Abstract
The L1 retrotransposon is the most highly successful autonomous retrotransposon in mammals. This prolific genome parasite may on occasion benefit its host through genome rearrangements or adjustments of host gene expression. In examining possible effects of L1 elements on host gene expression, we investigated whether a full-length L1 element inserted in the antisense orientation into an intron of a cellular gene may actually split the gene's transcript into two smaller transcripts: (1) a transcript containing the upstream exons and terminating in the major antisense polyadenylation site (MAPS) of the L1, and (2) a transcript derived from the L1 antisense promoter (ASP) that includes the downstream exons of the gene. Bioinformatic analysis and experimental follow-up provide evidence for this L1 “gene-breaking” hypothesis. We identified three human genes apparently “broken” by L1 elements, as well as 12 more candidate genes. Most of the inserted L1 elements in our 15 candidate genes predate the human/chimp divergence. If indeed split, the transcripts of these genes may in at least one case encode potentially interacting proteins, and in another case may encode novel proteins. Gene-breaking represents a new mechanism through which L1 elements remodel mammalian genomes.
Transposable elements are neither “junk DNA” nor mere curiosities to be categorized taxonomically and relegated to dusty catalogs; rather, they can affect gene expression in important ways and are a dynamic and significant part of our evolutionary history (Boissinot et al. 2000; Meischl et al. 2000; Myers et al. 2002; Brouha et al. 2003; Salem et al. 2003; Kazazian Jr. 2004). Transposons comprise nearly 45% of the human genome (International Human Genome Sequencing Consortium 2001) and include DNA transposons, LTR retrotransposons, and non-LTR retrotransposons such as LINEs and SINEs. Each class can be divided into several subgroups, each with a unique evolutionary history. Most elements are currently inactive, but the active elements shape the genome through continued expansion, transduction, chromosomal rearrangements (Gilbert et al. 2002; Symer et al. 2002), and even direct effects on gene expression (van de Lagemaat et al. 2003; Druker et al. 2004; Han and Boeke 2004). The related endogenous retroviruses have also been implicated in affecting gene expression, inserting new promoters and/or 3′ end forming sequences into many human genes (Landry 2003).
L1 elements, retrotransposons comprising nearly 17% of the human genome (Smit 1996; International Human Genome Sequencing Consortium 2001), possess not only the well-known 5′ forward promoter (Swergold 1990) long known to initiate transcription of the two open reading frames, but also a 5′ antisense promoter (ASP) that for unknown reasons drives outward transcription of adjacent flanking sequences. Speek (2001) and Nigumann et al. (2002) identified many cellular transcripts apparently produced by the ASP.
Han et al. (2004) identified another way in which L1 elements significantly affect transcription of their flanking sequences. When present in a gene in the antisense orientation, an L1 element can truncate transcripts from the gene's promoter by premature polyadenylation within the 3′end of the ORF2 region of the antisense L1. This polyadenylation can be relatively efficient, but still allows a certain amount of read-through. The major antisense polyadenylation signal (MAPS, Fig. 1) defined by those experiments is located at position 5584 in human L1rp (contained in the RP2 gene, accession no. AJ007590), on the antisense strand (Han et al. 2004). Although these experiments were performed on episomal gene fusions that lacked introns, Han et al. predicted that similar effects could potentially occur if antisense L1 elements were inserted into native chromosomal genes. We provide evidence here supporting this hypothesis.
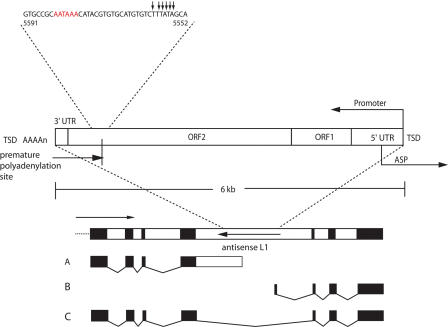
Figure 1.
Gene-breaking model. A generic gene is shown, containing an L1 element positioned in the antisense direction in an intron. Three transcripts could result from this arrangement: transcript A, an early-terminating upstream transcript containing the 5′ exons, part of the intron, and part of L1; transcript B, a transcript originating from the antisense promoter in L1 and containing part of L1 spliced to the downstream exons; and transcript C, the native (expected) transcript. Arrows show the direction of transcription; the arrow for the antisense L1 indicates transcription from the native L1 promoter. Red letters, the poly(A) signal; small arrows, polyadenylation sites; TSD, target site duplication; UTR, untranslated region; ORF, open reading frame; ASP, antisense promoter.
The “gene-breaking” model
Coupled together, the discoveries of the L1 ASP and the L1 MAPS sparked our consideration of the “L1 gene-breaking” hypothesis that is the subject of this communication. Gene-breaking refers to the situation in which an L1 element positioned in the anti-sense orientation, relative to the host gene, in any intron could in principle split what was originally a single transcription unit into two: an upstream gene (terminating at the MAPS) and a downstream gene (starting at the ASP). The full transcript may also be made, depending on the strength of the MAPS (Fig. 1).
Bioinformatic evidence for the predicted transcripts
To investigate our L1 “gene-breaking” hypothesis, we began by searching for evidence of L1 ASP-derived transcripts. We used the first 600 nucleotides of L1rp (Schwahn et al. 1998) as a query in a BLAST search (Altschul et al. 1997) against the human EST database. Over 60 ESTs met our criteria of having an antisense alignment ≥200 base pairs, >90% identity, an e-value <1e-50, and substantial amounts of both L1 and non-L1 sequence; therefore, they could be readily aligned to known or predicted genes and were unambiguously derived from L1. The genes corresponding to the candidate ESTs were aligned to the genome using Spidey (Wheelan et al. 2001) to determine whether the transcript commenced in an intronic antisense L1. Fifteen of these genes met the following stringent criteria: the ASP-derived EST aligned perfectly to the antisense 5′ L1 sequence situated in an intron, the gene's alignment with the EST spanned the entire length of the EST, with an e-value <1e-60, and the L1 was full-length (covering all 6019 bases of L1rp) and flanked by target site duplications. Five of these ESTs represent cases identified by Nigumann et al. (2002) as transcripts driven by the ASP, and 10 are newly identified.
Next, we identified upstream transcripts that terminate in the MAPS. The last 485 residues of L1 were used as a query in an unfiltered BLAST search against the human EST database. Over 4900 ESTs with BLAST alignment e-values >1e-10 were identified this way. Those for which the alignments terminated 3′ of the MAPS (between residues 5551 and 5562) were further analyzed. We identified eight transcripts (six EST sequences plus one gene record and a predicted gene record) that terminate exactly in the MAPS (Fig. 2). Six of these are clearly polyadenylated and thus unambiguously represent transcripts produced by termination at the MAPS. The other two come from libraries obtained by using an oligo dT primer and presumably also did contain polyA tails that were truncated in the database records.
Figure 2.
Alignments to the MAPS region. Alignments of six EST sequences, one predicted gene, and one mRNA sequence to the MAPS region of L1rp. In red is the L1 polyadenylation signal. The sequences terminating in uppercase “A”s have poly(A) tails in their database records. Termination sites, as described in Han et al. (2004), are shown with arrows. Five of the sequences have poly(A) tails starting at one of the known termination sites; these sites are labeled #1, #2, and #3 to correspond with the first, second, and third polyadenylation sites identified by Han et al. and marked with arrows on the L1 sequence. Sequence CN806550 terminates just 5′ of the first termination site; this may represent a previously unknown termination site.
Our search for 3′ antisense L1 sequence in the RefSeq database also revealed a cellular gene, kinetochore protein Spc25, that is normally polyadenylated at the MAPS of an L1 sequence lying distal to the last exon; its last 457 nucleotides are identical to the first 457 3′ antisense nucleotides of L1rp. The L1 element partially contained in Spc25 is 95% identical to L1rp. The existence of multiple Spc25 transcripts with this structure in the database provides additional evidence that the MAPS activity is present in endogenous L1 elements.
We performed another search of the EST database with the entire L1 sequence in order to detect any other mRNA termination sites. L1 positions 5170, 5172, and 5173 form a noticeable cluster of antisense EST termination sites (359 ESTs on the antisense strand). These positions are situated 68 bases downstream of a potential poly(A) signal, on the antisense strand. None of the ESTs contains any non-L1 sequence and therefore cannot be assigned to any particular genomic location. However, these data indicate that there may be more than one cryptic polyadenylation signal in the antisense L1 for premature termination of a cellular transcript.
Although the data presented thus far confirm that the L1 ASP and the L1 MAPS are used in human genes, they do not provide complete evidence for a true gene-breaking event; i.e., a single gene which gives rise to both an ASP transcript and a MAPS-terminated transcript. To address this, we searched for potential MAPS-terminated ESTs upstream of the 15 already identified ESTs originating from intronic L1 ASPs. To be considered a potential MAPS-terminated transcript, an EST would ideally extend from the adjacent exon upstream of the antisense L1, into intronic sequence, and terminate in the L1 MAPS. Realistically, it would be unlikely to find such an EST in the database since the sequencing reads generally are not long enough to contain all of this information.
The 15 genes identified, many of them named and characterized transcripts, were analyzed in detail (Table 1). Each gene contains a presumably young L1, nearly identical to L1rp, in the antisense orientation. For each gene we have identified one EST clearly initiating in the 5′ antisense L1 sequence and containing downstream exons. ESTs terminating upstream of the intron containing the L1 are present for all genes, but these may or may not represent MAPS-terminated sequences, as the recorded sequences do not extend into the intronic or antisense L1 sequences. All L1s in the 15 examples are full-length (just over 6 kb), have over 96% identity to L1rp, and are flanked by target site duplications, features indicating relatively recent insertion. All ESTs derived from the L1 ASP match canonical splice site sequences at the junction of the L1 sequence and the sequence from the joined downstream exon, consistent with the examples described by Nigumann et al. (2002)
Table 1.
Fifteen gene-breaking candidates studied in detail
| Accession number | Gene name | L1 EST | % id to L1rp | TSD lenb |
|---|---|---|---|---|
| NM_000245a | MET | CB988551 | 97% | 13 |
| NM_000740 | CHRM3: cholinergic receptor, muscarinic 3 | CD654050 | 98% | 6 |
| NM_002499 | Neogenin 1 | BX951091 | 96% | 8 |
| NM_004411a | Dynein | AA220950 | 96% | 6 |
| NM_004866a | SCAMP1 | BE566710 | 96% | 5 |
| NM_005816a | CD96 antigen | BE568884 | 98% | 13 |
| NM_014497 | NP220 nuclear protein | BG542212 | 98% | 7 |
| NM_014960 | Arylsulfatase G | BG994996 | 96% | 8 |
| NM_017679 | Breast carcinoma amplified sequence 3 (BCAS3) | AJ518836 | 97% | 15 |
| NM_022743 | SMYD3 | BM809976 | 97% | 8 |
| NM_024583a | Secernin 3 | AA226814 | 97% | 6 |
| NM_145259 | Activin A receptor | BE787024 | 98% | 9 |
| NM_175624 | RAB3A interacting protein (rabin3) | BE617461 | 99% | 11 |
| NM_182758 | BX957167 | 96% | 10 | |
| NM_198499 | CN272126 | 97% | 6 |
MAPS-terminated transcripts
Thus ASP-directed sequences were in hand for all the 15 candidate genes, but none had an unambiguous upstream transcript clearly demonstrating termination at the intronic antisense L1s. We designed and performed an RT-PCR screen to search for such transcripts (Fig. 3A). Total RNA extracts from four different kinds of human cells (Hela, HCT116, 293, and NCI-H69) were first subjected to cDNA synthesis by using a polyT-L1 chimeric primer capable of priming only on mRNAs polyadenylated at the MAPS. The resultant cDNA was amplified with a common intronic primer designed to hybridize to L1 sequences just upstream of the MAPS, and a gene-specific primer, which anneals to an exon upstream of the exon just 5′ to the L1-containing intron. The primers were designed to detect products containing antisense L1 sequence from the end of L1 up to the MAPS, the intronic segment upstream of the L1, and transcript sequence up to two exons upstream, so that sequencing will reveal whether the transcript has been spliced. This primer design, therefore, enables discrimination of PCR amplifications of the upstream mRNA of interest from genomic DNA that might contaminate the extracted RNA samples, as well as full-length mRNA.
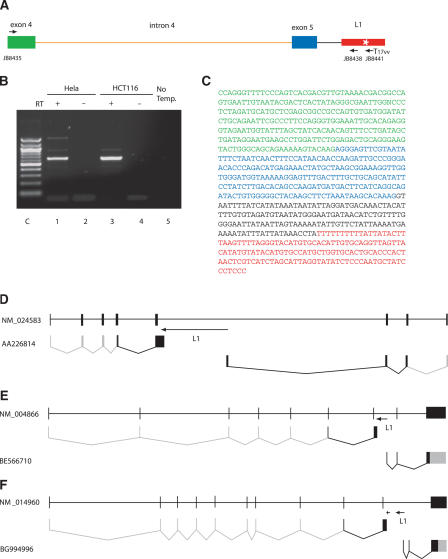
Figure 3.
RT-PCR cloning of the upstream run-on transcript from NM_024583 (secernin 3). (A) The RT-PCR strategy. The full-length L1 (red box) is located in intron 5 (black line) in the antisense orientation. After cDNA synthesis from the extracted total RNA with the L1-poly T primer (JB8441), the L1-antisense primer (JB8438), which hybridizes just upstream of the MAPS (white star), and the gene specific primer (JB8435), which anneals within exon 4 (green box), were used to amplify the upstream run-on transcript. Exon 5 is shown as a blue box. (B) Gel purification of the PCR product. From the two different human cell lines, Hela (lane 1) and HCT116 (lane 3), ∼650-base pair fragments were generated as major bands whereas no product was amplified without reverse transcriptase on cDNA synthesis (lanes 2,4) and template DNA on PCR (lane 5). The marker is a 100-base pair DNA ladder (lane C). (C) The sequence of the ∼650-base pair PCR product contains the 5′ end of intron 5 (in black) but not intron 4 (orange in A), which was spliced out. Colors as in Figure 2a. In D,E,F, gray boxes and lines indicate sequence and splicing not observed in the database or in our transcripts, as these short sequencing products do not contain the entire mRNA, but which are inferred from the sequence of the full-length mRNA. (D) NM_024583 (secernin 3). The L1 is 6 kb and is shown as an arrow; the direction of the arrow indicates the direction of transcription from the native L1 promoter. The first line shows the full-length gene structure as defined by Spidey. Underneath is the polyadenylated transcript discovered and sequenced here that terminates in the L1, and below that is a previously described transcript that starts in the 5′ end of the L1 and is potentially driven by the ASP. (E) NM_004866 (SCAMP1). The L1 is 6 kb and is shown as an arrow. As in (D), the first line shows the full-length gene structure as defined by Spidey, the second line shows the polyadenylated transcript described here, and the third line shows the database sequence BE566710, which is the ASP-derived product. (F) NM_014960 (arylsulfatase G). The full-length gene structure is shown in the first line. Here there are two arrows for the L1. The first arrow indicates a truncated antisense L1 situated upstream of the full-length antisense L1, indicated by the second arrow. The second line is the product described here that terminates at the MAPS in the truncated L1. The last line depicts the database sequence that appears to be derived from the ASP of the full-length L1 element.
Using this strategy, we successfully amplified upstream transcripts for three of the 15 L1-split candidate genes; the strategy was successful in these three cases most likely because the short distance from the MAPS to the nearest 5′ exon-intron junction facilitates an efficient RT-PCR reaction. Secernin 3 (NM_024583) has the shortest distance between the cellular exon and the L1 element. A full-length antisense L1 is located in intron 5 of this gene. In Hela and HCT116 cells, RT-PCR with the secernin 3-specific primer annealing to exon 4 provided a single major band migrating at the expected position (Fig. 3B). Sequencing clones of the PCR product showed that intron 4 had been removed by splicing but exon 4 and exon 5 sequences, as well as the 5′ segment of the intron upstream of the L1, were all present in the expected configuration (Fig. 3C). This, together with the previously found ASP-derived transcript (AA226814), demonstrates gene-breaking in the human secernin 3 gene (Fig. 3D).
RefSeq NM_004866 contains a full-length L1 element in intron 7; this element is ∼2 kb downstream of the 3′ end of exon 7. We amplified a transcript from NM_004866 that contains part of exon 6, all of exon 7, and intron 7 up to the L1 element (Fig. 3E).
RefSeq NM_014960 is split by an antisense L1 in intron 10. This gene actually contains two L1 elements in intron 10, one a 5′ truncated L1 (containing nucleotides 5393 onward) in the antisense orientation, and a full-length L1 element also in the antisense orientation downstream of the truncated L1. We identified an upstream transcript containing part of exon 9, all of exon 10 (and none of intron 9), and intron 10 up to the MAPS in the truncated L1. The ASP-derived transcript identified by database searches commences in the full-length L1 downstream (Fig. 3F). The existence of the transcript terminating at the MAPS of the truncated L1 demonstrates that even partial antisense L1 elements can truncate cellular mRNAs as predicted from transfection experiments.
The other 12 candidates contain L1 elements that are further away from the nearest upstream intron-exon boundary (7–100 kb). For technical reasons, these lengthy mRNAs are more difficult to amplify.
Confirming the ASP product
Although the EST data strongly support the existence of ASP-driven mRNA from the 15 candidate genes, we attempted to obtain a complete set of gene-breaking evidence in a single cell line by RT-PCR cloning of the ASP-driven product from Secernin 3. Our efforts resulted in a previously unpublished sequence containing the first 91 bases of the intronic L1 (in the antisense orientation), 25 bases of intron 5, and then splicing to exon 7 of secernin 3 (Fig. 4). Exon 6 is skipped, presumably spliced out. This case confirms that in HeLa cells, the secernin 3 coding region generates two transcription units split by the L1. The identification of exon skipping in the ASP-promoted transcript also raises the point that the L1-containing transcripts may be recognized differently by the splicing machinery and may be spliced in alternate ways, as the database transcript AA226814 is also an ASP-derived transcript and this sequence contains exon 6 as well as part of the intronic L1, part of intron 5, and part of exon 7.
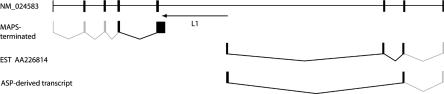
Figure 4.
The downstream product of the L1 ASP in Secernin 3 (NM_024583). The full gene product (top line) as well as the MAPS-terminated (line 2) and two ASP-derived (lines 3,4) products are shown; the ASP-derived product that we sequenced, in line 4, skips exon 6 of secernin 3, presumably due to splicing, whereas the database sequence contains exon 6 as well as the L1 sequence and part of exon 7. As in Figure 3, gray boxes and lines indicate the expected structure of the mRNAs shown; these sequences are inferred from the known sequence of the full-length mRNA.
Predicted protein products
We find strong evidence for gene-breaking in a variety of cell lines and tissues with both bioinformatics and experimental assays. The phenomenon clearly produces separate mRNA transcripts from at least three genes, and it is possible that the transcripts have functional significance apart from their RNA forms. Protein products from such split genes could in principle maintain separate structurally stable and functional domains, and it is possible that such proteins survive in the cellular environment long enough to play roles in the functions of their parental genes. The interactions that normally stabilize the folded structure of peptides could well facilitate the interaction of the two protein fragments to form a protein of normal or slightly altered function. In one of our examples, BCAS3 (NM_017679), gene-breaking has produced a subfragment of the original protein.
In another example, MET (NM_000245), gene-breaking could produce functionally interacting proteins.
The hepatocyte growth factor receptor gene, MET, contains an L1 in the antisense direction in its second intron, shown in Figure 5A. The EST, CB98851, confirms that this L1 does express the downstream exons of MET from its antisense promoter.
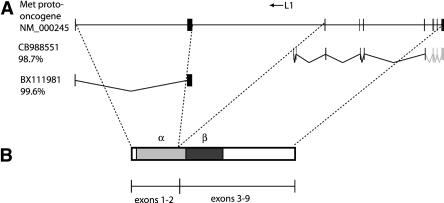
Figure 5.
(A) The MET gene, showing the location of the L1 element as well as the 5′ and L1-promoted ESTs. (B) The MET gene product, showing the cleavage point between the α and β subunits as well as the mapping of the gene's exons onto the protein product.
As seen in Figure 5B, the MET product is synthesized as a single protein which is predicted to be proteolytically cleaved into the α and β subunits and to function as a disulfide-linked heterodimer. The protease cleavage site defining the α and β subunits is located 25 codons upstream of the end of exon 2. Therefore, a 5′ MET transcript terminating in the L1 MAPS can create a protein similar to the α subunit, whereas the L1 ASP-derived transcript can create a protein similar to the β subunit. These slightly altered protein products could interact in a fashion analogous to the original MET protein.
We sought to identify more examples in which an antisense L1 element might affect the protein products of the host gene. This can happen because the mature transcripts terminated by L1 MAPS and produced by L1 ASP contain intronic sequence at the 3′ end and 5′ end, respectively. In both cases, this can cause the open reading frame to extend into intronic sequence, creating an altered protein isoform. We looked at all upstream transcripts to determine whether extension of the transcript into intronic sequence could affect the encoded amino acid sequence. This analysis revealed that all transcripts potentially encode cellular proteins truncated at their carboxy termini, containing the sequence from the upstream exons attached to adjacent intron-derived sequences; these extensions added 1–85 intron-encoded amino acids to the C-terminus of the encoded upstream proteins. The presence of part of an L1 element in either the upstream or downstream transcripts may alter splicing of the transcript to produce translation products not otherwise seen.
More novel protein structures produced by the L1 antisense promoter
In addition to the products described above, other novel protein segments could also be encoded by the downstream transcripts derived from the L1 ASP. Most of the L1 ASP-derived ESTs have ORFs encoding products ranging in size from four to 88 amino acids, most consisting entirely of L1 sequence. Alternatively, novel proteins could potentially result from the fusion of translated L1 sequences with cellular proteins. They could also be translated from ATG codons internal to the target gene, which would create a shorter gene product. For example, the transcript AJ518836, driven by the L1 ASP in the BCAS3 gene (NM_017679), is the product of splicing L1 antisense sequence to exon 3 of BCAS3. In the frame defined by the first ATG, it encodes a 113-amino acid protein derived partly from L1 and partly from BCAS3, but in the frame defined by the second ATG, it is in the same frame as the full gene transcript and encodes a C-terminal subfragment of the BCAS3 protein. Remarkably, this smaller protein is identical to the previously identified Maab3 protein (CAD57724), or “metastasis-associated antigen of breast cancer,” from an unpublished screen of overexpressed proteins in the metastatic breast cancer cell line MCF7. Both ATG sequences are embedded in reasonable Kozak sequences and could potentially be used in vivo. In this example, the L1 ASP-derived transcript encodes a protein, Maab 3, encoded by a transcript resulting from fusion of L1 sequence to that encoded by internal exons of a cellular gene.
The antisense promoter produces a variety of mRNA types
Nigumann et al. (2002) analyzed the L1 ASP in detail. Looking at the alignments of genes to this promoter region, they created six categories describing the different ways that L1 antisense sequence was included in the transcript. Some of these categories include spliced transcripts, in which a piece of the L1 antisense sequence is removed from the final transcript, joining two nonadjacent L1 sequences. We did a similar analysis of our 15 examples (Supplemental Fig. 1), and found that nearly all of our alignments involve splicing from the ASP to cellular exons, leaving out part of the intervening antisense L1 sequence as well as adjacent intron, and occasionally exon, sequences. All intronexon junctions observed occur at canonical splice sites.
To address whether L1 ASP or MAPS activities are tissue-specific, we noted the tissue distribution of the cells generating ESTs described here. The ESTs are derived from a wide range of cell types, spanning nearly every tissue as well as both cancerous and noncancerous cells.
Comparison of human and chimpanzee genes
In order to gauge the evolutionary timing of gene-breaking, we examined the chimpanzee orthologs of all 15 genes studied here. The chimpanzee orthologs were identified using the reciprocal best hit method: the human mRNA was aligned to the chimpanzee draft assembly (13 Nov. 2003, NCBI Build 1, Version 1, produced by the Chimpanzee Genome Sequencing Consortium), the putative chimpanzee gene was extracted using the alignment, and finally, the chimpanzee sequence was aligned to all human mRNA sequences. If the best hit in the final search was the same as the starting mRNA, the sequences were labeled orthologous. Twelve of the orthologous chimp genes contained L1 elements in the intron corresponding to the human L1 position, but three of the chimp orthologs contained no L1 sequences. Therefore, in most cases, the L1 elements involved in gene-breaking predate the chimp–human divergence.
Conclusions
From this analysis it is apparent that L1-mediated gene-breaking is a biologic reality. The cell type distributions of both the MAPS and ASP ESTs, as well as our results from two separate cell lines, indicate that expression from the L1 ASP and polyadenylation at the MAPS are global phenomena. Using bioinformatics and direct experimental approaches, we show that gene-breaking plays a part in the expression of at least three novel human genes. Over 150 full-length, nearly exact (>98%) matches to L1rp were found in the antisense orientation in introns of cellular genes, and thousands more very slightly degenerate L1s were found in these positions as well. Truncated L1 elements are hundreds of times more numerous, presenting many more MAPS that may affect gene expression. Thus, MAPS termination and gene-breaking potentially have a significant effect on gene expression. Whether these truncated transcripts are merely tolerated or are functionally important contributors remains to be seen; in at least one case a gene-breaking transcript may have its own cellular function. Many factors are expected to influence the relative abundance of the 5′ and 3′ transcripts of “broken” genes, including (1) the strength of the native promoter in the cell or tissue of interest, (2) RNA stabilities affected by 3′ and 5′ UTR sequences, (3) activity of the L1 ASP in the cell or tissue in question, and (4) possible effects of RNAi on L1-containing “broken transcripts.” Further studies will concentrate on finding ancient examples of gene-breaking in which what was once a single gene is now broken into two interacting genes separated by an L1, such that the original full-length transcript no longer exists at all, and on quantifying the impact of gene-breaking on cellular transcription.
Methods
Cell culture
Hela and 293 cells were gifts from J. Moran (The University of Michigan Medical School) and S. Blackshaw (The Johns Hopkins University School of Medicine), respectively, and were maintained in DMEM supplemented with 10% FBS and 0.5 mg/mL Normocin (InvivoGen). HCT116 and NCI-H69 cells were purchased from ATCC and maintained in McCoy's 5A and RPMI (Invitrogen) supplemented with 10% FBS and 0.5 mg/mL Normocin (InvivoGen).
RT-PCR cloning
Total RNA was extracted from Hela, 293, HCT116, and NCI-H69 cells with an RNeasy Mini Kit (Qiagen). To prevent genomic DNA contamination, an RNase-Free DNase Set (Qiagen) was utilized during the RNA extraction. The rest of the extraction process was carried out according to the manufacturer's instructions. In searching for transcripts terminating in L1 MAPS, the cDNA synthesis was performed with 2–5 μg/mL total RNA, 0.12 μM L1-polyT chimera primer (JB8441: 5′-TTTTTTTTTTTTTTTT TAAGACA), and SUPERSCRIPT II RNaseH-Reverse Transcriptase (Invitrogen). In searching for transcripts starting in the L1 ASP, instead of JB8441, 3′RACE RT primer (5′-GACTCGAGTCGA CATCGATTTTTTTTTTTTTTTTT) was used. One μL cDNA synthesis solution was used for PCR amplification with ExTaq polymerase HS (Takara), 1 × Buffer 1 or 3 (Roche), 0.4 mM dNTPs, 0.2 μM primers (JB8435: 5′-CCAGGGTGGAAATTGCACAG for Secernin 3 exon 3 and JB8437:5′-TCCCCAAAACAGATGTTATCATTCC or JB8438: 5′-GGGAGGGATAGCATTGGGAGA for Secernin 3 intron 5; JB9131: 5′-ATGGAAATGCAGAAATCACCCCTCTTC for Secernin 3 intron 5; JB9134: 5′-ATCCCACGATACATGAATCCAA TTCCA for Secernin 3 exon 8; JB8568: 5′-TGGGTCAAAAATGGG GATATGG for NM_004866 exon 6, JB8447: 5′-GGGGGTTAGGG GGAGAGAAA for NM_004866 intron 7; JB8501: 5′-GTGGAC GTCTCCGAGGTGCT for NM_014960 exon 9, JB8557: 5′-TGCC CACTGTCACCACTCCT for NM_014960 intron 10), and the following thermal conditions: 95°C, 2 min; 50 cycles of 95°C, 30 sec; 60°C, 30 sec; 72°C, 1.5–8 min; and 72°C, 10 min. The PCR products (major bands) were purified on agarose gels, cloned by using a TOPO TA Cloning Kit (Invitrogen), and sequenced at the DNA sequencing facility of The Johns Hopkins University High-Throughput Biology Center.
Acknowledgments
The authors wish to thank Dave Valle and Jeremy Nathans for helpful comments on the manuscript and Brian Greenlee for help with the figures. This work was supported in part by NIH grant CA16519 to J.D.B. and training grant 5 T32 CA09139 to S.J.W.
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3688905. Article published online before print in July 2005.
References
- Altschul, S., Madden, T., Schaffer, A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot, S., Chevret, P., and Furano, A. 2000. LINE-1 retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17: 915–928. [DOI] [PubMed] [Google Scholar]
- Brouha, B., Schustak, J., Badge, R., Lutz-Prigge, S., Farley, A., Moran, J., and Kazazian Jr., H. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. 100: 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker, R., Bruxner, T., Lehrbach, N., and Whitelaw, E. 2004. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 32: 5800–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., Lutz-Prigge, S., and Moran, J. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110: 315–325. [DOI] [PubMed] [Google Scholar]
- Han, J. and Boeke, J. 2004. A highly active synthetic mammalian retrotransposon. Nature 429: 314–318. [DOI] [PubMed] [Google Scholar]
- Han, J., Szak, S., and Boeke, J. 2004. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429: 268–274. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Kazazian Jr., H. 2004. Mobile elements: Drivers of genome evolution. Science 303: 1626–1632. [DOI] [PubMed] [Google Scholar]
- Landry, J., Mager, D., and Wilhelm, B. 2003. Complex controls: The role of alternative promoters in mammalian genomes. Trends Genet. 19: 640–648. [DOI] [PubMed] [Google Scholar]
- Meischl, C., Boer, M., Ahlin, A., and Roos, D. 2000. A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur. J. Hum. Genet. 8: 697–703. [DOI] [PubMed] [Google Scholar]
- Myers, J., Vincent, B., Udall, H., Watkins, W., Morrish, T., Kilroy, G., Swergold, G., Henke, J., Henke, L., Moran, J., et al. 2002. A comprehensive analysis of recently integrated human Ta L1 elements. Am J. Hum. Genet. 71: 312–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigumann, P., Redik, K., Mätlik, K., and Speek, M. 2002. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79: 628–634. [DOI] [PubMed] [Google Scholar]
- Salem, A., Myers, J., Otieno, A., Watkins, W., Jorde, L., and Batzer, M. 2003. LINE-1 preTa elements in the human genome. J. Mol. Biol. 326: 1127–1146. [DOI] [PubMed] [Google Scholar]
- Schwahn, U., Lenzner, S., Dong, J., Feil, S., Hinzmann, B., van Duijnhoven, G., Kirschner, R., Hemberger, M., Bergen, A., Rosenberg, T., et al. 1998. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat. Genet. 19: 327–332. [DOI] [PubMed] [Google Scholar]
- Smit, A. 1996. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6: 743–748. [DOI] [PubMed] [Google Scholar]
- Speek, M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell Biol. 6: 1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swergold, G. 1990. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 10: 6718–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symer, D., Connelly, C., Szak, S., Caputo, E., Cost, G., Parmigiani, G., and Boeke J. 2002. Human L1 retrotransposition is associated with genetic instability in vivo. Cell 110: 327–338. [DOI] [PubMed] [Google Scholar]
- van de Lagemaat, L., Landry, J., Mager, D., and Medstrand, P. 2003. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 19: 530–536. [DOI] [PubMed] [Google Scholar]
- Wheelan, S., Church, D., and Ostell, J. 2001. Spidey: A tool for mRNA-to-genomic alignments. Genome Res. 11: 1952–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]