Abstract
A region-specific ENU mutagenesis screen was conducted to elucidate the functional content of proximal mouse Chr 5. We used the visibly marked, recessive, lethal inversion Rump White (Rw) as a balancer in a three-generation breeding scheme to identify recessive mutations within the ∼50 megabases spanned by Rw. A total of 1003 pedigrees were produced, representing the largest inversion screen performed in mice. Test-class animals, homozygous for the ENU-mutagenized proximal Chr 5 and visibly distinguishable from nonhomozygous littermates, were screened for fertility, hearing, vestibular function, DNA repair, behavior, and dysmorphology. Lethals were identifiable by failure to derive test-class animals within a pedigree. Embryonic lethal mutations (total of 34) were overwhelmingly the largest class of mutants recovered. We characterized them with respect to the time of embryonic death, revealing that most act at midgestation (8.5–10.5) or sooner. To position the mutations within the Rw region and to guide allelism tests, we performed complementation analyses with a set of new and existing chromosomal deletions, as well as standard recombinational mapping on a subset of the mutations. By pooling the data from this and other region-specific mutagenesis projects, we calculate that the mouse genome contains ∼3479–4825 embryonic lethal genes, or about 13.7%–19% of all genes.
A long-term goal of the human genome project (HGP) is to define and understand the functions of all human genes and their relationships to health and disease. The HGP has already made a massive impact on genetic research, having yielded genomic sequences, comprehensive polymorphic genetic marker sets, and sundry molecular reagents for humans, mice, and many other experimental organisms. This has greatly simplified positional cloning of disease genes and genetic mapping of simple and complex traits. Concurrent efforts have identified thousands of expressed genes, and coupled with development and implementation of gene expression technologies, we are in the powerful position of knowing the sequence identity of many genes, their locations in the genome, their expression patterns, and which proteins interact with one another. While these data are valuable for deducing or suggesting the roles of genes in development and homeostasis, the functions of most genes remain unknown. Identifying their functions is a much more difficult challenge, and for most genes, direct experimentation in the context of a whole organism will be required.
The phenotype-driven approach of genetic analysis, also called “forward genetics,” is a classical strategy of randomly mutagenizing the entire genome, followed by a breeding scheme to evaluate progeny for phenotypes of interest without any preconception of what the underlying genes might be. Forward genetic strategies have long been implemented in model organisms such as yeast, worms, and fruit flies, enabling remarkable advances toward understanding the developmental genetics of those organisms. An effective way to induce germ-line mutations in mice is with the chemical N-ethyl-N-nitrosourea (ENU) (Shedlovsky et al. 1986, 1988). ENU induces mutations in the male germ line at a high frequency, enabling efficient screening of mutagenized pedigrees for aberrant phenotypes (Hitotsumachi et al. 1985). As a point mutagen, it produces not only null alleles, but also hypomorphs and gain-of-function mutations. This is useful for dissecting the functions of a protein and producing phenotypes that might be obscured by null mutations.
We have conducted a program to mutagenize the mouse genome with ENU and specifically recover mutations in the region spanned by the rump-white (Rw) inversion on Chr 5. Rw is a radiation-induced mutation causing depigmentation of the posterior and ventral abdomen in heterozygotes and embryonic lethality in homozygotes (Stephenson et al. 1994). Recombination in heterozygotes is effectively suppressed throughout the inversion region by elimination of nonviable recombinants, and thus, it serves as a balancer chromosome. These properties make it ideal for use in a classical 3 generation region-directed mutagenesis screen as originally demonstrated in mice by Roderick (1983) and more recently by Kile et al. (2003). Herein, we describe the screening of 1003 gametes using the inversion screen paradigm, and report the recovery of 43 mutations, 37 of which cause embryonic or perinatal lethality. Combining these data with those from other published screens, we estimate that ∼13.7%–19% of genes in the mammalian genome are required for survival through embryogenesis.
Results
ENU mutagenesis of proximal mouse chromosome 5
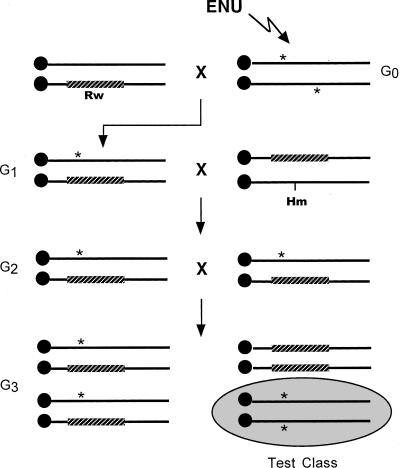
We (J. Schimenti and M. Bucan) previously outlined a strategy to perform saturation mutagenesis of the proximal region of Chr 5 using a series of chromosome deletions to uncover recessive ENU-induced mutations (Schimenti and Bucan 1998). This involved using a minimum tiling path of deletions to cover a ∼30-cM target region (spanned by the Rw inversion), using a two-generation breeding scheme. However, many deletions induced at the Huntington disease locus (Hdh) are subviable, subfertile, and caused serious haploinsufficient phenotypes reminiscent of Wolf Hirschhorn Syndrome (Naf et al. 2001). These traits would seriously impact the efficiency of the screen and obfuscate the detection of phenotypes. To overcome this problem, we adopted an alternative strategy of using the Rw inversion itself to conduct a three-generation screen for mutations within and near the inversion breakpoints (diagrammed in Fig. 1).
Figure 1.
Chr 5Rw Inversion screen. Male C57BL/6J mice (generation G0) were treated with ENU to induce point mutations (*) and mated to C3H-Rw/+ females (the Rw inversion is designated as a thick, striped bar). G1 offspring inheriting Rw (white spotting on hindquarters) were selected and crossed to mice carrying Rw in trans to Hm, a semidominant mutation causing webbed digits. This permits visual selection of G2 progeny that are Rw/+* (they have white spotting, but no webbed digits). G2 animals were intercrossed to produce the three types of offspring shown. Rw/Rw embryos die in utero. Non-Rw G3 animals are homozygous for the ENU-exposed Chr 5, and were screened for phenotypes.
The key feature of this strategy is that the mice used in the breeding scheme contain visibly marked chromosomes that allow the ENU-induced mutations to be followed visually. Pedigrees were founded by first generation (G1) males containing the Rw inversion in trans to an ENU-mutagenized chromosome (+*) of strain C57BL/6J (“B6”). We crossed G1 males to females of the genotype Rw +/+ Hm (“Rw/Hm”), and selected rump white, nonhammertoe offspring (G2 generation). Hammertoe (Hm) is a semidominant, fully penetrant mutation causing webbed digits. Rw homozygotes die in utero. Therefore, these rump white, nonhammertoe G2 animals must have inherited Rw from the Rw/Hm mother, and +* from the father. Importantly, the Rw inversion suppresses recombination with the wild-type meiotic partner in the region spanned by the inversion, be it +* or the Hm-containing chromosome. We then intercrossed G2 animals (Rw/+*) to produce test-class (+*/+*) mice for phenotyping. Since Rw/Rw is lethal, the G3 Rw animals are obligate heterozygotes (Rw/+*). We maintain the mutations in crosses dictating forced heterozygosity (Rw/+* × Rw/+*).
A key element of this strategy is that embryonic lethal mutations are easy to detect; pedigrees containing a lethal mutation in the Rw region fail to yield test-class G3 animals. One caveat is that nonlethal mutations detected in non-Rw G3 mice may actually map elsewhere in the genome, because these mice carry unlinked segregating mutations that can be rendered homozygous. However, such mutations are easily eliminated from the Rw region by virtue of the fact that some test-class animals will not have the phenotype and some Rw/+* animals will have the phenotype. For the most part, we elected not to pursue the majority of non-Chr 5 mutations. It should also be noted that mutations will be identified on Chr 5 that lie just outside the breakpoints of the Rw inversion and fall within the region of linkage disequilibrium.
Screening and identification of ENU-induced mutations in the Rw inversion region
A total of 1003 pedigrees were generated. Table 1 outlines the phenotypic screens conducted, and how many of the pedigrees we evaluated for each phenotype. Potential lethal mutations were flagged when intercrosses of G2 carriers failed to produce any non-Rw weanlings of 15–20 total. We considered them as being true mutations if non-Rw animals failed to appear after 30 progeny (P = .0001). In the fertility screens, we tested one or more males and females for nearly all pedigrees. In the behavioral screen, we selected families in which non-Rw progeny exhibited behavioral traits that were statistically different from values observed in Rw/+ progeny. For the remaining screens, we typically evaluated a single test-class animal.
Table 1.
Phenotype screens
| Phenotype | Pedigrees | Mutants |
|---|---|---|
| Lethality | 1003 | 37 |
| Fitness | 1003 | 1 |
| Fertility | 990 | 2 |
| Hearing | 949 | 1 |
| Vestibular | 989 | 0 |
| Micronucleus | 821 | 0 |
| Behavior | 83 | 2 |
| Eye | 25 | 0 |
Both of the infertility mutations are male specific. Males homozygous for Ste5Jcs1 produce low-to-normal numbers of sperm, with normal morphology and axoneme structure (as judged by EM—data not shown), but sperm motility is impaired. Ste5Jcs3 mutants exhibit no epididymal sperm. Histological analysis reveals that spermatogenesis proceeds through meiosis, but spermiogenesis is severely disrupted, in that spermatids do not form flagella as do controls (Supplemental Fig. 1a,b).
A semilethal dysmorphology mutant called Dumbo was identified as having unusually large or abnormally oriented earlobes (Supplemental Fig. 1c). We recovered one fitness mutation (Fit5Jcs1); test-class animals arose at lower than expected frequencies at weaning, and usually died of unknown causes before 6-wk-of-age. A single deafness mutant was found in screens of 949 pedigrees, but no vestibular mutants, despite the presence of the tilted (tlt) gene in the Rw interval (Ying et al. 1999). The deafness mutation was localized to the proximal Rw region by virtue of noncomplementation with the Dpp6df1J deletion (Fig. 3, below). No other overt phenotypic mutants mapping to the Rw region were observed.
Figure 3.
Map locations of lethal mutations. The proximal region of Chr. 5 is depicted as a horizontal line, with the centromere (filled circle) on the left. The region spanned by the Rw inversion is indicated above; its breakpoints are within Dpp6 proximally and near c-kit distally. Map positions (in megabases, accordingly to mouse genome Build 33) are indicated. Microsatellite loci are abbreviated by exchanging the prefix “M” for “D5Mit.” Deletions are indicated as horizontal rectangles, either solid or striped, and are color coded with the locus at which they were induced (red) Dpp6; (blue) Hdh; (green) Qdpr. The amount of DNA known to be absent in each deletion is spanned by the rectangles. The thin lines extending from the ends of the rectangles indicate the regions in which the deletion breakpoints reside. Those deletions that have been converted into stocks of mice are represented by solid rectangles, and associated allele names are given (the bracketed text corresponds to superscripting). New deletions at the Qdpr locus existing only in ES cells are represented by the striped or unfilled rectangles. In this latter case, the number of independent ES cell lines containing a particular class of deletion (currently indistinguishable with the markers used in this study) is indicated on the right. The intervals containing certain lethal mutations (abbreviated as “L. #”) are bracketed at the bottom of the map. Those that were deletion mapped are color coded. Those that were recombination mapped are in black. The yellow-shaded “ES cell haplolethal zone” is an interval that connot be deleted in ES cells. Originally described by Schimenti et al. (2000), its proximal end has been refined as described in the text, and the distal end was also refined by further analysis of deletions centered at the Gabrb1 (data not shown). (Shh) Sonic hedgehog homolog.
We used a flow cytometric micronucleus assay to identify mice with mutations causing genomic instability. This assay was used successfully in genome-wide recessive ENU screens (Shima et al. 2003). Though we did not find any mutations in the Rw region, two mutations causing elevated micronuclei on other chromosomes were isolated (Shima et al. 2003).
Preliminary characterization of lethal mutations
Lethal mutations were the most prevalent class of deviants recovered (total of 37, including Dumbo). Thirty-four of these act exclusively prenatally. To characterize the nature and timing of the lethalities, we intercrossed Rw/+* animals and dissected litters at various stages of development. Each embryo was inspected for abnormalities and PCR genotyped to identify those that were of the test class (homozygous for B6 at loci within the Rw inversion region). If no test-class embryos were identified after genotyping 15 or more of a particular gestational stage in a given pedigree, we concluded that the mutation acts at an earlier time point. We classified the embryonic lethal mutations into three broad groups as follows: early (preimplantation–E8.5), midgestation (E8.5–E12.5), and late gestation (E13.5–E17.5). The results are summarized in Table 2.
Table 2.
Summary of lethal and fitness mutations
| Group | Stage | Defect onset | Mutation | Phenotype | Rw/+*× Rw/+* | |
|---|---|---|---|---|---|---|
| Rw
|
nonRw
|
|||||
| Early | Pre/Peri-implantation | E3.5-E5.5 | L5Jcs4 | 78 | 0 | |
| L5Jcs7 | 92 | 0 | ||||
| L5Jcs10 | 123 | 0 | ||||
| L5Jcs14 | 104 | 1* | ||||
| L5Jcs22 | 60 | 0 | ||||
| L5Jcs24 | 122 | 0 | ||||
| L5Jcs25 | 72 | 0 | ||||
| L5Jcs27 | 59 | 0 | ||||
| L5Jcs30 | 82 | 0 | ||||
| L5Jcs31 | 40 | 0 | ||||
| L5Jcs35 | 33 | 0 | ||||
| Post-implantation | E5.5-E7.5 | L5Jcs6 | Gastrulation | 59 | 0 | |
| L5Jcs8 | 95 | 0 | ||||
| L5Jcs11 | Gastrulation | 64 | 0 | |||
| L5Jcs18 | Gastrulation | 82 | 1* | |||
| L5Jcs36 | 29 | 0 | ||||
| L5Jcs38 | Gastrulation | 63 | 0 | |||
| Undetermined | Pre E9.5-10.5 | L5Jcs5 | 48 | 0 | ||
| Middle | Midgestation | E8.5-E12.5 | L5Jcs2 | 64 | 0 | |
| L5Jcs3 | 97 | 0 | ||||
| L5Jcs13 | 112 | 0 | ||||
| L5Jcs15 | CF/CV/placenta | 120 | 0 | |||
| L5Jcs17 | Growth retardation | 55 | 0 | |||
| L5Jcs23 | NTD | 57 | 1 | |||
| L5Jcs28 | Growth retardation | 63 | 0 | |||
| L5Jcs32 | NTD; pericardial edema | 35 | 0 | |||
| L5Jcs33 | CF/CV | 32 | 2 | |||
| L5Jcs37 | CF/CV/placenta | 20 | 0 | |||
| Late | E10.5-birth | L5Jcs20 | 38 | 0 | ||
| L5Jcs34 | 30 | 1 | ||||
| Perinatal | E18.5-1dpp | L5Jcs1 | Skeletal, bone defects | 141 | 0 | |
| Postnatal | 1dpp-10dpp | L5Jcs16 | Fitness, runty | 69 | 0^ | |
| UNK | L5Jcs29 | 61 | 2 | |||
| L5Jcs26 | 33 | 0 | ||||
| L5Jcs12 | 53 | 0 | ||||
| L5Jcs19 | 73
|
0
|
||||
| Total | 2215 | 8 | ||||
Sickly; (^) no runts at wean age; (CF) craniofacial; (CV) cardiovascular; (NTD) neural tube defects.
Of the 34 pedigrees carrying embryonic lethal mutations, about half (18) fell into the early class. We classified them as such by the absence of test-class embryos in E8.5–E12.5 litters (concomitant with empty decidua), or the presence of severely underdeveloped embryos. Based on these observations, we placed 11 members into a pre- or peri-implantation (E3.5–E5.5) subcategory, and another six that failed to demonstrate normal gastrulation, or which were arrested at the egg cylinder stage, into a post-implantation lethal category (E5.5–E7.5). Of the remaining 14 lethals, 10 were classified as midgestation and four as late gestation, the latter including one acting perinatally (E18.5–1dpp) and another postnatally (pre-10 dpp).
While the causes of death are still under investigation, we assigned preliminary phenotype characterizations to 11 of the prenatal lethals in which we observed two or more test-class embryos (summarized in Table 2). Several mutants exhibit clearly abberant phenotypes, including craniofacial defects, cardiovascular defects, neural tube defects, gastrulation abnormalities, placental defects, and growth retardation. Selected examples of mutant phenotypes are shown in Figure 2. L5Jcs2/L5Jcs2 embryos show abnormal amnion development (Fig. 2B); L5Jcs11 mutants fail to gastrulate (Fig. 2D); L5Jcs15 mutants have craniofacial and cardiovascular defects (Fig. 2F); and embryos homozygous for L5Jcs32 exhibit a failure of anterior neural tube closure as seen in both whole mounts (Fig. 2H) and histological sections (Fig. 2G). L5Jcs1 mutants, which die around birth, often exhibit a skeletal patterning abberation, in which the eighth rib is attached to the sixth sternebra, possibly reflecting a posterior-to-anterior transformation (Fig. 2J).
Figure 2.

Phenotypes of embryonic lethal mutants. Shown are images of whole-mount (A–F,H) embryos. Except for G and H, age-matched control (wt) embryos are at left, alongside corresponding mutants (mut) at right. (B) L5Jcs2 at E9.5. The arrowhead highlights a defective placenta. (D) L5Jcs11 at E8.5. (F) L5Jcs15 at E12.5. The arrowhead is pointing to a ballooned pericardium. (H) L5Jcs32 at E10.5. The arrow is highlighting the defect in neural tube closure, and G is a hematoxylin+eosin-stained transverse section obtained from the same embryo. The black arrows on the left (dorsal) indicate the open neural tube, and the asterisks mark the telencephalic vesicle. (I,J) Alcian blue and alizarin red-stained E19.5 L5Jcs1/L5Jcs1 embryos showing overgrowth of the eighth rib attaching abnormally to the 6th sternebra. The white or black bars indicate 1 mm.
The Rw inversion enables simple maintenance of the lethal mutations by conducting balanced lethal matings of Rw/+* × Rw/+*. If Rw acts as a true balancer (i.e., completely blocks recombination in heterozygotes), then the only progeny that can result from such a cross are rump white (Rw/+*). Thus far, of the 37 mutations, only six lines have yielded non-Rw offspring (Table 2). In the cases of L5Jcs14 and 18, the lone non-Rw offspring were runted and probably represented rare escapers with <100% expressivity. The non-Rw, normal-appearing survivors of the remaining four lines may be explained in three ways as follows: (1) the mutations actually reside just outside of the Rw inversion, and recombined onto Rw chromosome; (2) these mutations are incompletely penetrant; or (3) they are the product of double-crossover events between the Rw and mutation-bearing chromosomes in one of the Rw/+* parents. As we map these mutations and conduct additional breeding, these issues should resolve. Overall, we found only eight such exceptions in 2380 offspring, demonstrating that the Rw inversion acts as a reliable balancer.
Mapping of mutations and assessment of allelism
The ultimate value of this mutant collection hinges on identifying the genes underlying the mutant phenotypes. This first requires genetic localization of the mutations. Two strategies have been taken, standard meiotic recombination mapping and deletion mapping. With respect to the former, we have been generating sets of chromosomal deletions across much of the Rw region for use in complementation tests with the ENU-induced mutations to facilitate their localization. We previously described the generation of deletion complexes centered at the Dpp6, Hdh, and Gabrb1 loci using the technique of ES cell irradiation (Schimenti et al. 2000). However, this collection contained a large gap between Hdh and an ES cell haplolethal locus, the deletion of which was incompatible with survival of ES cells (Schimenti et al. 2000).
To fill this gap, we generated a new panel of deletions centered at the Qdpr locus (Fig. 3). We isolated several dozen ES cell clones containing deletions, and genotyped them to identify approximate breakpoint locations. As expected, no deletions were recovered that extended past D5Jcs58, a newly identified marker that better defines the proximal end of the haplolethal locus. We selected a subset for blastocyst injection and creation of mouse lines. These collectively provided a set of spaced, nested breakpoints between Hdh and D5Jcs58. The Qdpr deletions that were transmitted through the germ line are shown in Figure 3 along with some of the other previously reported deletions centered at Hdh and Dpp6 that will be useful for mutation mapping (Schimenti et al. 2000).
Given this deletion collection, we adopted a stratified approach to genetic characterization of the ENU-induced mutations. First, the mutations are crossed to three deletions (Dpp6df1J, Hdhdf7J, and Qdprdf3J) to assess complementation. Collectively, these span ∼21–33 Mb of the 51.5 Mb Rw region. While Hdh deletions larger than Hdhdf7J are available, in practice, they are very difficult to breed due to the reduced viability associated with deletions of the mouse ortholog of the Wolf-Hirschhorn critical region (Naf et al. 2001). Second, the mutations mapping within these deletions are mated to other deletions with breakpoints in the vicinity to refine the map location and thereby aid in eventual gene identification. Third, those mutations failing to be complemented by the same deletion, and which display similar phenotypes, are intercrossed to identify possible allelism.
In addition to deletion mapping, we conducted traditional recombinational mapping for nine of the lethals (see Methods). Mice heterozygous for the lethal mutations (C3H/+*) were intercrossed, and the live offspring were genotyped with microsatellite markers across the Rw region. The locations of mutations were deduced indirectly as those regions that could be rendered homozygous for B6 alleles of Chr5, based on analysis of ∼50 F2 progeny.
A summary of the first tier of deletion and genetic mapping is presented in Table 3. Complementation tests between nearly all of the lethals and the Dpp6df1J and Qdprdf3J deletions were performed, but the Hdhdf7J crosses remain to be completed (this deletion is subviable). For the Dpp6df1J and Hdhdf7J deletions, which are maintained in trans to Rw, the results of these complementation tests could be ascertained by visual inspection of offspring in crosses to Rw/+* mates. The presence of a single non-Rw offspring (+*/Del) indicates complementation, i.e., the lethal does not map within the deletion region. Failure to derive such animals indicates the mutation maps within the deletion region. Because Qdprdf3J could not be placed in trans to Rw (see below), we genotyped non-Rw offspring of Qdprdf3J/+ × Rw/+* to determine whether Qdprdf3J/+* progeny could not be obtained.
Table 3.
Deletion and recombination mapping of lethals
| Mutation | Dpp6df1J | Qdprdf3J | Hdhdf7J | Recomb map |
|---|---|---|---|---|
| L5Jcs1 | Yes | NO | N/A | Y |
| L5Jcs2 | Yes | N/A | N/A | Y |
| L5Jcs3 | Yes | Yes | N/A | Y |
| L5Jcs4 | NO | N/A | N/A | Y |
| L5Jcs5 | Yes | Yes | – | |
| L5Jcs6 | Yes | Yes | – | |
| L5Jcs7 | Yes | Yes | – | |
| L5Jcs8 | Yes | N/A | N/A | Y |
| L5Jcs10 | Yes | N/A | N/A | Y |
| L5Jcs11 | Yes | Yes | – | |
| L5Jcs12 | Yes | – | – | |
| L5Jcs13 | – | – | Yes | |
| L5Jcs14 | Yes | Yes | NO | Y |
| L5Jcs15 | Yes | N/A | N/A | Y |
| L5Jcs16 | Yes | Yes | – | |
| L5Jcs17 | Yes | Yes | Yes | |
| L5Jcs18 | Yes | NO | N/A | Y |
| L5Jcs19 | Yes | NO | N/A | |
| L5Jcs20 | NO | N/A | N/A | |
| L5Jcs22 | Yes | Yes | Yes | |
| L5Jcs23 | Yes | Yes | – | |
| L5Jcs24 | Yes | Yes | Yes | |
| L5Jcs25 | Yes | Yes | – | |
| L5Jcs26 | Yes | Yes | – | |
| L5Jcs27 | Yes | – | – | |
| L5Jcs28 | Yes | Yes | – | |
| L5Jcs29 | Yes | Yes | – | |
| L5Jcs30 | – | Yes | Yes | |
| L5Jcs31 | Yes | Yes | Yes | |
| L5Jcs32 | Yes | NO | Yes | |
| L5Jcs33 | Yes | Yes | – | |
| L5Jcs34 | Yes | Yes | – | |
| L5Jcs35 | Yes | Yes | Yes | |
| L5Jcs36 | NO | N/A | N/A | |
| L5Jcs37 | Yes | Yes | NO | |
| L5Jcs38 | Yes | Yes | – | |
| Dumbo | Yes | – | – |
(Yes) Complementation between the deletion and the mutation; (No) noncomplementation; (N/A) complementation test was not done, since the mutation was mapped to another deletion or by recombination mapping; (–) test was not done. In the Recomb(ination) map column, Y indicates the mutation was mapped by meiotic recombination.
An example of the second tier of mapping is illustrated by the case of L5Jcs4. Dpp6df1J failed to complement this lethal, which was then mated to a smaller deletion, Dpp6df5J. This also failed to complement, thereby narrowing the L5Jcs4 gene to the <3.7 Mb region spanned by this deletion (Fig. 3). We corroborated this localization by low-resolution recombinational mapping (data not shown). The deafness mutation was also mapped to the Dpp6df1J deletion interval. An overview of map positions obtained by both recombinational and deletion mapping is presented in Figure 3. Of the 37 lethals, nine failed to complement one of the three deletions, and nine were recombination mapped. Since four were mapped by both methods, overall, 14 have been mapped to some degree.
The third tier of genetic analyses—complementation testing the lethal collection for different alleles of the same genes—is in early stages and will take advantage of the mapping data (see Discussion). Given the possibility of mutation “clusters,” in which the identical spermatogonial mutation in a treated male is passed to multiple offspring, we complementation tested two lethal pairs that were derived from a common mutagenized male, L5Jcs22/23 and L5Jcs27/30. No evidence for identical mutations was found. The L5Jcs13/14 pair arose in the same pedigree, but allelism was genetically excluded by the observation that L5Jcs13 complements Hdhdf7J, but L5Jcs14 does not.
Rw lethality is not due to disruption of Dpp6
The proximal breakpoint of the Rw inversion occurs within the Dpp6 gene (Hough et al. 1998). Since a deletion (W19H) of the distal inversion breakpoint complements Rw, it was suggested that the Rw lethality might be due either to the disruption of Dpp6, or mutation of another gene within the inversion (Lyon et al. 1984; Hough et al. 1998). In the course of setting up balanced lethal crosses to maintain deletions in trans to Rw, we found that the Dpp6df1J, Dpp6df4J, and Dpp6df5J deletions all complement Rw. Therefore, it can be concluded that mutation of Dpp6 is not the Rw lethal factor, and furthermore, given the disruption of this gene by Rw, that Dpp6 is not required for viability. Superficially, the Rw/Del animals are indistinguishable from mice heterozygous for the deletion. Thus, the function of this gene is unclear.
At this stage, it is not possible to determine the origin of the Rw lethal factor. The Rw mutation was recovered in the course of radiation mutagenesis experiments, identifiable by the dominant pigmentation phenotype, and also noted to be recessive lethal in the initial report (Batchelor et al. 1966). Since the lethality appears not to be due to disruption of sequences at the inversion breakpoints, it may be caused by an intragenic mutation, or small deletion, that arose concurrently from radiation treatment. Notably, the ENU experiments conducted here cannot produce a null allele of the Rw lethal, since pedigrees were initiated by males carrying the ENU-mutagenized Chr 5 in trans to Rw. We have found that at least two of the larger Qdpr deletions (Qdprdf3J and Qdprdf11J) are unable to complement Rw. Thus, it appears that the Rw inversion carries at least one recessive mutation in the region around Qdpr.
Screen for behavioral mutants
We screened 83 of the 1003 pedigrees for behavioral anomalies. A total of 534 G3 progeny (290 Rw/+ heterozygotes and 244 non-Rw test class) were tested in several behavioral paradigms spanning 1–2 mo per animal. Phenotypic analyses included tests for neuromuscular anomalies using a rotarod, anomalies in sensorimotor gating assessed by prepulse inhibition of a startle response, and anxiety-like behavior by monitoring animals in a zero maze. In addition to these tests, 35 pedigrees were also examined for anomalies in exploratory behavior and activity in a hole-board apparatus, as well as for abnormal circadian wheel-running behavior. We examined 5–10 progeny from each pedigree, and selected as potential mutants non-Rw progeny that exhibited, as a group, behavioral values that were different from their Rw/+ littermates, parental strains (B6 and C3H), and their combinations (F1 and F2). We outcrossed non-Rw phenodeviants to C3H/HeJ inbred mice to generated F1 progeny, then intercrossed F1s to produce F2 animals for phenotyping and genetic mapping. This led to the identification and mapping of two presumptive behavioral mutations linked to the Rw target region (Fig. 4; Supplemental Fig. 2). One (Beh5Jcs1) showed decreased activity in the hole board. Based on phenotype and genotype analysis of 71 F2 progeny, we assigned a provisional map location around the D5Mit10–D5Mit314 interval, distal to the Rw region (Fig. 4B). The other behavioral mutation, Beh5Jcs2, showed reduced strength of circadian cycle measured by Fast Fourier Transform analysis (FFT; Supplemental Fig. 2). Five G3 non-Rw progeny had a lower FFT than their Rw/+ littermates. The strength of circadian cycling in these mice was also lower than in wild-type parental strains (B6, C3H/HeJ, and F1 and F2 progeny of these strains). We performed genetic and phenotypic analysis of an F2 cohort, which showed that mice homozygous for the B6 (mutagenized) chromosome between markers D5Mit210 and D5Mit314 (also distal to Rw) had a lower FFT than heterozygotes or noncarriers (data not shown).
Figure 4.
The Beh5Jcs1 mutant. (A) Activity level in the hole board is presented for C57BL/6J (B6) (n = 16), C3H/HeJ (n = 9), B6xC3H F1 (n = 25), nonaffected G3 progeny (n = 118), Rw/+* (n = 5) and +*/+* test class Beh5Jcs1 littermates (n = 10). There is significant difference between nonaffected G3 and Beh5Jcs1 +*/+* using the two-tailed t-test (P-value 0.0018). (B) Genetic mapping of the activity phenotype in 71 F2 progeny from a Beh5Jcs1+*/+ X Beh5Jcs1+*/+ intercross using seven microsatellite markers (abbreviated by exchanging the prefix “M” for “D5Mit”) along Chr 5: M146(8.5Mb), M387(26.6Mb), M183(52.6Mb), M201(74.5Mb), M309(78.2Mb), M312(93.7Mb), M10(101.6Mb), M314(108.4Mb), and M95(123.5Mb). Black bars represent values observed in mice homozygous for C57BL/6J (B/B), white bars for homoygous C3H/HeJ loci (C/C), and striped heterozygous for the two strains (B/C). The significant P-values from the two-tailed t-test for M10 B/B vs B/C and B/B vs C/C (n = 37) are 0.0027 and 0.0006, respectively.
Discussion
Random mutagenesis of the mouse genome as a functional genomics strategy has become more popular as burgeoning genomic resources have facilitated positional cloning. There are two basic ENU mutagenesis paradigms, genome-wide and regional. Genome-wide screens can be conducted for both dominant and recessive mutations. For genome-wide recessive screens, three generations of breeding are required to produce animals (or embryos) homozygous for induced mutations. The major advantage of genome-wide screens is that they cast a wide net, enabling the recovery of mutations on all of the autosomes. The disadvantage is that the location of the mutations underlying phenotypes are unknown, necessitating linkage mapping. In situations where homozygotes are inviable or sterile, this can create particular difficulties and added work, as discussed below.
Region-specific saturation mutagenesis has been utilized in Drosophila melanogaster for quite some time (Hoogwerf et al. 1988; Underwood et al. 1990; Clegg et al. 1993; Heitzler et al. 1993; Littleton and Bellen 1994). The major advantages are that the locations of the induced mutations are known, and since some kind of visual marking scheme is typically used, only the subset of animals that is homozygous for the ENU-mutagenized target region needs to be phenotype screened. The major drawbacks of this approach are that only a small portion of the genome is scanned, and special chromosome reagents are required.
The availability of deletion and balancer stocks in flies made regional mutagenesis commonplace and accessible. Until recently, region-specific mutagenesis endeavors in mice were rather limited, due to the lack of these reagents. However, the development of strategies for making chromosomal rearrangements and deletions (Ramirez Solis et al. 1995; You et al. 1997) has eliminated this restriction.
Region-specific mutagenesis is especially well suited for the isolation of genes which, when mutated, cause embryonic lethality or infertility. While there have been impressive reports of successful genome-wide ENU screens for recessive lethal mutations (Kasarskis et al. 1998; Herron et al. 2002), they required the dissection of carrier females to identify the mutations, and were necessarily directed at specific gestational time points. Region-directed screens enable the detection of lethal mutations acting at any stage of development by “loss-of-class” phenotype as we report here; no experimental intervention is required to identify pedigrees harboring a lethal mutation in the target region. There are also important advantages over genome-wide screens for identifying sterility mutants. Although we have used genome-wide screens to isolate and clone new infertility mutations, (Libby et al. 2002, 2003; Ward et al. 2003), one of the difficulties of this approach is that until they are mapped, maintenance of a sterile mutation is difficult. Carriers must be progeny tested, a process in which potential heterozygotes are mated to known or other potential heterozygotes to see whether any offspring are sterile, indicating that both parents are indeed heterozygotes. This problem is eliminated with regional mutagenesis.
With respect to using a deletion or inversion for regional mutagenesis, there are pros and cons. The two disadvantages of an inversion are (1) it requires an extra generation of breeding; and (2) because mice are intercrossed at the G2 generation, ENU-induced mutations throughout the genome can be rendered homozygous. Thus, when the non-Rw G3 animals are phenotyped, it is very commonplace that new variants are detected that do not map to the target region. While it is not an overwhelming problem, it can add extra work in validating the genetic basis of new variant phenotypes. The silver lining of this “problem” is that some of the mutations that map elsewhere in the genome are of sufficient interest to pursue, as Kile and colleagues have shown (Kile et al. 2003). For the most part, the advantages of the inversion screen more than offset these disadvantages. While an extra generation is required, a larger region of DNA is screened in every mutation family, compared with individual deletions (although this is offset to some degree by a lower resolution map position).
A resource of nested deletions provides a valuable tool for mapping the ENU-induced point mutations induced in a regional inversion screen. However, in the first step of obtaining a low-resolution map position of a mutation, standard recombination is about as effective. Using the formula of Durrett et al. (2002), about 50 F2 offspring are required to obtain a 10-Mb mapping resolution at 95% confidence using the current value of 2932 Mb comprising the mouse genome and a genetic length of 1355 cM (Rowe et al. 2003). Scanning a 50-Mb target region using five deletions of 10 Mb each would use a similar number of mice (∼40; on average, four to rule out a deletion region, plus 24 to implicate a deletion at P < 0.05), but this assumes 100% viability and Mendelian transmission of the deletions, which is often not the case. Whereas recombinational mapping requires genotyping of all offspring to localize breakpoints, making useful deletion sets in mice requires substantial up-front effort. Furthermore, as illustrated here in the case of the haplolethal region demarcated proximally by D5Jcs58 at 60.7 Mb and distally by D5Mit110 at 65.4 Mb, certain regions of the genome are refractory to segmental haploidy. Nevertheless, once the rough map location of a mutation is known, the availability of nested deletion sets become very useful for rapid, higher resolution mapping. The ∼30 Mb spanned by the 12 germ-line-transmitted deletions shown in Figure 3 represents 24 breakpoints with an average (albeit uneven) spacing of 1.25 Mb, and only the relevant deletions in the target deletions need be complementation tested. Achieving such resolution recombinationally at 95% confidence would require 835 F2 offspring (Durrett et al. 2002), each of which would require genotyping to identify recombinants.
The generation of multiple alleles of single genes is a typical outcome of regional mutagenesis experiments, and some genes are much more frequent targets than others (Rinchik and Carpenter 1999). One approach to determine allelism would be to perform complementation tests in all possible combinations with each mutant. This would entail 666 crosses [(372–37)/2] for all pairwise combinations of the 37 lethals reported here. Alternatively, the mapping of mutations into different regions automatically eliminates possible allelism between sets of mutations, thus subdividing those mutations that are potentially allelic to a much smaller number. This may also help determine whether any of the lethal mutations in our collection are allelic to any of the previously identified 16 lethal genes in the Rw interval. For example, the gastrulation mutant L5Jcs8 maps to the same small interval and has a similar phenotype as the lazy mesoderm ENU-induced mutant in Ugdh (Garcia-Garcia and Anderson 2003).
This project also served as a pilot screen for identification of behavioral mutations in a battery of assays. We performed behavioral assessment on only 8% of the pedigrees (83/1003). Progeny testing on the initially selected 10 outliers resulted in the validation of the abnormal phenotype in only two pedigrees, the Beh5Jcs1 mutation, which causes decreased activity in the hole board, and Beh5Jcs2, which causes decreased strength of the circadian cycle. We presume that a mixed genetic background (C57BL/6J and C3H/HeJ) contributed to subtle and variable phenotypes in the observed behavioral mutants. Therefore, it was necessary to perform progeny testing in the absence of a balancer chromosome and genetic mapping of abnormal phenotypes at several loci along Chr 5 to confirm the presence of mutant alleles. Notably, both mutations are distal to the Rw inversion.
Whole-genome ENU mutagenesis screens performed in this lab using the same treatment regimens (Ward et al. 2003), plus an ongoing large-scale screen for reproductive mutants using identical ENU mutagenesis protocols (http://reprogenomics.jax.org/), reveal that about 12%–13% of pedigrees on average contain an infertility mutation. The screen of the Rw region, which contains only 1/75th of all the estimated genes in the genome, produced two infertility mutants, or 15% of the pedigrees. Thus, there is no indication that this region is relatively enriched or devoid of fertility loci.
This and other region-directed mutagenesis projects, in which lethals were detectible as a loss-of-class, and therefore immune to phenotyping bias, offer insight into the number of genes required for embryonic development in mammals. Table 4 summarizes the results of this and three other regional ENU mutagenesis screens. Extrapolating these data to estimate the number of such genes in the entire genome depends on knowing the ENU mutagenesis efficiency. While one classical-specific locus test study indicated a mutation rate of 1/700/gamete (Hitotsumachi et al. 1985), the rates at individual loci in the tester strain ranged widely, from no mutations at the a locus to 24 at the p locus. Other studies have indicated much lower mutation rates, but they involved different strains of mice, different test loci, and different ENU treatment regimens (Bode 1984; Rinchik and Carpenter 1999; Barnett et al. 2002). Based on the low frequency of multiple alleles obtained in the Kile et al. (2003) study, the mutation rate is estimated at about 1/1500/locus (K. Hentges and M. Justice, pers. comm.). Considering these various studies, we estimated the number of embryonic lethals genes in the four mutagenized regions of the mouse genome based on a 1/1500/gamete value (Table 4).
Table 4.
Embryonic lethal mutation densities in regional mutagenesis screens
| Region | Chr | Size (Mb) | # gametes | # genes | Lethal compl. groups | Est. # lethal genes | % lethal genes |
|---|---|---|---|---|---|---|---|
| Rw | 5 | 49.1 | 1003 | 336 | ≤34 | 70 | 20.8 |
| Albino | 7 | 13.2 | 4557 | 89 | 7 | 7.3 | 8.2 |
| t complex | 17 | 21.3 | 423 | 429 | ≤17 | 69 | 16.1 |
| Trp53—Wnt3 | 11 | 34.3 | 735 | 764 | ∼30 | 75 | 9.8 |
The lethal genes include embryonic lethal mutations only, not those causing post- or perinatal lethality. The 423 gametes reported in the t complex study represent a composite of multiple breeding paradigms covering the region spanned by T proximally and either tufted (tf) or H-2 distally. The albino target region, and corresponding gene count, corresponds to the interval between D7Mit350 and D7Mit351 (D. Johnson, pers. comm.). The size of the target regions and gene numbers are taken from Emsembl as of February, 2005. The “Lethal compl. groups” category is the number of complementation groups recovered, where known. The number of lethal mutations that were not recovered as mutants was estimated based on a per-locus mutation rate of 1/1500/gamete and calculated as [(1499/1500)P] [L/1-(1499/1500)P] where P = # gametes; L = # complementation groups. This was added to the number of complementation groups to obtain the “Est. # lethal genes.” The “% lethal genes” is calculated as the “Est. # lethal genes”/“# genes”)*100. The references for the albino, t complex, and Trp53—Wnt3 studies are Rinchik and Carpenter (1999), Shedlovsky et al. (1988), and Kile et al. (2003), respectively. The latter study uncovered 55 lethal mutations in total, representing 51 complementation groups (K. Hentges and M. Justice, pers. comm.), 30-32 of which acted embryonically.
The percentage of embryonic lethal genes ranged from 8.2% to 20.8%, suggesting variation in the nature of gene-function content between regions. Combined, an estimated 13.7% of all of the genes in these regions (221 of 1618) are required for embryonic development to term. Given the estimated 25,397 active protein-coding mouse genes (February 2005 Ensembl tabulation, release 28.33d.1), which is roughly consistent with human gene number estimates (International Human Genome Sequencing Consortium 2004), our calculations from these mutagenesis studies estimates that the mouse genome contains 3479 embryonic lethal genes.
How accurate is our estimate? To explore this question, we queried the Mouse Genome Database to determine the fraction of targeted mutations that resulted in embryonic lethality (GO term = “survival: embryonic lethality”). In March 2004, the database contained records of at least one targeted allele in 1759 different genes. There were 383 genes (21.8%) in which at least one of these targeted alleles caused embryonic lethality. There are several possibilities for the difference between this measure and our estimate of 13.7%. First, a perusal of the MGD embryonic lethal genes includes several instances where lethality occurs only in certain strain backgrounds. Second, some knockouts were classified as lethal in MGD, but in reality, displayed reduced penetrance (frequency of homozygotes was less than Mendelian expectations). Third, the four regions subjected to ENU mutagenesis may have an underrepresentation of lethal genes. Fourth, there may be a bias in genes that investigators have subjected to gene targeting, in that such genes may be more likely to be evolutionarily conserved and have essential functions. Fifth, the experimental ENU mutagenesis frequencies may be lower than 1/1500. Finally, 49 of the MGD lethal genes were also classified as perinatal lethals (Go term = “survival: perinatal lethality”). If these are eliminated from the embryonic lethal class, this would lead to a downwardly revised calculation of gene-targeted lethals of 19%. Taking these issues into consideration, it seems reasonable to estimate that 13.7%–19% of all mouse genes result in fully penetrant embryonic lethality when mutated, corresponding to 3479–4825 genes. This is more than the estimated 1400–2400 embryonic-essential genes in zebrafish (6%–10% of 23,524 Ensemble genes in zebrafish genome release 30.4c), which might be attributed partly to the genomic duplication in that organism (Haffter et al. 1996; Taylor et al. 2003; Amsterdam et al. 2004). Mutational analysis of the yeast Saccharomyces cerevisiae indicates that 19% of protein-coding genes are required for viability (Giaever et al. 2002), while 7% of Caenorhabditis elegans genes, as determined by RNAi analysis, are required for embryonic viability (Kamath et al. 2003). The higher percentage of embryonic lethal genes in mice may reflect an inherently more complex or sensitive process of development.
Methods
ENU mutagenesis
Male C57BL/6J (B6) mice, 8–10 wk old, obtained from The Jackson Laboratory, were given three weekly intraperitoneal injections of 80 or 85 mg/kg body weight of ENU. The preparation and quantitation of ENU was done as described (Nolan et al. 1997). Treated animals that proved to be sterile for at least 10 wk following the last injection were mated to C3HeB/FeJ-Rw/+ females, and resulting male G1 progeny were used to establish independent pedigrees that were bred according to the scheme shown in Figure 1 and discussed in the Results. Any G1 offspring displaying overt phenotypic abnormalities, such as causing coat color spotting, were not used.
Notably, Nolan et al. (1997) and Kile et al. (2003) used a 3 × 100 mg/kg regimen on C57BL/6J mice with success. In our hands, however, this dosage was excessive. A total of 55 of 58 C57BL/6J males so treated were rendered permanently sterile. The 80–85 mg/kg level used in our program resulted in 2/3 of the males regaining fertility.
Infertility screen
For each family that was tested for infertility, at least one non-Rw G3 male and female was test mated to determine whether any progeny resulted. If either animal failed to produce offspring, additional G3 animals were fertility tested to confirm this result.
Hearing tests
A click box from the Institute for Hearing Research was used to test for the Preyer reflex (twitch or flick of the pinnae) in non-Rw G3 animals as described in the supplier's manual. The instrument emits a sound burst of 18–20 kHz, to which mice are most sensitive.
Vestibular test
Test-class animals were placed in a beaker of water to assess their ability to swim. Failure to remain upright with their snout above water is an indicator of possible defective vestibular function.
Timed matings and genotyping embryos
To determine the time at which embryos homozygous for lethal mutations succumb, pregnant Rw/+* mothers, which were mated to males of the same genotype, were dissected first at midgestation (E10.5), and the litter was examined for defective, dying, or resorbed embryos. The yolk sac of each was removed for genotyping. Since the mutations were induced on strain B6, and Rw is C3H-derived, the specimens were genotyped with D5Mit148, which is polymorphic between these strains.
Micronucleus assay for genomic instability
The flow cytometric assay for detection of micronuclei in peripheral blood is based on published methods (Dertinger et al. 1996; Torous et al. 2001) and was implemented for the identification of genomic instability mutants as previously described (Shima et al. 2003).
Induction and characterization of deletions at the Qdpr locus
Radiation-induced deletions of the Qdpr locus in ES cells were made using the plasmids and strategy of You et al. (1997). First, a cassette containing the herpes simplex virus thymidine kinase (tk) and neomycin resistance (neo) was inserted into the Qdpr locus of v6.4 F1 hybrid ES cells (C57BL/6J × 129/Jae) by homologous recombination. The targeting vector was constructed as follows. A λ phage clone containing Qdpr was isolated from a C57BL/6N genomic library. Two homology arms were sequentially cloned into the vector pBTΔBam. The 5′ arm was 2.9 kb and contained exon 2 plus most of intron 2. The second homology arm was 4.1 kb and contained most of intron 4 and exon 5. The tk-neo cassette, excised from pBANTKcass, was cloned into a BamHI site between the two arms. This plasmid was linearized with NotI, electroporated into the ES cells, and selected in Geneticin (300 μg/mL, Life Technologies). Clones targeted at Qdpr were identified by Southern blotting. The vector design was such that exons 3 and 4 were entirely deleted. Analysis of mice derived from a targeted line revealed lack of Qdpr function (data not shown).
Second, targeted clones were maintained under G418 selection (300 μg/mL) on neomycin-resistant feeder cells and were trypsinized and exposed to 4 Gy of radiation from a 137Cs source.
Approximately 1 × 106-irradiated cells were plated onto 150-mm plates without feeders, and FIAU selection was begun after 72 h. As in similar experiments involving other loci (Bergstrom et al. 2003), the parental Qdpr-targeted ES cell line selected for making deletions initially exhibited a high background of FIAU-resistant clones, which would complicate the identification of deletions. Hence, deletions were induced from a subclone (targeted on the 129 allele) exhibiting no background FIAU resistance (robust tk expression). DNA was extracted from these FIAU-resistant clones to characterize the extent of the deletions, using microsatellite markers polymorphic between the parental alleles. Primers for the D5Jcs58 locus are, from 5′ to 3′, TTTTCTC CCAGTGTTCTTTGC and CCTGACCAACCATGTTTTCC. This locus maps to Mb position 60.7 in the Mouse Genome Build 33.
Generation of chimeras by microinjection
Ten to 15 cells from deletion-bearing ES cell clones were injected into 3.5 dpc C57BL/6J blastocysts. In some cases, resulting chimeras were mated to C3H-a/a females. Since the deletion-bearing ES cells were of the genotype (129/S4SvJae × C57BL/6J)F1, offspring produced from matings of chimeras to C57BL/6J females produced were either agouti (and thus ES-cell derived) or black (ES-cell derived or from the host blastocysts). Typically, the agouti offspring were typed with a microsatellite marker within the deletion region that was polymorphic between C3H and B6. Those displaying only the C3H allele contain the deletion. In other cases, chimeras with high percent ES cell contribution (nearly 100% agouti) were mated to C3H-Rw/+ females, and all of the offspring (agouti) were typed for the presence of the deletion.
Behavioral tests
The behavioral screen was performed at the University of Pennsylvania on members from two to four pedigrees at a time, composed of both Rw/+* and +*/+*progeny. The order of behavioral tests was as follows: (1) zero maze, (2) accelerating rotarod, (3) acoustic startle and prepulse inhibition (320 G3 mice); or (1) hole board, (2) accelerating rotarod, (3) acoustic startle and prepulse inhibition, and (4) wheel-running activity (214 G3 mice). Each test was separated by 72 h. Mice, 8–12 wk-of-age, were tested in the morning (9:00 a.m.–12:00 p.m.) or afternoon (1:00 p.m.–4:00 p.m.). Equipment used for behavioral testing and the procedures for zero maze, rotarod, and acoustic startle response tests have been described (Tarantino et al. 2000). The Hole board (San Diego Instruments) was used to assess exploratory behavior and home-cage activity (File and Wardill 1975). This apparatus consists of a box with 16 evenly spaced cylindrical holes. Infrared beams located around the perimeter of the board measure activity in the center versus the periphery, whereas infrared beams inside the holes record nose “pokes.” The hole board tests involve recording of three measures over a 10-min period as follows: (1) the number of rearings, (2) exploratory attempts (nose pokes), and (3) total activity. Finally, circadian wheel-running activity was measured by placing mice into individual cages equipped with running wheels within ventilated chambers with controlled lighting. Mice were initially monitored in a 12 h light/12 h dark cycle for 7 d to assess entrainment to light. Subsequently, locomotor activity was recorded in constant darkness (DD) for several weeks to assess the circadian period and the amplitude of circadian periodicity. The amplitude of the Fourier periodogram over 24 h for 15 d in DD was used as an indicator of the strength of the circadian cycle. The FFT analysis was performed using Clock Lab (Actimetrics) with the circadian peak expressed as relative power of the entire curve normalized to 1.
Statistical analysis of behavioral phenotypes
The statistical software in Excel (Microsoft) was used for analysis of behavioral data. G3 mice (Rw/+* and +*/+*) were compared with inbred baseline strains (C57BL/6J, C3H/HeJ, and their F1 and F2 progeny) to ascertain aberrant behaviors. Significance of results was analyzed using the t-test using P < 0.05 as the threshold for significant deviations from baseline in pairwise group comparisons. To identify anomalies of circadian amplitude, mice with amplitude of periodicity greater than three standard deviations from the mean in wild-type mice (C3H/HeJ/C57BL/6J F1 mice) were considered as potential mutants.
Recombinational mapping
Meiotic mapping was conducted for nine lethal (Table 3) and two behavioral (Beh5Jcs1 and Beh5Jcs2) mutations. Rw/+* mice were crossed to C3H/HeJ (or C3H/FeJ), and progeny from the crosses were intercrossed to generate F2 progeny. Tails DNAs were genotyped with an initial set of 20 simple sequence length polymorphism (SSLP) markers that are polymorphic between B6 and C3H. PCR amplification was performed with the following fluorescently labeled markers: D5MIT146, D5MIT387, D5MIT183, D5MIT201, D5MIT10, D5MIT314, D5MIT95 (ABI PRISM Mouse Mapping Set), and PCR products were sized on an ABI 3100, while PCR products generated by nonfluorescent markers were separated on NuSieve agarose gels. Linkage analysis was performed using MapManager QTX (Manly et al. 2001).
Acknowledgments
This work was supported by grants HD35984 to J.S., HD24180 to M.B., and a Postdoctoral Fellowship from The Jackson Laboratory to Y.C. The mutant lines, while extant, will be made available to academic researchers upon request. We thank Amy Sears for excellent technical assistance.
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.3826505. Article published online before print in July 2005.
References
- Amsterdam, A., Nissen, R.M., Sun, Z., Swindell, E.C., Farrington, S., and Hopkins, N. 2004. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. 101: 12792–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett, L., Tyl, R., Shane, B., Shelby, M., and Lewis, S. 2002. Transmission of mutations in the lacI transgene to the offspring of ENU-treated Big Blue male mice. Environ. Mol. Mutagen. 40: 251–257. [DOI] [PubMed] [Google Scholar]
- Batchelor, A.L., Phillips, R.J., and Searle, A.G. 1966. A comparison of the mutagenic effectiveness of chronic neutron- and γ-irradiation of mouse spermatogonia. Mutat. Res. 3: 218–229. [DOI] [PubMed] [Google Scholar]
- Bergstrom, D.E., Bergstrom, R.A., Munroe, R.J., Lee, B.K., Browning, V.L., You, Y., Eicher, E.M., and Schimenti, J.C. 2003. Overlapping deletions spanning the proximal two-thirds of the mouse t complex. Mamm. Genome 14: 817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode, V.C. 1984. Ethylnitrosourea mutagenesis and the isolation of mutant alleles for specific genes located in the T region of mouse chromosome 17. Genetics 108: 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg, N., Whitehead, I., Brock, J., Sinclair, A., Mottus, R., Stromotich, G., Harrington, M., and Grigliatti, T. 1993. Cytogenetic analysis of chromosomal region 31 of Drosophila melanogaster. Genetics 134: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dertinger, S.D., Torous, D.K., and Tometsko, K.R. 1996. Simple and reliable enumeration of micronucleated reticulocytes with a single-laser flow cytometer. Mutat. Res. 371: 283–292. [DOI] [PubMed] [Google Scholar]
- Durrett, R.T., Chen, K.Y., and Tanksley, S.D. 2002. A simple formula useful for positional cloning. Genetics 160: 353–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- File, S.E. and Wardill, A.G. 1975. The reliability of the hole-board apparatus. Psychopharmacologia 44: 47–51. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia, M.J. and Anderson, K.V. 2003. Essential role of glycosaminoglycans in Fgf signaling during mouse gastrulation. Cell 114: 727–737. [DOI] [PubMed] [Google Scholar]
- Giaever, G., Chu, A.M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Haffter, P., Granato, M., Brand, M., Mullins, M.C., Hammerschmidt, M., Kane, D.A., Odenthal, J., van Eeden, F.J., Jiang, Y.J., Heisenberg, C.P., et al. 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123: 1–36. [DOI] [PubMed] [Google Scholar]
- Heitzler, P., Coulson, D., Saenz-Robles, M., Ashburner, M., Roote, J., Simpson, P., and Gubb, D. 1993. Genetic and cytogenetic analysis of the 43A-E region containing the segment polarity gene costa and the cellular polarity genes prickle and spiny-legs in Drosophila melanogaster. Genetics 135: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron, B.J., Lu, W., Rao, C., Liu, S., Peters, H., Bronson, R.T., Justice, M.J., McDonald, J.D., and Beier, D.R. 2002. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat. Genet. 30: 185–189. [DOI] [PubMed] [Google Scholar]
- Hitotsumachi, S., Carpenter, D.A., and Russell, W.L. 1985. Dose-repetition increases the mutagenic effectiveness of N-ethyl-N-nitrosourea in mouse spermatogonia. Proc. Natl. Acad. Sci. 82: 6619–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogwerf, A., Akam, M., and Roberts, D. 1988. A genetic analysis of the rose-gespleten region (68C8–69B5) of Drosophila melanogaster. Genetics 118: 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough, R.B., Lengeling, A., Bedian, V., Lo, C., and Bucan, M. 1998. Rump white inversion in the mouse disrupts dipeptidyl aminopeptidase-like protein 6 and causes dysregulation of Kit expression. Proc. Natl. Acad. Sci. 95: 13800–13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kasarskis, A., Manova, K., and Anderson, K.V. 1998. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc. Natl. Acad. Sci. 95: 7485–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile, B.T., Hentges, K.E., Clark, A.T., Nakamura, H., Salinger, A.P., Liu, B., Box, N., Stockton, D.W., Johnson, R.L., Behringer, R.R., et al. 2003. Functional genetic analysis of mouse chromosome 11. Nature 425: 81–86. [DOI] [PubMed] [Google Scholar]
- Libby, B.J., De La Fuente, R., O”Brien, M.J., Wigglesworth, K., Cobb, J., Inselman, A., Eaker, S., Handel, M.A., Eppig, J.J., and Schimenti, J.C. 2002. The mouse meiotic mutation mei1 disrupts chromosome synapsis with sexually dimorphic consequences for meiotic progression. Dev. Biol. 242: 174–187. [DOI] [PubMed] [Google Scholar]
- Libby, B.J., Reinholdt, L.G., and Schimenti, J.C. 2003. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc. Natl. Acad. Sci. 100: 15706–15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton, J. and Bellen, H. 1994. Genetic and phenotypic analysis of thirteen essential genes in cytological interval 22F1–2; 23B1–2 reveals novel genes required for neural development in Drosophila. Genetics 138: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon, M.F., Glenister, P.H., Loutit, J.F., Evans, E.P., and Peters, J. 1984. A presumed deletion covering the W and Ph loci of the mouse. Genet Res. 44: 161–168. [DOI] [PubMed] [Google Scholar]
- Manly, K.F., Cudmore Jr., R.H., and Meer, J.M. 2001. Map Manager QTX, cross-platform software for genetic mapping. Mamm. Genome 12: 930–932. [DOI] [PubMed] [Google Scholar]
- Naf, D., Wilson, L.A., Bergstrom, R.A., Smith, R.S., Goodwin, N.C., Verkerk, A., van Ommen, G.J., Ackerman, S.L., Frankel, W.N., and Schimenti, J.C. 2001. Mouse models for the Wolf-Hirschhorn deletion syndrome. Hum. Mol. Genet. 10: 91–98. [DOI] [PubMed] [Google Scholar]
- Nolan, P.M., Kapfhamer, D., and Bucan, M. 1997. Random mutagenesis screen for dominant behavioral mutations in mice. Methods 13: 379–395. [DOI] [PubMed] [Google Scholar]
- Ramirez Solis, R., Liu, P., and Bradley, A. 1995. Chromosome engineering in mice. Nature 378: 720–724. [DOI] [PubMed] [Google Scholar]
- Rinchik, E.M. and Carpenter, D.A. 1999. N-ethyl-N-nitrosourea mutagenesis of a 6- to 11-cM subregion of the Fah-Hbb interval of mouse chromosome 7: Completed testing of 4557 gametes and deletion mapping and complementation analysis of 31 mutations. Genetics 152: 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderick, T. 1983. Using inversions to detect and study recessive lethals and detrimentals in mice. In Utilization of mammalian specific-locus studies in hazard evaluation and estimation of genetic risk (eds. F. deSerres and W. Sheridan), pp. 135–167. Plenum, New York.
- Rowe, L.B., Barter, M.E., Kelmenson, J.A., and Eppig, J.T. 2003. The comprehensive mouse radiation hybrid map densely cross-referenced to the recombination map: A tool to support the sequence assemblies. Genome Res. 13: 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimenti, J. and Bucan, M. 1998. Functional genomics in the mouse: Phenotype-based mutagenesis screens. Genome Res. 8: 698–710. [DOI] [PubMed] [Google Scholar]
- Schimenti, J., Libby, B., Bergstrom, R., Wilson, L., Naf, D., Tarantino, L., Alavizadeh, A., Lengeling, A., and Bucan, M. 2000. Interdigitated deletion complexes on mouse chromosome 5 induced by irradiation of embryonic stem cells. Genome Res. 10: 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlovsky, A., Guenet, J.-L., Johnson, L., and Dove, W. 1986. Induction of recessive lethal mutations in the T/t-H-2 region of the mouse genome by a point mutagen. Genet. Res. Camb. 47: 135–142. [DOI] [PubMed] [Google Scholar]
- Shedlovsky, A., King, T.R., and Dove, W.F. 1988. Saturation germ line mutagenesis of the murine t region including a lethal allele at the quaking locus. Proc. Natl. Acad. Sci. 85: 180–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima, N., Hartford, S., Duffy, T., Wilson, L., Schimenti, K., and Schimenti, J.C. 2003. Phenotype based identification of mouse chromosome instability mutants. Genetics 163: 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson, D.A., Lee, K.-H., Nagle, D., Yen, C.-H., Morrow, A., Miller, D., Chapman, V., and Bucan, M. 1994. Mouse rump-white mutation associated with an inversion of chromosome 5. Mamm. Genome 5: 342–348. [DOI] [PubMed] [Google Scholar]
- Tarantino, L.M., Gould, T.J., Druhan, J.P., and Bucan, M. 2000. Behavior and mutagenesis screens: The importance of baseline analysis of inbred strains. Mamm. Genome 11: 555–564. [DOI] [PubMed] [Google Scholar]
- Taylor, J.S., Braasch, I., Frickey, T., Meyer, A., and Van de Peer, Y. 2003. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 13: 382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torous, D.K., Hall, N.E., Dertinger, S.D., Diehl, M.S., Illi-Love, A.H., Cederbrant, K., Sandelin, K., Bolcsfoldi, G., Ferguson, L.R., Pearson, A., et al. 2001. Flow cytometric enumeration of micronucleated reticulocytes: High transferability among 14 laboratories. Environ. Mol. Mutagen. 38: 59–68. [DOI] [PubMed] [Google Scholar]
- Underwood, E., Briot, A., Doll, R., Ludwiczak, R., Otteson, D., Tower, J., Vessey, K., and Yu, K. 1990. Genetics of 51D-52A, a region containing several maternal-effect genes and two maternal-specific transcripts in Drosophila. Genetics 126: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, J.O., Reinholdt, L.G., Hartford, S.A., Wilson, L.A., Munroe, R.J., Schimenti, K.J., Libby, B.J., O'Brien, M., Pendola, J.K., Eppig, J., et al. 2003. Toward the genetics of mammalian reproduction: Induction and mapping of gametogenesis mutants in mice. Biol. Reprod. 69: 1615–1625. [DOI] [PubMed] [Google Scholar]
- Ying, H.C., Hurle, B., Wang, Y., Bohne, B.A., Wuerffel, M.K., and Ornitz, D.M. 1999. High-resolution mapping of tlt, a mouse mutant lacking otoconia. Mamm. Genome 10: 544–548. [DOI] [PubMed] [Google Scholar]
- You, Y., Bergstrom, R., Klemm, M., Lederman, B., Nelson, H., Ticknor, C., Jaenisch, R., and Schimenti, J. 1997. Chromosomal deletion complexes in mice by radiation of embryonic stem cells. Nat. Genet. 15: 285–288. [DOI] [PubMed] [Google Scholar]
Web site references
- http://reprogenomics.jax.org/; The Jackson Laboratory.