Abstract
The standard model of eukaryotic ribosomal RNA (rRNA) genes involves tandem arrays with hundreds of units in clusters, the nucleolus organizer regions (NORs). A first genomic overview for human cells is reported here for these regions, which have never been sequenced in their totality, by using molecular combing. The rRNA-coding regions are examined by fluorescence on single molecules of DNA with two specific probes that cover their entire length. The standard organization assumed for rDNA units is a transcribed region followed by a nontranscribed spacer. While we confirmed this arrangement in many cases, unorthodox patterns were also observed in normal individuals, with one-third of the rDNA units rearranged to form apparently palindromic structures (noncanonical units) independent of the age of the donors. In cells from individuals with a deficiency in the WRN RecQ helicase (Werner syndrome), the proportion of palindromes increased to one-half. These findings, supported by Southern blot analyses, show that rRNA genes are a mosaic of canonical and (presumably nonfunctional) palindromic units that may be altered by factors associated with genomic instability and pathology.
The basic organization of NORs (nucleolus organizer regions) has been described in terms of 43-kb repeats that contain the transcribed regions of the 18S, 5.8S, and 28S rRNA genes, plus an intergenic spacer (IGS) (Sylvester et al. 1986; Gonzalez and Sylvester 1995) as represented schematically in Figure 1A. The NORs are located on the short arms of the five acrocentric human chromosomes in a telomere-to-centromere orientation (Henderson et al. 1972; Worton et al. 1988). Such redundant structures imply highly efficient gene conversion (Elder Jr. and Turner 1995), which is not readily reconciled with the similar amounts of active and silent genes observed within close proximity (Conconi et al. 1989; Dammann et al. 1995; French et al. 2003). The precise structure of such highly repeated regions cannot be analyzed by sequencing, due to difficulties in cloning (Labella and Schlessinger 1989; Hattori et al. 2000). We have therefore investigated the NORs using single DNA molecule analysis by molecular combing (Bensimon et al. 1994; Michalet et al. 1997), which is particularly well suited for studying genomic regions rich in repeated sequences (Pasero et al. 2002). A substantial fraction (one-third) of the rRNA-coding sequences exist in an unorthodox arrangement appearing as palindromic structures, interpreted as twinned, partial pseudogenes, in normal adult and fetal cells. Experiments were also performed to evaluate the extent of palindromic pseudogenes in Werner Syndrome (WS) because of the well-characterized nature of the lesion involving a RecQ DNA helicase (Yu et al. 1996). In addition, the WRN protein is localized in the nucleolus (Gray et al. 1998; Marciniak et al. 1998), the compartment of the nucleus where the rDNA is stored, and it is homologous to the sgs1 gene in yeast (Johnson et al. 1999). The sgs1 gene was shown to be involved in rearrangements of the rDNA locus, resulting in the accumulation of extrachromosomal circles that are associated with the aging of the cells (Sinclair and Guarente 1997). When DNA samples were also examined from cells of WS patients, the level of palindromic pseudogenes increased to 50%, indicating the fraction of pseudogenes reflects a dynamic process that can be altered under pathophysiological conditions. The presence of unorthodox arrangements of rRNA-coding sequences in presumably nonfunctional pseudogenes is also consistent with findings from Southern blots presented here.
Figure 1.
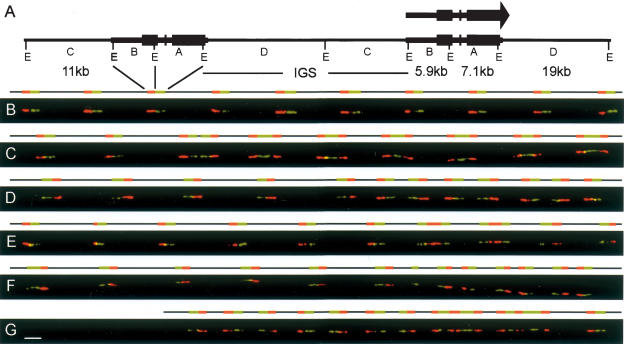
Structural analysis of the human rDNA locus by molecular combing. (A) Schematic representation of two canonical rDNA units. Restriction with EcoRI (sites E) yields four distinct fragments spanning the transcribed region (thick line) and the intergenic spacer or IGS (thin line). The orientation of the rRNA transcript and the positions of the ribosomal genes (black boxes) are shown. (B) Two-color hybridization on combed human DNA. The red probe is the 5′ EcoRI fragment B detected with Texas Red. The green probe is the 3′ fragment A detected with FITC. The image displays 10 canonical rDNA units in tandem, each composed of a dual fluorescent signal and the adjacent nonhybridizing spacer segments. (C) Hybridizations of the probes on human DNA that illustrates noncanonical units. The image displays a region containing two canonical units (left) followed by seven palindromic units, with each half joined by its 3′ region (3′–3′ palindromes) and separated by short IGS segments. (D) Hybridizations illustrating successive inverted units with gaps. The image displays five canonical units, followed by a series of seven palindromic units separated by short gaps. (E) Hybridizations illustrating 5′–5′ palindromic units. The image displays six canonical units (left), followed by two gapless palindromic units joined at their 5′ extremities and two canonical units (separated by a short IGS) in an inverted orientation with respect to the canonical units on the left. (F) Hybridizations illustrating 5′–5′ palindromic units with gaps. The image displays six canonical units, followed by a single 3′ fragment and three 5′–5′ palindromes with short central gaps separated by short IGS segments. (G) Hybridization illustrating complex recombinant structures. The image displays closely spaced, inverted units (left), followed by two complex units with alternating 5′ and 3′ coding sequences, separated by short IGS sequences, and one canonical unit (right). Scale bar, 10 kb, applicable to all images.
Results
Visualization of extended arrays of rRNA genes
Two-color hybridization of rRNA genes was performed using two adjacent fluorescent probes, a red 5′ probe (B, 5.9 kb) and a green 3′ probe (A, 7.1 kb), spanning the transcribed region of the rRNA-coding units (Fig. 1A). For a large fraction of the signals analyzed, the basic canonical structure of the rRNA gene repeats with an intervening IGS (nontranscribed spacer) was confirmed (see Fig. 1B), but with greater variability in the length than previously reported (Naylor et al. 1983; Erickson and Schmickel 1985; Schofer et al. 1998); data on several hundred units indicated an average IGS length of 34.2 ± 5.4 kb, with individual IGS units ranging from 9 to 72 kb.
A more striking divergence from the canonical units was observed for about one-third of the signals, which were detected in clusters of genes with novel structures, notably symmetric assemblies of 5′ and 3′ coding sequences that form palindromic structures, probably partial pseudogenes. The appearance of fluorescent signals for typical palindromes is shown in Figure 1C, adjacent to canonical units. In these palindromic examples, designed 3′–3′ palindromes (to indicate that the peripheral 5′ coding sequences are joined by central 3′ sequences) the length of the 5′ peripheral sequences are roughly standard length, but the 3′ central units vary along the length of the cluster presented. Examples of other palindromic patterns are presented in Figure 1, D and E. Figure 1E presents examples of 5′–5′ palindromes (with 3′ peripheral regions joined via central 5′ regions), which are flanked by canonical units, but with the sequence inverted between the units on the left and on the right, indicating that all canonical units are not necessarily in the same (telomere-to-centromere) orientation previously reported (Worton et al. 1988). An intermediate situation is presented in Figure 1D, with canonical units on the left, followed by alternating orientations on the right that appear as successive palindromes with gaps, although with the 3′ units in closer proximity. A more extreme example of this pattern is presented in Figure 1F, with palindromic units of the 5′–5′ type with a small gap. In a limited number of cases, more complex arrangements are also observed, with multiple contiguous 5′ and 3′ regions, as illustrated in Figure 1G. In rare instances, complex patterns arising from partial overlapping of two individual DNA molecules were observed, but these anomalies were readily identified and excluded from further analysis.
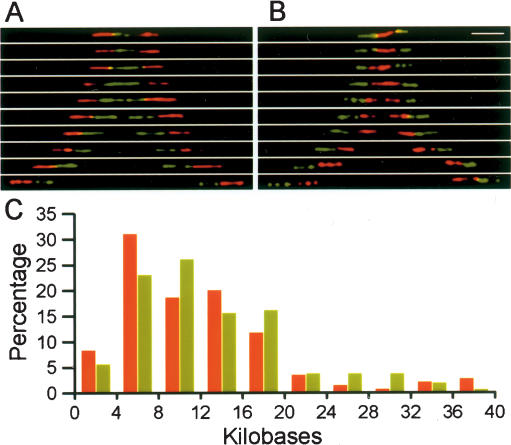
The overall analysis of the large sample of data obtained for the palindromic structures indicates a relatively continuous variation in the size of the central hybridizing region, ranging from compact 3′–3′ or 5′–5′ palindromes (<4 kb between the outer probes) to large gaps between the adjacent inverted units (>36 kb between the inner probes). A sampling of the distribution is presented in “Eiffel Tower” form for 3′–3′ palindromes in Figure 2A and for 5′–5′ palindromes in Figure 2B, with a summary of all data presented in bar graph form in Figure 2C. The distributions for the 5′–5′ and 3′–3′ palindromes are not significantly different, with both showing a large concentration in the range of from 4 to 20 kb for the spacing between the outer probes. Beginning in the middle of this range, gaps appear between the central probes and increase in size as the spacing between the outer probes increases. Overall, analysis of thousands of ribosomal RNA gene hybridizing sequences revealed nearly one-third in palindromic arrangements, in control cells from both adult and fetal samples (Figure 3).
Figure 2.
Variability in rDNA spacer length. (A) A series of hybridization signals depicting the variability in the length for 3′–3′ palindromes. (B) Signals for 5′–5′ palindromes. (C) Bar graphs of the distribution of palindromic signals with respect to the total length between the outer probes, for 5′–5′ palindromes in red and 3′–3′ palindromes in green. The data represent a compilation of measurement on 306 individual palindromes.
Figure 3.
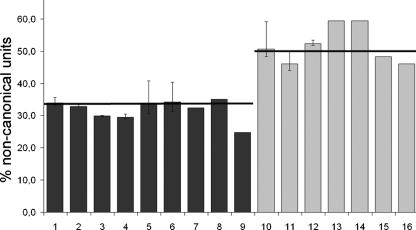
Noncanonical units in human cell lines. A bar chart showing the total percentage of noncanonical rDNA units in control cell lines in dark gray (with the total number of counted rDNA units for each cell line in parenthesis): (1) AG12657 (1948); (2) AG11561 (1554); (3) AG05283 (1640); (4) AG13077 (1549); (5) AG07898 (998); (6) AG07897 (2522); (7) D1; (8) MRCV (3649); (9) IMR90 (6357); and in Werner syndrome cell lines in light gray: (10) AG07896 (1269); (11) AG03141 (2421); (12) AG03829 (2206); (13) AG12797 (1275); (14) AG06300 (1275); (15) AG11395 (1381); (16) WV1 (1003). Horizontal bars are set at the average percentage for control and WS cells. Error bars are shown for the mean of three experiments; the other values are each the mean of two experiments.
Rearrangements of rRNA genes in Werner Syndrome cells
We investigated changes in the structural organization of the rDNA locus in cell lines from seven WS patients, as summarized in Figure 3. These cell lines were chosen on the basis of a normal karyotype in order to avoid additional chromosomal changes or imbalances that could further modify rRNA gene stability. We analyzed the rDNA locus in three primary fibroblast cultures from patients sampled at ages 30 (AG03141), 36 (AG12797A), and 37 yr (AG06300A), in two SV-40 transformed fibroblast cell lines (AG11395 and WV1), and in two lymphoblastoid cell lines derived from a 42-yr-old patient (AG03829) and a 57-yr-old patient (AG07896). The latter cell line was chosen for comparison with cell lines derived from two unaffected family members, the 90-yr-old father and a 55-yr-old sibling (lymphoblastoid controls, AG07897 and AG07898), both of whom were indistinguishable from other control cells (Fig. 3). The ribosomal DNA of the WS cell lines and cultures displayed a significant increase in the level of noncanonical arrangements; the mean incidence of such units is very close to one-half (Fig. 3). In particular, the cell line derived from the 57-yr-old patient (AG07896) displays a higher percentage of palindromic units than cell lines from the father (AG07897) and an age-matched sibling (AG07898). Thus, the elevated proportion of noncanonical units appears to be caused by the Werner syndrome affecting these patients and not by distinct biological origins or age.
Southern blotting analysis
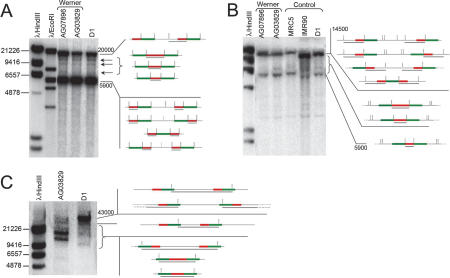
Southern blotting was performed on DNA from a control adult cell line (D1), two normal primary fetal cells (IMR90 and MRC5), and two Werner cell lines (AG03829 and AG07896). D1 is a cell line established with EBV, for which the percentage of rDNA rearranged units has been quantified at 33% (Fig. 3, lane 7), in the range of other control cells. Following digestion with EcoRI, HindIII, and I-PpoI, samples were hybridized with the 32P-labeled 5.9-Kb probe (fragment B) corresponding to the 5′ portion of the transcribed region (see Fig. 1). EcoRI cleaves three times in the canonical units and the hybridization revealed the two previously described major bands, 5.9 and 21 kb, together with three supplemental discrete bands, around 6.5 kb and between 9.5 and 20 kb. (Fig. 4A, arrows). Upon digestion with HindIII, Southern blotting showed the expected band at ∼15 kb (Romao-Correa et al. 2004). The bands previously described, ranging from 13.3 to 16.7, are not visible, because they require overexposure of the membrane and a higher separation (Krystal and Arnheim 1978). Conversely, additional bands of lower molecular weight are apparent (Fig. 4B). In view of the rearrangements observed on individual DNA molecules, we suggest that the additional minor bands obtained with EcoRI and HindIII restriction are a consequence of the palindromic structures. A schematic representation of the structures that could give rise to the supplemental bands is presented alongside the Southern blot (Fig. 4). In the EcoRI Southern blotting, the lack of EcoRI restriction sites (Wellauer and Dawid 1979; Erickson et al. 1981), which results in the major band at 21 kb, can be due either to a true polymorphism of the sequence or to loss of the site because of the rearrangements.
Figure 4.
Southern blotting of human DNA hybridized with the 5.9-kb probe (fragment B, Fig. 1). DNA was extracted from a control (D1) and two Werner (AG03829 and AG07896) cell lines, in addition to two primary fetal cells (IMR90 and MRC5). (A) EcoRI digestion. The two major expected bands are apparent, at 5.9 and 20 kb, after a complete and partial enzymatic digestion due to a polymorphism of an EcoRI site (Wellauer and Dawid 1979). The arrows indicate the supplemental bands. On the right of the gels, a schematic representation presents the genomic structures that can account for the different bands after EcoRI digestion. Red lines represent the 5.9-kb probe, whereas green bars indicate the 7.1-kb probe (Fig. 1), black bars correspond to the DNA fiber. Gray bars show the regions recognized by the radioactive probe. Vertical black dotted lines show the restriction sites. The vertical gray dotted lines signify a position of digestion that can be lost after the rearrangement. (B) HindIII digestion. The band at ∼14.5 kb is expected from the digestion of canonical units (at the top of the scheme). Additional bands are visible, between 5.9 and 14.5 kb. The scheme on the right shows the noncanonical units that could generate those bands upon HindIII digestion. (C) I-PpoI digestion, run on a PFEG. The control and Werner sample (D1 and AG03829, respectively) show the expected band at 43 kb and several additional bands. One band of the Werner sample shows a different size, suggesting that a type of rearrangement is more represented in this cell type. The scheme explains the canonical and noncanonical units that could generate the supplementary bands. Numbers on the right of the gels indicate the approximate size of the bands, expressed in base pairs.
Finally, the presence of abnormal structures was supported by Southern blotting after digestion with I-PpoI. This enzyme cleaves only once in the rDNA canonical unit, generating a unique product of ∼43 Kb that is visible in both normal and Werner samples. As for the EcoRI and HindIII digests, additional bands of lower molecular weight are apparent (Fig. 4C). The proportion of those bands is different between the control and the Werner DNA, but the data are not sufficiently quantitative to permit correlations of the intensities of the minor bands with the increased frequency of palindromic sequences observed by combing (Fig. 3). The sequence of rDNA genes, which have many simple repeats, high G+C content, and pyrimidine-rich regions, can influence denaturation and hybridization conditions, rending difficult a comparison based on feasible quantitative analysis. It is interesting to note that most of the bands have the same molecular weight in the two samples, except for the smallest one. According to the scheme in Figure 4C, this band corresponds to noncanonical units with the 5′ (red) segment in the middle. The different size of this band in the control and Werner cells suggests that palindromes with a longer red segment are more abundant in Werner's.
Discrete bands were detected by Southern blotting (Fig. 4), rather than a continuous smear, suggesting that breaks and recombination occur at specific positions. This series of fragments evokes the length variability measured on the palindromic units observed on individual DNA molecules (Fig. 1). A subgroup of palindromic units that could give rise to the supplemental bands carries a 5′ (red) signal between two 3′ (green) signals (see Fig. 1). As the promoter is located just before the red segment, the 5′ end, units with the continuous red segment at the center of the palindrome would lack the promoter and cannot be transcribed. In support of these considerations, we did not observe new additional transcripts by Northern blotting from the adult cell lines (data not shown).
Discussion
The results presented here provide a first overview of extended regions of rRNA gene clusters, revealing a totally unanticipated degree of complexity. A substantial fraction of the coding sequences are found in palindromes, whose percentage is not related to age in control cells. These noncanonical units are probably not transcribed, particularly for the examples with the 5′ at the center of the palindrome; the unit has presumably lost the promoter, and the palindrome is not transcribed. The aberrant structures of the palindromic units also make it unlikely that they were formed by retrotransposition, as in the case of certain well-characterized processed pseudogenes (Mighell et al. 2000). Although some retrotransposition events may have been involved at some stage, as in the case of 5S rRNA pseudogenes (Frederiksen et al. 1997), the fact that the various palindromic forms are found in clusters with similar structures clearly implies amplification steps at the DNA level.
Concerning the origin of the palindromes and the maintenance of the functional rRNA-coding sequences, gene conversion (Elder Jr. and Turner 1995) has been invoked to counteract disrupting recombination events (Ji et al. 2000; Schiestl and Bishop 2000), for example in primates via Alu elements (Batzer and Deininger 2002), but genetic rearrangements in the NORs are shown here to occur on a scale not previously envisaged. It had been anticipated that units of repeating rRNA genes would be found exclusively in a telomere-to-centromere orientation (Henderson et al. 1972; Worton et al. 1988), but our results indicate that there are exceptions to this rule, as described for the sequences presented in Figure 1E. A precise estimate of the number of genes in each orientation will be difficult to obtain, however, since molecular combing does not provide an absolute orientation, so that the opposite orientation of sets of rDNA genes can only be ascertained if they are on the same continuous DNA fragment visualized by combing. With respect to the observations of similar levels of active and inactive genes within close proximity (Conconi et al. 1989; Dammann et al. 1995; French et al. 2003), methylases and deacetylases in conjunction with the nucleolar remodeling complexes have been implicated (Bird 2002; Moss and Stefanovsky 2002; Santoro and Grummt 2002; Grummt and Pikaard 2003), as well as more complex phenomena such as nucleolar dominance (Pikaard 2000), but the genetic rearrangements presented here must also be taken into account.
Variations in structure of the rRNA genes from cells of WS patients shed additional light on the origin of the varied forms of palindromes. The WRN gene responsible for this disorder (Yu et al. 1996) encodes a RecQ DNA helicase that is unique in possessing an additional 3′–5′ exonuclease activity (Shen and Loeb 2001). WRN helicase is implicated in DNA replication and DNA repair pathways, possibly by unwinding DNA aberrant structures that occur during DNA replication or DNA damage-induced recombination (Shen and Loeb 2001). Phenomena interpreted as premature aging, changes in nucleolar structure, and hyperrecombination of the rRNA gene locus resulting in extrachromosomal circles were described in yeast cells mutated in the sgs1 gene that encodes a nucleolar DNA helicase of the RecQ family (Gangloff et al. 1994; Watt et al. 1996; Sinclair and Guarente 1997; Heo et al. 1999). The sgs1 gene is the human homolog of the WRN gene, although there is no evidence that aging in humans involves the accumulation of rDNA circles, as observed in yeast (Heo et al. 1999). The higher level of palindromic structures observed in WS samples could also be related to an increase in the methylation state of the rRNA genes of fibroblast cultures from WS patients, which may be related to inactivation of transcription units following rearrangements (Machwe et al. 2000). These authors also described additional, slow-migrating linear ribosomal DNA species resulting from EcoRI digestion, which were more easily detected in the most senescent passages of fibroblasts from older individuals and WS patients. Such results are consistent with the various rDNA structures that we detect, especially units putatively lacking an EcoRI site, and thus, yielding a longer fragment upon cleavage with the restriction enzyme (see below).
The human rDNA region has been extensively analyzed using restriction assays and Southern blotting for the purpose of studying its sequence organization and evolution compared with other rDNA species. The length variations found among rDNA copies have been documented to include the presence or absence of restriction sites, as well as differences in the number of internal short and long-repeat motifs scattered through the gene (Arnheim and Southern 1977; Krystal and Arnheim 1978; Schmickel et al. 1980). In human cells, two classes of rRNA genes within the same individual were observed, one of which was missing the EcoRI restriction site at the 5′ extremity of the 18S region. This polymorphism was revealed by Southern blotting with the 18S probe, resulting in a supplemental band of about 20 kb. Later studies focused on the variability of intra- and interspecies (Nelkin et al. 1980), indicating that individuals of a species present very similar EcoRI restriction patterns, which are simple and consist of a small number of bands containing >90% of the total amount of rDNA. Minor bands were also observed, and they were attributed to the length variability of the intergenic spacer. When the human rDNA unit was cloned and analyzed (Erickson and Schmickel 1985), it was confirmed that the transcribed region is highly conserved, in agreement with previous studies, except for a HincII site; 50% of genomic DNA lacked one HincII site, in contrast to the cloned samples, which all contained it. This discrepancy was explained by DNA modifications occurring in the genomic DNA or by other nondefined eukaryotic DNA modifications. The Southern analysis presented in Figure 4 also shows some unorthodox patterns for a substantial fraction of the rDNA. Data from single combed molecules and the observation of palindromic rearrangements described in this report now provide a new explanation for the Southern results.
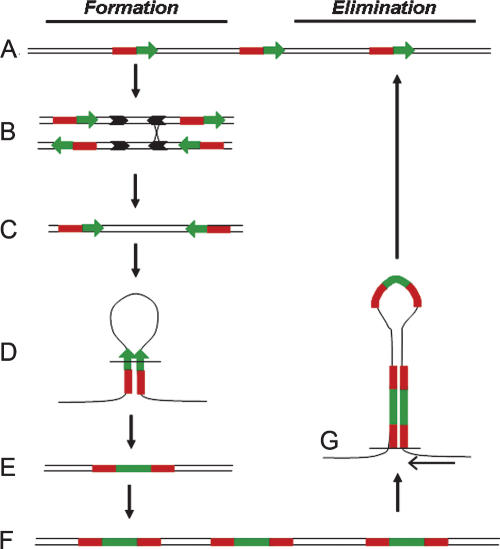
The findings presented here are in agreement with the view that rDNA genes are heterogeneous and dynamic (Gonzalez and Sylvester 1995, 2001; Kuo et al. 1996; Grozdanov et al. 2003; Romao-Correa et al. 2004). Gonzalez and Sylvester analyzed the type of exchange that occurs in the rDNA region, and they reported the presence of very divergent IGS sequence classes (Gonzalez and Sylvester 1995). According to our results, a high level of genetic instability is likely to prevail, involving the genes and the IGS, with constant readjustments between homogenization by gene conversion, production, and elimination of palindromes. Both inter- and intrachromosomal events are likely to be involved (Elder Jr. and Turner 1995). As a result, small changes in factors involved in DNA replication and repair may lead to variations in the level of palindromic sequences, as observed for several lines of cancer cells (Caburet 2002). Under any given set of conditions, the competing processes of homogenization and elimination would establish the steady-state level of palindromic sequences in what may be called a “balanced instability” as represented in Figure 5. The competing pathways of palindrome formation and expansion (Fig. 5, left) versus elimination (Fig. 5, right) are depicted schematically for a canonical sequence (Fig. 5A). In this case, inversions are generated by hypothetical recombination with crossover at Alu sequences (Fig. 5B), but other triggering could involve other recombinogenic sequences. In the example presented, single-stranded loop formation with extension of the opposite strand (Fig. 5D) generates a 3′–3′ palindrome (Fig. 5E). The same principles could be used to generate 5′–5′ palindromes from the other product of the cross-over in Figure 2B. Once created, gene duplication of the palindrome generates multiple copies (Fig. 5F). In the elimination pathway, formation of a loop between palindromic sequences can lead to their elimination (Fig. 5G). Similar mechanisms could lead to the production of either shorter or longer palindromes, which may then be expanded to create homogeneous clusters, thereby suggesting a mechanism for the formation of the full range of palindromic sizes observed. In WS cells, recombination may be increased at blocked forks due to the absence WRN helicase, leading to increased breakage. Considerable future work will be required to investigate more fully the individual steps for this series of genetic rearrangements and the changes in probability of palindrome formation as a function of the specific conditions in the cellular environment. In addition, instabilities in the extended clusters of rRNA genes observed in eukaryotic cells may provide a highly sensitive detection system for modification of cellular functions involved in DNA processing, such as aging or cancer, or for specific genetic lesions, as in the case of WS or the related Bloom syndrome (Schawalder et al. 2003).
Figure 5.
Competing processes underlying palindrome formation, variation, expansion, and elimination. The pathway of formation on the left and the pathway of elimination on the right. (A) Schematic representation of canonical rDNA genes (5′ in red, 3′ in green) separated by IGS sequences. (B) One of many possible mechanisms, inversions are presented based on recombination between Alu repeats (represented by black arrows) of opposite orientation present in the IGS sequences. (C) One of the two products of recombination with crossover in B producing inverted rDNA genes. (D) Single-strand loop formation (possibly with cleavage at the base). (E) A 3′–3′ palindrome generated by the preceding reactions. (F) Expansion of the palindromic sequence. (G) Competing reaction of elimination of the palindromic sequence, analogous to those presented in D or DNA replication slippage, shown by the arrow coming from the right.
Methods
Cells
The two control lymphoblastoid cell lines AG07897 and AG07898, the four control primary fibroblast cell cultures (AG12657, AG11561, AG05283, and AG13077), and the four WS cell cultures (AG03141, AG07896, AG03829, and AG11395) were obtained from the Aging Cell Culture Repository (NIA, Coriell Institute for Medical Studies). The primary fetal cells IMR90 and MRC5 were obtained from the ATCC repository. These cell lines were cultivated according to the recommendations of the Repository. D1 was kindly provided by Dr. Mounira Amor-Guéret, UMR 8126, Villejuif (Paris). WV1 is a gift of Yannick de Santigny, CEA, Saclay.
Hybridization and fluorescent detection
High-molecular weight genomic DNA was prepared from cells embedded in PGFE plugs essentially as described (Michalet et al. 1997). Probes A and B (see Fig. 1), spanning the transcribed region, were kindly provided by G. Brock. Probe A was labeled with digoxigenin and probe B with biotin by overnight random priming. Hybridization was performed overnight on combed genomic DNA denatured for 20 min in 1 M NaOH (see Supplemental material). Fluorescent detection and signal amplification were performed using five successive layers of fluorochrome-conjugated antibodies. Images were acquired on an epifluorescence Zeiss microscope with a CCD camera, using Smart Capture 2 software (Digital Scientific). Measurements were performed using a dedicated lab-made software, CartographiX.
Southern blotting
EcoRI digestion
A total of 100 μL was diluted with 100 μL of water and digested with 5 μL of EcoRI (10 U/μL) for 3 h (or overnight) at 37°C.
HindIII digestion
A total of 11 μg of DNA was digested with 5 μL of enzyme (10 U/μL) ON at 37°C. Five micrograms of EcoRI-digested DNA and 10 μg of HindIII-digested DNA were loaded on a standard 0.8% agarose gel and run 20 h at 50V in 1× TBE.
I-PpoI digestion
A total of 11 μg of DNA was digested with 5 μL of enzyme (200 U/μL) ON at 37°C. Ten micrograms were run by PFGE (0.9%) on a CHEF apparatus (BioRad). The program was as follows: two states, angle 120°, 6 V/cm, pulses 1–6 sec, rampin 0, 11 h. Gels were washed 10 min in 0.25N HCl; twice for 30 min in 0.5 M NaOH, 1.5 M NaCl; and twice for 30 min in 1.5 M NaCl, 0.5 M Tris-HCl, 0.5 M Tris base. DNA was transferred onto a Hybond N+ membrane in 20× SSC for 20 h and hybridized following the protocol proposed by the supplier, with some modifications (Supplemental materials). After hybridization, the membrane was incubated for 12–48 h in a cassette for PhosphorImager.
Acknowledgments
We thank J. Weitzman for helpful comments on the manuscript. This work was supported by grants from the NIH, the AFM, and the ARC. S.C. was supported by a Ph.D. fellowship from ARC. C.C. was supported by a post-doc fellowship from CANAM (Caisse Nationale d'Assurance Maladie et Maternité des Travailleurs Non Salariés des Professions Non Agricoles) and from Fondation Louis D–Institut de France.
[Supplemental material is available at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: M. Amor-Guéret, Y. de Santigny, and G. Brock.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3970105. Article published online before print in July 2005.
References
- Arnheim, N. and Southern, E.M. 1977. Heterogeneity of the ribosomal genes in mice and men. Cell 11: 363–370. [DOI] [PubMed] [Google Scholar]
- Batzer, M.A. and Deininger, P.L. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370–379. [DOI] [PubMed] [Google Scholar]
- Bensimon, A., Simon, A., Chiffaudel, A., Croquette, V., Heslot, F., and Bensimon, D. 1994. Alignment and sensitive detection of DNA by a moving interface. Science 265: 2096–2098. [DOI] [PubMed] [Google Scholar]
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes & Dev. 16: 6–21. [DOI] [PubMed] [Google Scholar]
- Caburet, S. 2002. Structure mosaïque et instabilité de l'ADN ribosomal humain: Implications dans la sénescence et la cancérogènese. Ph.D. thesis, Université de Paris VII, Paris, France.
- Conconi, A., Widmer, R.M., Koller, T., and Sogo, J.M. 1989. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57: 753–761. [DOI] [PubMed] [Google Scholar]
- Dammann, R., Lucchini, R., Koller, T., and Sogo, J.M. 1995. Transcription in the yeast rRNA gene locus: Distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol. 15: 5294–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder Jr., J.F. and Turner, B.J. 1995. Concerted evolution of repetitive DNA sequences in eukaryotes. Quart. Rev. Biol. 70: 297–320. [DOI] [PubMed] [Google Scholar]
- Erickson, J.M. and Schmickel, R.D. 1985. A molecular basis for discrete size variation in human ribosomal DNA. Am. J. Hum. Genet. 37: 311–325. [PMC free article] [PubMed] [Google Scholar]
- Erickson, J.M., Rushford, C.L., Dorney, D.J., Wilson, G.N., and Schmickel, R.D. 1981. Structure and variation of human ribosomal DNA: Molecular analysis of cloned fragments. Gene 16: 1–9. [DOI] [PubMed] [Google Scholar]
- Frederiksen, S., Cao, H., Lomholt, B., Levan, G., and Hallenberg, C. 1997. The rat 5S rRNA bona fide gene repeat maps to chromosome 19q12→qter and the pseudogene repeat maps to 12q12. Cytogenet. Cell Genet. 76: 101–106. [DOI] [PubMed] [Google Scholar]
- French, S.L., Osheim, Y.N., Cioci, F., Nomura, M., and Beyer, A.L. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 23: 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff, S., McDonald, J.P., Bendixen, C., Arthur, L., and Rothstein, R. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14: 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, I.L. and Sylvester, J.E. 1995. Complete sequence of the 43-kb human ribosomal DNA repeat: Analysis of the intergenic spacer. Genomics 27: 320–328. [DOI] [PubMed] [Google Scholar]
- ———. 2001. Human rDNA: Evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics 73: 255–263. [DOI] [PubMed] [Google Scholar]
- Gray, M.D., Wang, L., Youssoufian, H., Martin, G.M., and Oshima, J. 1998. Werner helicase is localized to transcriptionally active nucleoli of cycling cells. Exp. Cell. Res. 242: 487–494. [DOI] [PubMed] [Google Scholar]
- Grozdanov, P., Georgiev, O., and Karagyozov, L. 2003. Complete sequence of the 45-kb mouse ribosomal DNA repeat: Analysis of the intergenic spacer. Genomics 82: 637–643. [DOI] [PubMed] [Google Scholar]
- Grummt, I. and Pikaard, C.S. 2003. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell Biol. 4: 641–649. [DOI] [PubMed] [Google Scholar]
- Hattori, M., Fujiyama, A., Taylor, T.D., Watanabe, H., Yada, T., Park, H.S., Toyoda, A., Ishii, K., Totoki, Y., and Choi, D.K. 2000. The DNA sequence of human chromosome 21. Nature 405: 311–319. [DOI] [PubMed] [Google Scholar]
- Henderson, A.S., Warburton, D., and Atwood, K.C. 1972. Location of ribosomal DNA in the human chromosome complement. Proc. Natl. Acad. Sci. 69: 3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, S.J., Tatebayashi, K., Ohsugi, I., Shimamoto, A., Furuichi, Y., and Ikeda, H. 1999. Bloom's syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells 4: 619–625. [DOI] [PubMed] [Google Scholar]
- Ji, Y., Eichler, E.E., Schwartz, S., and Nicholls, R.D. 2000. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 10: 597–610. [DOI] [PubMed] [Google Scholar]
- Johnson, F.B., Sinclair, D.A., and Guarente, L. 1999. Molecular biology of aging. Cell 96: 291–302. [DOI] [PubMed] [Google Scholar]
- Krystal, M. and Arnheim, N. 1978. Length heterogeneity in a region of the human ribosomal gene spacer is not accompanied by extensive population polymorphism. J. Mol. Biol. 126: 91–104. [DOI] [PubMed] [Google Scholar]
- Kuo, B.A., Gonzalez, I.L., Gillespie, D.A., and Sylvester, J.E. 1996. Human ribosomal RNA variants from a single individual and their expression in different tissues. Nucleic Acids Res. 24: 4817–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella, T. and Schlessinger, D. 1989. Complete human rDNA repeat units isolated in yeast artificial chromosomes. Genomics 5: 752–760. [DOI] [PubMed] [Google Scholar]
- Machwe, A., Orren, D.K., and Bohr, V.A. 2000. Accelerated methylation of ribosomal RNA genes during the cellular senescence of Werner syndrome fibroblasts. Faseb J. 14: 1715–1724. [DOI] [PubMed] [Google Scholar]
- Marciniak, R.A., Lombard, D.B., Johnson, F.B., and Guarente, L. 1998. Nucleolar localization of the Werner syndrome protein in human cells. Proc. Natl. Acad. Sci. 95: 6887–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet, X., Ekong, R., Fougerousse, F., Rousseaux, S., Schurra, C., Hornigold, N., van Slegtenhorst, M., Wolfe, J., Povey, S., Beckmann, J.S., et al. 1997. Dynamic molecular combing: Stretching the whole human genome for high-resolution studies. Science 277: 1518–1523. [DOI] [PubMed] [Google Scholar]
- Mighell, A.J., Smith, N.R., Robinson, P.A., and Markham, A.F. 2000. Vertebrate pseudogenes. FEBS Lett. 468: 109–114. [DOI] [PubMed] [Google Scholar]
- Moss, T. and Stefanovsky, V.Y. 2002. At the center of eukaryotic life. Cell 109: 545–548. [DOI] [PubMed] [Google Scholar]
- Naylor, S.L., Sakaguchi, A.Y., Schmickel, R.D., Woodworth-Gutai, M., and Shows, T.B. 1983. Organization of rDNA spacer fragment variants among human acrocentric chromosomes in somatic cell hybrids. J. Mol. Appl. Genet. 2: 137–146. [PubMed] [Google Scholar]
- Nelkin, B., Strayer, D., and Vogelstein, B. 1980. Divergence of primate ribosomal RNA genes as assayed by restriction enzyme analysis. Gene 11: 89–96. [DOI] [PubMed] [Google Scholar]
- Pasero, P., Bensimon, A., and Schwob, E. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes & Dev. 16: 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard, C.S. 2000. The epigenetics of nucleolar dominance. Trends Genet. 16: 495–500. [DOI] [PubMed] [Google Scholar]
- Romao-Correa, R.F., Ruiz, I.R., Maria, D.A., Neto, C.F., and Sanches, J.A. 2004. Ribosomal DNA exhibits few alterations in human skin cancers. J. Dermatol. Sci. 34: 109–111. [DOI] [PubMed] [Google Scholar]
- Santoro, R., Li, J., and Grummt, I. 2002. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32: 393–396. [DOI] [PubMed] [Google Scholar]
- Schawalder, J., Paric, E., and Neff, N.F. 2003. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell. Biol. 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl, R.H. and Bishop, A.J. 2000. Homologous recombination as a mechanism for genome rearrangements: Environmental and genetic effects. Mutat. Res. 454: 53–62. [DOI] [PubMed] [Google Scholar]
- Schmickel, R.D., Waterson, J.R., Knoller, M., Szura, L.L., and Wilson, G.N. 1980. HeLa cell identification by analysis of ribosomal DNA segment patterns generated by endonuclease restriction. Am. J. Hum. Genet. 32: 890–897. [PMC free article] [PubMed] [Google Scholar]
- Schofer, C., Weipoltshammer, K., Almeder, M., and Wachtler, F. 1998. Arrangement of individual human ribosomal DNA fragments on stretched DNA fibers. Histochem. Cell Biol. 110: 201–205. [DOI] [PubMed] [Google Scholar]
- Shen, J. and Loeb, L.A. 2001. Unwinding the molecular basis of the Werner syndrome. Mech. Age. Dev. 122: 921–944. [DOI] [PubMed] [Google Scholar]
- Sinclair, D.A. and Guarente, L. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sylvester, J.E., Whiteman, D.A., Podolsky, R., Pozsgay, J.M., Respess, J., and Schmickel, R.D. 1986. The human ribosomal RNA genes: Structure and organization of the complete repeating unit. Hum. Genet. 73: 193–198. [DOI] [PubMed] [Google Scholar]
- Watt, P.M., Hickson, I.D., Borts, R.H., and Louis, E.J. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer, P.K. and Dawid, I.B. 1979. Isolation and sequence organization of human ribosomal DNA. J. Mol. Biol. 128: 289–303. [DOI] [PubMed] [Google Scholar]
- Worton, R.G., Sutherland, J., Sylvester, J.E., Willard, H.F., Bodrug, S., Dube, I., Duff, C., Kean, V., Ray, P.N., and Schmickel, R.D. 1988. Human ribosomal RNA genes: Orientation of the tandem array and conservation of the 5′ end. Science 239: 64–68. [DOI] [PubMed] [Google Scholar]
- Yu, C.E., Oshima, J., Fu, Y.H., Wijsman, E.M., Hisama, F., Alisch, R., Matthews, S., Nakura, J., Miki, T., and Ouais, S. 1996. Positional cloning of the Werner's syndrome gene. Science 272: 258–262. [DOI] [PubMed] [Google Scholar]