenopus transgenesis protocol and for reagents, T. Becker for his comments, and J. Modolell for his support and help with the manuscript. We thank R. Grainger, P.A. Krieg, E. Bellefroid, J.L. Mullor, and Z. Gong for reagents. We acknowledge grants from Spanish Ministry of Education and Science (BFU2004-00310) and Junta de Andalucía to J.L.G.-S.; Spanish Ministry of Education and Science (BMC2002-03558) and EMBO YIP programme to M.M.; Fondecyt (1031003), ICM (P02-050), UNAB (10-03), and ICGEB (CRP/CHI03-03c) to M.A.; Universidad de Chile-CSIC collaboration grant (2004CL0018) to M.A. and J.L.G.-S.; Spanish Ministry of Education and Science (BMC2002-411) to J. Modolell; Spanish Ministry of Education and Science (GEN2001-4846-C05-0) to G. Morata; and an institutional grant from Fundación Ramón Areces to the CBMSO. A predoctoral I3P fellowship from the CSIC to E. Rodríguez-Seguel is also acknowledged.
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: E. Amaya, A. Leticia, P.A. Krieg, E. Bellefroid, J.L. Mullor, and Z. Gong.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.4004805. Article published online before print in July 2005.
References
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310. [DOI] [PubMed] [Google Scholar]
- Bagheri-Fam, S., Ferraz, C., Demaille, J., Scherer, G., and Pfeifer, D. 2001. Comparative genomics of the SOX9 region in human and Fugu rubripes: Conservation of short regulatory sequence elements within large intergenic regions. Genomics 78: 73–82. [DOI] [PubMed] [Google Scholar]
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S., and Haussler, D. 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325. [DOI] [PubMed] [Google Scholar]
- Bellefroid, E.J., Kobbe, A., Gruss, P., Pieler, T., Gurdon, J.B., and Papalopulu, N. 1998. Xiro3 encodes a Xenopus homolog of the Drosophila Iroquois genes and functions in neural specification. EMBO J. 17: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., Elgar, G., Sandford, R., Macrae, A., Venkatesh, B., and Aparicio, S. 1993. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature 366: 265–268. [DOI] [PubMed] [Google Scholar]
- Briscoe, J. and Ericson, J. 2001. Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 11: 43–49. [DOI] [PubMed] [Google Scholar]
- Dildrop, R. and Ruther, U. 2004. Organization of Iroquois genes in fish. Dev. Genes Evol. 214: 267–276. [DOI] [PubMed] [Google Scholar]
- Duboule, D. 1998. Hox is in the hair: A break in colinearity? Genes & Dev. 12: 1–4. [DOI] [PubMed] [Google Scholar]
- Feijóo, C.G., Manzanares, M., de la Calle-Mustienes, E., Gomez-Skarmeta, J.L., and Allende, M.L. 2004. The Irx gene family in zebrafish: Genomic structure, evolution and initial characterization of irx5b. Dev. Genes Evol. 214: 277–284. [DOI] [PubMed] [Google Scholar]
- Frazer, K.A., Elnitski, L., Church, D.M., Dubchak, I., and Hardison, R.C. 2003. Cross-species sequence comparisons: A review of methods and available resources. Genome Res. 13: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, K.A., Pachter, L., Poliakov, A., Rubin, E.M., and Dubchak, I. 2004. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriock, R.J., Vokes, S.A., Small, E.M., Larson, R., and Krieg, P.A. 2001. Developmental expression of the Xenopus Iroquois-family homeobox genes, Irx4 and Irx5. Dev. Genes Evol. 211: 257–260. [DOI] [PubMed] [Google Scholar]
- Gawantka, V., Delius, H., Hirschfeld, K., Blumenstock, C., and Niehrs, C. 1995. Antagonizing the Spemann organizer: Role of the homeobox gene Xvent-1. EMBO J. 14: 6268–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem, N., Jarinova, O., Amores, A., Long, Q., Hatch, G., Park, B.K., Rubenstein, J.L., and Ekker, M. 2003. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 13: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Skarmeta, J.L. and Modolell, J. 2002. Iroquois genes: Genomic organization and function in vertebrate neural development. Curr. Opin. Genet. Dev. 12: 403–408. [DOI] [PubMed] [Google Scholar]
- Gómez-Skarmeta, J.L., Glavic, A., de la Calle-Mustienes, E., Modolell, J., and Mayor, R. 1998. Xiro, a Xenopus homolog of the Drosophila Iroquois complex genes, controls development of the neural plate. EMBO J. 17: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Skarmeta, J.L., Campuzano, S., and Modolell, J. 2003. Half a century of neural prepatterning: The story of a few bristles and many genes. Nat. Rev. Neurosci. 4: 587–598. [DOI] [PubMed] [Google Scholar]
- Gottgens, B., Barton, L.M., Gilbert, J.G., Bench, A.J., Sanchez, M.J., Bahn, S., Mistry, S., Grafham, D., McMurray, A., Vaudin, M., et al. 2000. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 18: 181–186. [DOI] [PubMed] [Google Scholar]
- Harland, R. 1991. In situ hybridization: An improved whole mount method for Xenopus embryos. Methods Cell Biol. 36: 685–695. [DOI] [PubMed] [Google Scholar]
- Houweling, A.C., Dildrop, R., Peters, T., Mummenhoff, J., Moorman, A.F.M., Rüther, U., and Christoffels, V.M. 2001. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech. Dev. 107: 169–174. [DOI] [PubMed] [Google Scholar]
- Jowett, T. and Lettice, L. 1994. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labelled probes. Trends Genet. 10: 73–74. [DOI] [PubMed] [Google Scholar]
- Kroll, K.L. and Amaya, E. 1996. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122: 3173–3183. [DOI] [PubMed] [Google Scholar]
- Lee, K.J. and Jessell, T.M. 1999. The specification of dorsal cell fates in the vertebrate central nervous system. Annu. Rev. Neurosci. 22: 261–294. [DOI] [PubMed] [Google Scholar]
- Manzanares, M., Cordes, S., Ariza-McNaughton, L., Sadl, V., Maruthainar, K., Barsh, G., and Krumlauf, R. 1999. Conserved and distinct roles of kreisler in regulation of the paralogous Hoxa3 and Hoxb3 genes. Development 126: 759–769. [DOI] [PubMed] [Google Scholar]
- Muller, F., Blader, P., and Strahle, U. 2002. Search for enhancers: Teleost models in comparative genomic and transgenic analysis of cis regulatory elements. BioEssays 24: 564–572. [DOI] [PubMed] [Google Scholar]
- Nobrega, M.A., Ovcharenko, I., Afzal, V., and Rubin, E.M. 2003. Scanning human gene deserts for long-range enhancers. Science 302: 413. [DOI] [PubMed] [Google Scholar]
- Ogura, K., Matsumoto, K., Kuroiwa, A., Isobe, T., Otoguro, T., Jurecic, V., Baldini, A., Matsuda, T., and Ogura, T. 2001. Cloning and chromosomal mapping of human and chicken Iroquois (Irx) genes. Cytogenet. Cell Genet. 92: 320–325. [DOI] [PubMed] [Google Scholar]
- Ovcharenko, I., Loots, G.G., Nobrega, M.A., Hardison, R.C., Miller, W., and Stubbs, L. 2005. Evolution and functional classification of vertebrate gene deserts. Genome Res. 15: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, T., Dildrop, R., Ausmeier, K., and Rüther, U. 2000. Organization of mouse Iroquois homeobox genes in two clusters suggests a conserved regulation and function in vertebrate development. Genome Res. 10: 1453–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin, A., Bailey, P., Bruce, S., Engstrom, P.G., Klos, J.M., Wasserman, W.W., Ericson, J., and Lenhard, B. 2004. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini, S., Boore, J.L., and Meyer, A. 2003. Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters. Genome Res. 13: 1111–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow, D.B., Latinkic, B., and Mohun, T.J. 2000. A simplified method of generating transgenic Xenopus. Nucleic Acids Res. 28: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz, F., Gonzalez, F., and Duboule, D. 2003. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113: 405–417. [DOI] [PubMed] [Google Scholar]
- Sumiyama, K., Irvine, S.Q., and Ruddle, F.H. 2003. The role of gene duplication in the evolution and function of the vertebrate Dlx/distal-less bigene clusters. J. Struct. Funct. Genomics 3: 151–159. [PubMed] [Google Scholar]
- Tan, J.T., Korzh, V., and Gong, Z. 1999. Expression of a zebrafish Iroquois homeobox gene, Ziro3, in the midline axial structures and central nervous system. Mech. Dev. 87: 165–168. [DOI] [PubMed] [Google Scholar]
- Taylor, J.S., Van de Peer, Y., Braasch, I., and Meyer, A. 2001. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356: 1661–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa, M., Ishida, Y., Takemoto, T., Kamachi, Y., and Kondoh, H. 2003. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4: 509–519. [DOI] [PubMed] [Google Scholar]
- Wang, X., Emelyanov, A., Speptsova-Friedrich, I., Korzh, V., and Gong, Z. 2001. Expression of two novel zebrafish Iroquois homologues (ziro1 and ziro5) during early development of axial structures and central nervous system. Mech. Dev. 105: 191–195. [DOI] [PubMed] [Google Scholar]
- Woolfe, A., Goodson, M., Goode, D.K., Snell, P., McEwen, G.K., Vavouri, T., Smith, S.F., North, P., Callaway, H., Kelly, K. et al. 2005. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]