Abstract
We report on our initial genetic linkage studies of schizophrenia in the genetically isolated population of the Afrikaners from South Africa. A 10-cM genomewide scan was performed on 143 small families, 34 of which were informative for linkage. Using both nonparametric and parametric linkage analyses, we obtained evidence for a small number of disease loci on chromosomes 1, 9, and 13. These results suggest that few genes of substantial effect exist for schizophrenia in the Afrikaner population, consistent with our previous genealogical tracing studies. The locus on chromosome 1 reached genomewide significance levels (nonparametric LOD score of 3.30 at marker D1S1612, corresponding to an empirical P value of .012) and represents a novel susceptibility locus for schizophrenia. In addition to providing evidence for linkage for chromosome 1, we also identified a proband with a uniparental disomy (UPD) of the entire chromosome 1. This is the first time a UPD has been described in a patient with schizophrenia, lending further support to involvement of chromosome 1 in schizophrenia susceptibility in the Afrikaners.
Introduction
Schizophrenia (MIM 181500) is a severe mental disorder affecting 1% of the world population and is characterized by psychotic symptoms and by cognitive, affective, and psychosocial impairment. There is substantial evidence that schizophrenia has a genetic basis (Gottesman and Shields 1982; Tsuang and Faraone 1994). For example, concordant diagnosis of schizophrenia is more than two times higher for MZ twins (41%–65% concordance) than for DZ twins (0%–28% concordance) and other first-degree relatives of schizophrenic individuals (for a review, see Cardno and Gottesman [2000]). It seems clear that multiple gene loci and environmental factors influence susceptibility to schizophrenia. Understanding of the role of individual genes in the disorder, however, remains limited.
Over the past few years, several searches for susceptibility loci have been undertaken. Genomewide linkage scans have suggested that susceptibility loci may be present on several chromosomes (Coon et al. 1994; Moises et al. 1995; Blouin et al. 1998; Faraone et al. 1998; Levinson et al. 1998; Hovatta et al. 1999; Rees et al. 1999; Williams et al. 1999; Bailer et al. 2000; Brzustowicz et al. 2000; Ekelund et al. 2000; Camp et al. 2001; Gurling et al. 2001; Paunio et al. 2001; DeLisi et al. 2002a, 2002b; Devlin et al. 2002; Straub et al. 2002). For most implicated regions, both positive and negative replication studies have been reported. In addition to locus heterogeneity, differences in diagnostic and ascertainment procedures, analytical methods, and sample configuration and size may help explain differences in the results. Ethnic heterogeneity might also be a contributing factor, since families studied in the initial genome scans were drawn mostly from the genetically diverse populations of Europe and the United States.
More recently, genetic studies of schizophrenia have employed genetically isolated populations from Finland (Hovatta et al. 1999; Ekelund et al. 2000; Paunio et al. 2001), Sweden (Lindholm et al. 2001), Iceland (Stefansson et al. 2002), Quebec (Maziade et al. 2002), Costa Rica (DeLisi et al. 2002a), and two Micronesian islands: Palau (Camp et al. 2001; Devlin et al. 2002) and Kosrae (Wijsman et al. 2003). It is possible that, in these populations, there will be fewer susceptibility loci and alleles (de la Chapelle and Wright 1998; Ober and Cox 1998). In addition, founder populations often exhibit less environmental heterogeneity than do other populations and allow detailed genealogical research and reconstruction of extended multigenerational pedigrees (Peltonen et al. 2000). Availability of such large pedigrees should facilitate the detection of linkage (Wijsman and Amos 1997; Dyer et al. 2001). Despite these potential advantages, it must be noted that the outcome of studies of complex traits in an isolated population will depend greatly on the particular demographic characteristics and historical circumstances surrounding the founding of the population.
Here, we report the results of our initial genetic linkage studies of schizophrenia in the genetically isolated population of the Afrikaners from South Africa. The Afrikaners descended from a small number (1,000–2,000) of immigrants, primarily of Dutch origin, who settled in the Cape in 1652. These immigrants spread inland, with splinter groups forming small, geographically isolated communities. Because of language differences (the Afrikaans language is derived from Dutch) and religious practices (most Afrikaners were members of the Dutch Reformed Church), the Afrikaners remained isolated. The population expanded over 13–15 generations almost entirely through reproduction, since immigration subsequent to the founding was minimal. In the early generations, consanguinity was common. The Afrikaner population, which is currently 3 million, has expanded in relative genetic isolation, as is evident from the unusually high frequency of certain rare Mendelian disorders (e.g., variegate porphyria), the low diversity of the associated allelic variants, and the large extent (8–11 cM) of conserved haplotypes around disease genes (Hayden et al. 1980; Brink et al. 1987; Rosendorff et al. 1987; Leitersdorf et al. 1989; Torrington and Viljoen 1991; Brink et al. 1995; Pronk et al. 1995; Goldman et al. 1996; Warnich et al. 1996; Groenewald et al. 1998; Roby et al. 1999).
In a previous study, we described our ongoing efforts to document the genealogical history of a sample of Afrikaner individuals with schizophrenia (Karayiorgou et al. 2004). We traced the ancestry of 98 probands with schizophrenia. It was remarkable that we found that 87 of them descended from a single couple who immigrated to South Africa ∼12.5 generations ago. This particular line of descent has not, to our knowledge, been described previously for any other disorder studied in the Afrikaner population (M.T., unpublished data). These results suggest that the affected individuals in this population may share a small number of disease alleles. In this setting, linkage and association scans hold exceptional promise, and the present article reports results from an initial scan for linkage in this population.
Material and Methods
Sample Recruitment
A collaborative project between the Laboratory of Human Neurogenetics at the Rockefeller University and two major psychiatric hospitals in South Africa was undertaken, with the goal of ascertaining and evaluating Afrikaner probands with a history of psychotic illness and their families for a genetic study. The participating hospitals were the Weskoppies Hospital in Pretoria, an academic hospital affiliated with the University of Pretoria, and the Valkenberg Hospital, which is affiliated with the University of Capetown. The Afrikaner families in our study were recruited from the regions of Tshwane (formerly known as Pretoria), Limpopo (formerly Northern Transvaal), and Mpumalanga (formerly the Eastern Transvaal), as well as from the greater Cape area. The study was approved by institutional review boards at all sites (Rockefeller University, University of Pretoria, and University of Capetown). Appropriate written informed consent was obtained from all study participants. A detailed description of our sample is available elsewhere (Karayiorgou et al. 2004).
Clinical Evaluation
The Diagnostic Interview for Genetic Studies (DIGS) (Nurnberger et al. 1994), after being translated and back-translated into Afrikaans, was the diagnostic instrument of choice. The DIGS was developed at the National Institute of Mental Health Diagnostic Centers for Psychiatric Linkage Studies and was specifically designed for the diagnosis of schizophrenia, schizoaffective disorder, major affective disorders, and anxiety disorders in genetic research. All diagnostic interviews were conducted in person by specially trained clinicians. Specifically, two psychiatrists and one clinical psychologist with a minimum of 10 years of clinical experience each were specially trained in the use of the DIGS and in the research application of the Diagnostic and Statistical Manual–4th Edition (DSM-IV) (American Psychiatric Association 1994). On the basis of information gathered in the DIGS, the clinical interviewers assigned appropriate diagnoses according to the DSM-IV. After each interview, diagnosticians completed (a) DSM-IV checklists for each category in which positive symptoms were identified in the DIGS; (b) a narrative chronologic summary of prodromal traits, symptom onset, and functional impairment; and (c) an English-language data summary form of selected DIGS items.
Ongoing reliability studies between the Afrikaner clinicians, the U.S. clinicians who conducted the diagnostic evaluations for our U.S.-based genetic schizophrenia sample, and the U.S. investigator who trained all the clinicians show >90% agreement for DSM-IV diagnoses. Reliability exercises consisted of yearly reliability interviews between all the clinicians, as well as review of videotapes of an additional two interviews per interviewer.
Affected Status Definitions
For the genetic linkage analysis, we used three phenotypic liability classes (LCs) (Straub et al. 1995; Kendler et al. 1996). LC I comprises the “core schizophrenic phenotypes” of schizophrenia and depressed-type-only schizoaffective disorder (family data suggest that the two diagnoses are alternative expressions of the same genotypes [Cloninger 1989]). LC II combines the LC I cases with all psychotic disorders, including primarily cases with schizoaffective disorder of mainly affective course, as well as some cases with bipolar disorder with psychotic symptoms, psychotic depression, and psychosis not otherwise specified (NOS). LC III combines the LC II cases with all psychiatric disorders, including depression, anxiety disorders, and ethanol dependence.
Family Sample
The entire genotyped sample includes 173, 205, and 253 individuals in LCs I, II, and III, respectively. These individuals are organized into 143 small families and >137 relative pairs (including 79 parent-child pairs, 35 sib pairs, 15 avuncular pairs, 4 grandparent-grandchild pairs, and 4 cousin pairs). Thirty-four of these families are informative for linkage (fig. 1). The remaining 110 are useful for allele frequency estimation and transmission disequilibrium testing. This analysis will be described elsewhere. The 34 families that are informative for linkage include 62, 81, and 100 individuals in LCs I, II, and III, respectively. Of the 19 cases included in LC II but not LC I, 10 meet criteria for schizoaffective disorder of mainly affective course, and 9 meet the criteria for other psychotic disorders.
Figure 1.
Informative pedigrees included in linkage analyses. Black coloring indicates affected individuals in LC I, and dark and light gray coloring indicate individuals in LCs II and III, respectively.
Genomewide Screening
DNA from all study participants was extracted from 24 ml of EDTA-treated blood, according to standard procedures (Ciulla et al. 1988). In addition to affected individuals, we collected DNA samples for genotyping from first-degree relatives (parents and siblings) whenever possible. A genomewide screen was performed at the Center for Inherited Disease Research (CIDR). A total of 602 samples was screened using 388 fluorescent microsatellite markers. The marker set was composed primarily of trinucleotide and tetranucleotide repeat markers and was based on the Marshfield Genetics version 8 screening set, available for purchase from Research Genetics (∼10% of the marker loci are different between the CIDR marker set and Marshfield version 8) (Center for Medical Genetics Web site; CIDR Human Marker Set Web site). The average spacing of the markers used was 9 cM, with one gap >20 cM. The average heterozygosity of the markers used was 0.76. CEPH samples 1331-1 and 1331-2 were included as size standards in all PCRs and gel runs. Internal standards were also included in each gel lane. Gel electrophoresis was performed on an ABI Prism 377 DNA sequencer instrument (Perkin Elmer), and genotypes were assigned using Genotyper 2.0 software (Perkin Elmer). Two individuals scored the alleles independently. Discrepancies were flagged and resolved.
Ten samples did not amplify for >25% of the markers and were dropped from the analysis. Of these 10, only one sample was from an affected proband; the other 9 were samples from unaffected relatives or parents. The overall error rate was 0.09%, and the inconsistency rate was 0.30%. The missing data rate was 6.6%.
Error Checking
We verified familial relationships through use of the GRR (Abecasis et al. 2001b) and RELPAIR (Epstein et al. 2000) programs. Four families showed segregation problems that were consistent with a sample switch in two of the families and with nonpaternity in the other two. After resolving problems in family structure, we used MERLIN (Abecasis et al. 2002) to check for Mendelian inconsistencies and to identify genotypes associated with an excessive number of recombination events.
Nonparametric Linkage Analysis
We performed genomewide, nonparametric linkage analyses using the SALL statistic (Whittemore and Halpern 1994), as recommended by Sengul et al. (2001). This statistic combines information from all affected individuals in a pedigree and can detect regions of excess identity-by-descent sharing. We conducted these analyses with the MERLIN computer program (Abecasis et al. 2002) and calculated LOD scores as described elsewhere (Kong and Cox 1997). We performed both multipoint and single-point analyses throughout the genome. Although multipoint linkage analysis is theoretically more powerful, it is also more sensitive to errors in genotype data or genetic map specification (Abecasis et al. 2001a; Sullivan et al. 2003). In contrast, single-point analysis is less efficient and more prone to random stochastic fluctuations, but it is more robust to genotyping error and does not require specification of genetic maps (Abecasis et al. 2001a; Sullivan et al. 2003).
Parametric Linkage Analysis
We repeated the linkage analyses using a set of four parametric models for schizophrenia, as described by Brzustowicz et al. (2000), corresponding to dominant and recessive models for LCs I and II. For LC I, when a dominant model was assumed, we set the allele frequency at p=0.0045 with penetrance vector f=[0.75,0.50,0.001]. If we assume a recessive model, p=0.065 and f=[0.50,0.0015,0.0015]. Elements of the penetrance vector f are ordered to denote the probability of disease for genotypes with frequency p2, 2(1-p)p, and (1-p)2, respectively. For LC II, when a dominant model was assumed, p=0.007 and f=[0.90,0.80,009]. For the recessive model, p=0.10 and f=[0.60,0.01,0.01]. These analyses were conducted with the Allegro computer program (Gudbjartsson et al. 2000).
Empirical Significance Levels
We calculated genomewide significance levels through use of 5,000 gene-dropping simulations. In each simulation, we retained the original phenotypes and generated a new data set with the same allele frequencies, marker spacing, phenotypes, and missing data pattern. Any evidence for linkage in these simulated data is due to chance. We analyzed each replicate and recorded the highest peak for each chromosome, to evaluate the false-positive rate. These simulations allowed us to account for uneven marker spacing and informativeness and for the diversity of family structures in our study, as well as to calculate the probability of observing multiple linkage peaks of a certain height. For a discussion of the utility of empirical significance levels in linkage analysis, see Kruglyak and Daly (1998).
Fine Mapping
A denser marker map was generated for the three chromosomal regions where markers showed a LOD score >1.5 in the original scan. The additional markers analyzed were selected from the Marshfield genetic maps, and their order was cross checked with the current genome assembly prior to analysis. CEPH samples 1331-1 and 1331-2 were included as size standards in all PCRs and gel runs. Internal standards were also included in each gel lane. Gel electrophoresis was performed on an ABI 3700 DNA sequencer instrument at the Starr Center Genotyping Facility at the Rockefeller University. Genotypes were assigned using Genotyper 2.5 software (Perkin Elmer). Two individuals scored the alleles independently. Discrepancies were flagged and resolved.
Overall, 19 additional markers (D1S2893, D1S2795, D1S2694, D1S1160, D1S503, D1S2736, D1S434, D1S507, D9S175, D9S153, D9S152, D13S174, D13S248, D13S778, D13S1295, D13S1315, D13S261, D13S293, and D13S1825) were investigated in regions showing evidence of linkage on chromosomes 1, 9, and 13. Marker D1S2694 showed clear evidence for departure from Hardy-Weinberg equilibrium (P<.0007) and was excluded from further analyses.
Results
Nonparametric Linkage Analysis
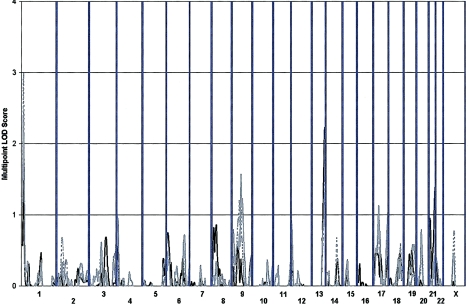
We initially performed single-point, nonparametric linkage analysis (Whittemore and Halpern 1994) using the NPL-ALL statistic (as recommended by Sengul et al. 2001). NPL Z scores were converted into LOD scores, as described elsewhere (Kong and Cox 1997). A summary of all estimated LOD scores is given in figure 2. In addition, observed LOD scores of ⩾1.0 are summarized in table 1. Overall, we observed the strongest evidence for linkage on chromosome 1 at marker D1S1612 (LOD score 2.28, asymptotic P=.0006 for LC I; LOD score 3.21, P=.00006 for LC II; and LOD score 3.30, P=.00005 for LC III). Markers D9S1122 on chromosome 9 (LOD score 1.84, P=.002 for LC II; and LOD score 2.20, P=.0007 for LC III) and D13S1265 on chromosome 13 (LOD score 1.88, P=.002 for LC I) also exhibit strong evidence for linkage.
Figure 2.
Single-point analysis results. Single-point Kong and Cox (1997) nonparametric LOD scores calculated using the SALL scoring statistic are shown. Results are summarized throughout the genome for three phenotypic LCs: LC I (circles), LC II (triangles), and LC III (squares).
Table 1.
Peak Single-Point LOD Scores Observed in the Nonparametric Linkage Analysis
|
LOD Score for |
||||
| Chromosomeand Markera | Locationb(cM) | LC I | LC II | LC III |
| 1: | ||||
| D1S2660 | 11 | … | 1.04 | … |
| D1S1612 | 16 | 2.28 | 3.21 | 3.30 |
| D1S1653 | 164 | 1.14 | … | … |
| 3: | ||||
| D3S2406 | 103 | … | … | 1.00 |
| 4: | ||||
| D4S2366 | 13 | … | … | 1.04 |
| 6: | ||||
| D6S2436 | 155 | 1.08 | … | … |
| 8: | ||||
| D8S1469 | 16 | 1.06 | … | … |
| D8S1130 | 22 | 1.46 | … | … |
| D8S136 | 44 | 1.16 | … | … |
| 9: | ||||
| D9S1122 | 76 | … | 1.84 | 2.20 |
| D9S1826 | 160 | … | … | 1.10 |
| 13: | ||||
| D13S1265 | 99 | 1.88 | 1.04 | … |
| D13S285 | 111 | 1.98 | … | … |
| 17: | ||||
| D17S974 | 22 | 1.41 | … | … |
| 18: | ||||
| D18S858 | 80 | 1.12 | … | 1.12 |
| 21: | ||||
| D21S1446 | 58 | 1.07 | … | … |
| X: | ||||
| DXS6800 | 93 | … | 1.40 | 1.24 |
All markers with a LOD score (Kong and Cox 1997) >1.0 in the single-point analyses are shown.
Marker locations are given according to the Marshfield genetic map.
Empirical Significance Levels
We used gene-dropping simulations to estimate genomewide P values for our observed linkage signals. The results of these simulations are summarized in table 2. In our sample, the probability of observing one or more chromosomes with a LOD score ⩾3.30 for LC III (the LOD score we observed at D1S1612) by chance is P=.007 (see table 2). Less than 1 such signal is expected to occur by chance in every 100 genome scans. Even after accounting for our use of three different phenotypic liability classes, the probability of observing a LOD score of ⩾3.30 is only .012.
Table 2.
Empirical Genomewide P Values for Markers with Observed Nonparametric Peak LOD Scores >1.0
|
Expectedc |
||||||
| LC andRank (r)a | Marker | Locationb | Observed LOD Score (Z) | E(No. of LODScores ⩾Z) | SD | P (r LODs ⩾Z)d |
| LC I: | ||||||
| 1 | D1S1612 | 16 | 2.28 | .07 | .27 | .072 |
| 2 | D13S285 | 111 | 1.98 | .40 | .62 | .060 |
| 3 | D8S1130 | 22 | 1.46 | 1.67 | 1.23 | .230 |
| 4 | D17S974 | 22 | 1.41 | 1.94 | 1.32 | .122 |
| 5 | D18S858 | 80 | 1.12 | 4.16 | 1.81 | .406 |
| 6 | D6S2436 | 155 | 1.08 | 4.56 | 1.88 | .294 |
| 7 | D21S1446 | 58 | 1.07 | 4.66 | 1.90 | .163 |
| LC II: | ||||||
| 1 | D1S1612 | 16 | 3.21 | .01 | .08 | .006 |
| 2 | D9S1122 | 76 | 1.84 | .75 | .84 | .171 |
| 3 | D13S1265 | 99 | 1.04 | 4.74 | 1.89 | .887 |
| LC III: | ||||||
| 1 | D1S1612 | 16 | 3.30 | .01 | .08 | .007 |
| 2 | D9S1122 | 76 | 2.20 | .20 | .54 | .035 |
| 3 | D18S858 | 80 | 1.12 | 3.85 | 1.74 | .771 |
| 4 | D4S2366 | 13 | 1.04 | 4.54 | 1.83 | .698 |
| 5 | D3S2406 | 103 | 1.00 | 4.93 | 1.89 | .571 |
For each phenotypic liability class, the table ranks markers exhibiting the strongest evidence for linkage.
Chromosomal locations are given in centimorgans only; chromosome assignments are implicit in the marker name.
The expected number of chromosomes exhibiting equal or greater LOD scores (and its SD).
The probability that ⩾r LOD scores of equal or greater magnitude would be observed by chance. The probabilities and expected values were estimated from 5,000 sets of genotypes generated under the null (see text). This simulation considered only autosomes 1–22.
For LCs I and II, the highest observed LOD scores are 2.28 and 3.21, respectively, and the probability of observing one or more chromosomes with equal or greater LOD scores by chance is P=.072 and P=.006, respectively. It is noteworthy that, for LC I, we observed evidence for linkage in more chromosomes than expected by chance. For example, at LOD score thresholds of 2.28, 1.98, 1.41, and 1.08, we observed evidence for linkage in one, two, four, and six chromosomes, respectively. In contrast, we would expect to observe evidence for linkage in only 0.07, 0.40, 1.97, and 4.56 chromosomes at each of these thresholds. Although this excess of linkage peaks does not reach genomewide significance levels, it is consistent with an oligogenic basis for schizophrenia, which would produce modestly increased allele sharing at a number of loci rather than large increases in sharing at a few loci (Cookson et al. [2001] present a similar analysis for eczema).
Table 2 also illustrates the utility of calculating analysis- and data-specific thresholds when evaluating genomewide significance levels. For example, at threshold 1.12, 4.16 linkage peaks are expected to occur by chance for LC I, but only 3.85 such peaks are expected for LC III. This is the result of differences in informativeness between the two phenotypic liability classes in our sample (for example, there are more affected individuals per family for LC III, and the proportion of different types of relative pairs is different for each LC).
Multipoint Nonparametric Linkage Analysis
Multipoint results are largely consistent with the single-point analyses (table 3; fig. 3). Strong evidence for linkage is observed on chromosome 1 (near marker D1S1612 at 16 cM), with LOD scores of 1.48 (P=.005), 2.99 (P=.0001), and 2.44 (P=.0004), for LCs I, II, and III, respectively. As in the single-point analyses, two other modest linkage peaks were observed on chromosomes 9 and 13, with peak LOD scores of 1.57 (P=.004) for LC III and 2.23 (P=.0007) for LC I, respectively. Concordance between the multipoint and single-point analyses increases our confidence that these linkage signals are real and not artifacts generated by genotyping or map error.
Table 3.
Peak Multipoint LOD Scores from the Nonparametric Linkage Analysis[Note]
| LC I |
LC II |
LC III |
|||||||
| Chromosomeand NearestMarker | LOD | Locationa(cM) | Intervalb(cM) | LOD | Locationa(cM) | Intervalb(cM) | LOD | Locationa(cM) | Intervalb(cM) |
| 1: | |||||||||
| D1S1612 | 1.48 | 16 | 13–21 | 2.99 | 16 | 9–25 | 2.44 | 16 | 9–25 |
| 9: | |||||||||
| D9S1118 | … | … | … | 1.19 | 61 | 59–64 | … | … | … |
| D9S1122 | … | … | … | 1.32 | 76 | 69–78 | 1.57 | 76 | 71–92 |
| 13: | |||||||||
| D13S285 | 2.23 | 109 | 93–111 | 1.53 | 110 | 101–111 | 1.31 | 109 | 102–111 |
| 17: | |||||||||
| D17S2196 | … | … | … | … | … | … | 1.13 | 45 | 42–46 |
| D17S928 | … | … | … | … | … | … | 1.04 | 123 | 120–126 |
| 21: | |||||||||
| D21S1446 | 1.40 | 56 | 46–58 | … | … | … | … | … | … |
Note.— The table lists all linkage peaks with a LOD score (Kong and Cox 1997) >1.0 in the multipoint analyses.
LOD score location (according to the Marshfield genetic map) for each peak.
Interval in which the LOD score is >1.0.
Figure 3.
Multipoint analysis results. Multipoint Kong and Cox (1997) nonparametric LOD scores calculated using the SALL scoring statistic are shown. Results are summarized throughout the genome for three phenotypic LCs: LC I (solid black line), LC II (dashed line), and LC III (solid gray line).
In inbred samples, LOD scores may be inflated (Genin and Clerget-Darpoux 1996). This is a phenomenon that could result in misleading inferences about linkage. To ensure that this was not the case in the sample of nuclear families we selected for genotyping, we calculated average nonparametric LOD scores (Kong and Cox 1997) at 1-cM intervals throughout the genome, through use of the LC III phenotype definition (data not shown). At each location, we set the sign of the LOD score to positive if there was evidence for excess sharing among affected individuals and to negative otherwise. We observed an average LOD score of 0.03, suggesting that inbreeding is not a serious cause for concern in interpreting our results.
Parametric Linkage Analysis
We first calculated parametric LOD scores for markers on chromosomes 1, 9, and 13, which exhibited the strongest evidence for linkage in multipoint and single-point nonparametric analyses. We considered four previously described disease models (Brzustowicz et al. 2000) and performed analyses both with and without a heterogeneity parameter. We speculated that these analyses might help evaluate different modes of action for susceptibility genes that may be segregating in our population.
Our peak LOD scores in single-marker analyses were 3.26 for marker D1S1612 (chromosome 1, 16 cM, LC II), 2.17 for marker D9S1122 (chromosome 9, 76 cM, LC II), and 1.79 for marker D13S285 (chromosome 13, 111 cM, LC I). Multipoint results also confirmed the results of our nonparametric analyses, although observed LOD scores were somewhat higher on chromosome 13, where the peak LOD score increased to 3.11 (chromosome 13, 106 cM, LC I), and somewhat lower on chromosomes 1 and 9, with peak LOD scores of 2.95 (chromosome 1, 16 cM) and 1.59 (chromosome 9, 75 cM). Analyses allowing for heterogeneity raised observed LOD scores only slightly.
A summary of our parametric analyses, including details of linkage peaks for each chromosome and analysis model, is provided in table 4. Although we observed the strongest evidence for linkage when we assumed a recessive disease model with high penetrance, it is important to note that small pedigrees have very limited utility for distinguishing between different disease models (for a detailed discussion of the issue, see Göring and Terwilliger [2000]), and one must be cautious in assuming that this model is closest to the truth.
Table 4.
Multipoint and Single-Point Parametric Linkage Analysis Results for Chromosomes 1, 9, and 13[Note]
|
Homogeneity |
Heterogeneity |
|||||||||
| Single-Point |
Multipoint |
Single-Point |
Multipoint |
|||||||
| Chromosome,LC, and Model | Marker(Position in cM) | LODa | Position(cM) | LODa | Marker(Position in cM) | αb | HLODc | Position(cM) | αb | HLODc |
| 1: | ||||||||||
| LC I: | ||||||||||
| Dominant | D1S1653 (164) | 1.94 | 14 | .26 | D1S1653 (164) | .00 | 1.94 | 14 | .37 | .86 |
| Recessive | D1S1612 (16) | 2.97 | 16 | 1.34 | D1S1612 (16) | .11 | 3.04 | 16 | .33 | 2.17 |
| LC II: | ||||||||||
| Dominant | D1S1612 (16) | 1.67 | 14 | 1.49 | D1S1612 (16) | .20 | 1.98 | 14 | .21 | 1.74 |
| Recessive | D1S1612 (16) | 3.26 | 16 | 2.95 | D1S1612 (16) | .00 | 3.26 | 16 | .13 | 3.04 |
| 9: | ||||||||||
| LC I: | ||||||||||
| Dominant | D9S1122 (76) | −.24 | 50 | −2.15 | D9S1122 (76) | .34 | 1.06 | 76 | .73 | .28 |
| Recessive | D9S1826 (160) | .69 | 51 | −.35 | D9S1826 (160) | .27 | 1.27 | 164 | .51 | 1.42 |
| LC II: | ||||||||||
| Dominant | D9S1122 (76) | .84 | 50 | −1.99 | D9S1122 (76) | .28 | 1.50 | 76 | .62 | .55 |
| Recessive | D9S1122 (76) | 2.17 | 75 | 1.59 | D9S1122 (76) | .08 | 2.20 | 75 | .24 | 1.88 |
| 13: | ||||||||||
| LC I: | ||||||||||
| Dominant | D13S1265 (99) | .64 | 105 | 1.84 | D13S1265 (99) | .32 | 1.48 | 104 | .12 | 1.86 |
| Recessive | D13S285 (111) | 1.79 | 106 | 3.11 | D13S1265 (99) | .34 | 2.26 | 106 | .09 | 3.15 |
| LC II: | ||||||||||
| Dominant | D13S894 (33) | −1.78 | 106 | −.59 | D13S1265 (99) | .60 | .66 | 104 | .54 | .78 |
| Recessive | D13S1265 (99) | 1.78 | 107 | 2.48 | D13S1265 (99) | .28 | 2.15 | 107 | .13 | 2.54 |
Note.— Boldface italic type indicates the combinations of LC and disease model that resulted in the strongest evidence for linkage.
Parametric LOD score calculated under the assumption of no heterogeneity.
Estimated value at each location.
Parametric LOD score maximized over a heterogeneity parameter α.
We also performed parametric linkage analyses for the rest of the other autosomes. In single-marker analyses, the three markers exhibiting the strongest evidence for linkage were D3S2460 (heterogeneity LOD score [HLOD] 2.34 with α=0.29, using LC I and the recessive model), D18S858 (HLOD 2.28 with α=0.10, using LC II and the recessive model), and D8S1130 (HLOD 1.70 with α=0.33, using LC I and the recessive model). Parametric multipoint analyses did not identify any regions outside chromosomes 1, 9, and 13 where LOD scores were >1.5 or HLODs were >1.5. Detailed tables, including all our parametric linkage results, are available online (authors' Web site).
Fine Mapping
We genotyped additional markers around our chromosome 1, 9, and 13 linkage peaks. We reasoned that these markers might help extract additional linkage information in these regions. In total, we genotyped eight new markers around each of our two highest peaks, on chromosomes 1 and 13, and three additional markers around the chromosome 9 linkage peak. These chromosomes were selected because they provided suggestive evidence for linkage (LOD score >1.5) in parametric and nonparametric analyses, whether single-point or multipoint.
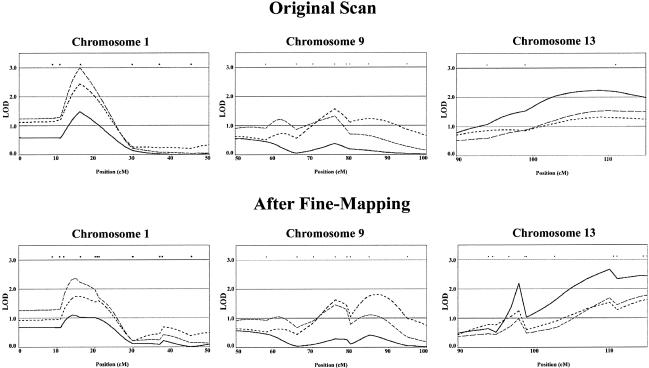
Overall, evidence for linkage to chromosome 1 weakened somewhat, with the peak nonparametric multipoint LOD score decreasing from 2.99 (for LC II at 16 cM) to 2.36 (also for LC II, but at 15 cM). In contrast, evidence for linkage on chromosomes 9 and 13 increased. On chromosome 9, the peak nonparametric LOD score increased from 1.57 (for LC III at 76 cM) to 1.83 (also for LC III, but at 87 cM). On chromosome 13, the peak nonparametric LOD score increased from 2.23 (for LC I at 109 cM) to 3.01 (also for LC I, at 111 cM). A comparison of our linkage peaks in the original and fine-mapping panels is provided in figure 4. We cautiously interpret these results as corroboration of our linkage peaks.
Figure 4.
Comparison of original and fine-mapping results. Multipoint Kong and Cox (1997) nonparametric LOD scores calculated using the SALL scoring statistic are shown. Results are summarized for three regions exhibiting strongest evidence for linkage, on chromosomes 1, 9, and 13. The three phenotypic LCs are LC I (solid black lines), LC II (lines with long dashes), and LC III (lines with short dashes).
Chromosome 1 Isodisomy
Close inspection of the Mendelian inconsistencies in our sample revealed that 13 of them occurred in a single family, all for markers on chromosome 1. Mendelian consistency for all markers on the X chromosome and for autosomes 2–22 suggests that nonpaternity (or nonmaternity) is not an issue. In this family, the proband is homozygous for all chromosome 1 markers and does not carry a maternally derived allele at 13 of these markers (fig. 5A). The genotypes for 30/32 chromosome 1 markers are consistent with a paternal isodisomy for the entire chromosome; genotypes are not available for the other two markers. This observation is intriguing, in light of the fact that our strongest linkage signal is also on chromosome 1 and our parametric analyses suggest a recessive mode of inheritance (all our peak LOD scores were observed with the recessive model). It is interesting that genealogical tracing revealed that the isodisomic individual belongs to a previously described cluster (Karayiorgou et al. 2004) of 87 individuals meeting our strict definition for schizophrenia (LC I) and descending from a single founder couple. The same cluster currently includes 12 of the 34 families we used in our linkage analyses (fig. 5B).
Figure 5.
Isodisomic individual. A, Chromosome 1 genotypes for isodisomic individual and two parents. The isodisomic chromosome is shaded in black, and asterisks (*) indicate markers where mother and offspring genotypes are incompatible. B, Shortest connection between individuals in LC I and a single ancestor who emigrated to South Africa ∼12 generations before present (Karayiorgou et al. 2004). Most affected individuals in our sample are connected to this founder through multiple descent routes. Twelve small pedigrees suitable for linkage analysis and included in our sample are boxed in the diagram. The cluster includes additional informative families when LCs II and III are used. The isodisomic individual is indicated by an arrow (↑).
Discussion
Current attempts to identify genetic loci for schizophrenia include the use of founder populations as a way to limit the genetic and environmental heterogeneity. Isolated populations established by a limited number of founders have proven useful for mapping genes of rare monogenic disorders (Hastbacka et al. 1994; Kalaydjieva et al. 1996; Nystuen et al. 1996; Visapaa et al. 2002). Their usefulness in mapping complex traits is still largely untested. Although it is perhaps unreasonable to expect that there be one major (monogenic disorder–like) susceptibility locus in genetic studies of complex traits such as schizophrenia, one can reasonably expect a smaller number of contributing loci and a smaller number of disease alleles at each locus for at least some founder populations.
Here, we report initial results from a 10-cM genomewide scan of families with schizophrenia from the Afrikaner population. Thirty-four two- to four-generation pedigrees with at least one individual meeting strict DSM-IV criteria for schizophrenia or schizoaffective disorder were informative for linkage. Using both nonparametric and parametric linkage analyses, we obtained evidence for a small number of disease loci on chromosomes 1, 9, and 13. These results are consistent with the notion that genes of substantial effect exist for schizophrenia in the Afrikaner population.
Our chromosome 9 and 13 findings do not reach genomewide significance levels but do show suggestive overlap with other studies. The locus on chromosome 9 overlaps with the locus described by Hovatta et al. (1999) in families from Finland. In the Finnish study, a maximum LOD score of 1.95 was obtained with marker D9S922 (3.4 cM telomeric to our most significant marker, D9S1122). It is interesting that positive findings in this region have been reported in the genome scans of Moises et al. (1995) and Levinson et al. (1998), 6 cM centromeric and 16 cM telomeric to our marker D9S1122, respectively.
The locus on chromosome 13 is slightly distal to the locus described by Blouin et al. (1998). Our most positive marker, D13S285, maps 14 cM telomeric to D13S174, the most positive marker in the Blouin et al. (1998) study. Recent results from a linkage disequilibrium–based positional cloning effort centered around marker D13S174 suggest an association between schizophrenia susceptibility and the G72 gene, which encodes a protein that may interact and modulate the activity of D-amino-acid-oxidase (Chumakov et al. 2002). Recent association studies suggest a potential contribution of G72 to bipolar disorder as well (Hattori et al. 2003). G72 is located 3.2 Mb and 7 Mb proximally from markers D13S1265 and D13S285, respectively, which gave the best linkage results in our studies. Association studies in an extended sample of Afrikaner families revealed a weak contribution of the G72 gene in our sample (D.H., J.A.G., and M.K., unpublished data), but it is not yet clear whether this can account for the observed linkage signal.
We also obtained strong evidence for an apparently novel locus on chromosome 1. Although other studies have implicated chromosome 1 in schizophrenia, the region identified in the present study is quite distant from the previously implicated regions, ∼148–154 cM from the locus described by Brzustowicz et al. (2000) and ∼208 cM from the locus described by Hovatta et al. (1999), both of which are in the q arm of chromosome 1. Our study demarcates a region on the p arm of chromosome 1 at ∼16 cM, which has not been implicated previously, with the possible exception of a recent genome scan in Icelandic families (Stefansson et al. 2002), in which a region on chromosome 1 that appears to overlap with ours provided a weak linkage signal.
It is most interesting that we identified one proband with uniparental disomy (UPD) for paternal chromosome 1. UPD, which was first described by Engel (1980), is a rare condition in which a diploid individual inherits two copies of a specific chromosome from one parent and no copy from the other parent. UPD may lead to rare recessive disorders or to developmental disturbances due to aberrant imprinting effects (Ledbetter and Engel 1995), or it may have no apparent phenotypic effects. There are very few reported cases of UPD of chromosome 1 (UPD 1), involving full or partial isodisomy. These include a small number of cases of maternal UPD 1 (Pulkkinen et al. 1997; Field et al. 1998; Dufourcq-Lagelouse et al. 1999), as well as a few cases of paternal UPD 1 (Gelb et al. 1998; Chen et al. 1999; Miura et al. 2000; Takizawa et al. 2000; Thompson et al. 2002). Among the latter, UPD 1 has been associated with (a) retinal dystrophy due to mutations in the RPE65 gene (Thompson et al. 2002), (b) congenital insensitivity to pain with anhidrosis due to mutations of the TRKA gene (Miura et al. 2000), (c) Herlitz junctional epidermolysis bullosa due to mutation in LAMC2 (Takizawa et al. 2000), or (d) pycnodysostosis due to mutations in the cathepsin K gene (Gelb et al. 1998).
All described individuals with UPD 1 developed normally, suggesting that chromosome 1 does not carry developmentally important maternally imprinted genes, although the existence of maternally imprinted genes with subtler effects on brain function and behavior cannot be excluded. The proband described here also had normal development (J.L.R. and M.K., unpublished data). To our knowledge, this is the first time a UPD of any chromosome has been described in a patient with schizophrenia and the second time a UPD has been discovered during a genome screen linkage study (the other one was described by Field et al. [1998]). On the basis of the striking coincidence of linkage and UPD on chromosome 1 and the genealogical evidence, it is tempting to speculate that this individual might be homozygous for a chromosome 1 schizophrenia-susceptibility locus segregating in the Afrikaner population. If this were the case, we might be able to exploit this fortuitous chromosomal abnormality in our attempts to identify and refine an Afrikaner risk haplotype on chromosome 1. Specifically, additional genotyping might allow us to identify regions of this individual’s haplotype that are shared by other individuals with schizophrenia. This focused search for sharing of a particular haplotype is simpler and more powerful than a search for any shared haplotype because it reduces multiple testing. This work is currently in progress.
It has been suggested that genes influencing susceptibility to schizophrenia and other psychiatric disorders may have pleiotropic effects (Collier and Sham 1997). In addition to the standard definitions of schizophrenia used in genetic studies (LC I and LC II), we also considered other psychiatric disorders found in relatives of schizophrenic individuals in the sample used in our study (LC III). The latter classification results in small increases in evidence for linkage for the chromosome 1 and 9 loci. This observation is compatible with the hypothesis that schizophrenia-susceptibility genes might lead to other psychiatric disorders in the relatives of patients with schizophrenia and that the clinical manifestation of these genes depends on interactions with the environment, other genes, and/or stochastic factors.
In summary, in the current study, we obtained evidence for schizophrenia loci on three chromosomes: 1, 9, and 13. Genealogical evidence shows that the affected individuals in our sample shared a relatively recent ancestor and are likely to share schizophrenia-susceptibility genes. If any schizophrenia-susceptibility alleles were inherited through the shared line of descent we have identified in our sample (Karayiorgou et al. 2004), they are likely to be surrounded by a relatively large haplotype, which will facilitate fine-mapping efforts. Nevertheless, it is possible that different lines of descent are important and that identifying the susceptibility alleles we are seeking will require extensive fine mapping with hundreds of markers. It is fortunate that these approaches are rapidly becoming feasible (Cardon and Abecasis 2003). Although we are cautious about overinterpreting the significance of these loci, at a minimum, our findings warrant further studies using our data set and independent data sets from the Afrikaner population. It is most important that, given the advantage of accurate genealogical tracing in this population, future studies will attempt to identify any extensive shared haplotypes by using denser 1–2-cM microsatellite maps, both at the identified loci and at the genomewide level.
Acknowledgments
The authors thank all the families that participated to the study; the clinicians Christina Sobin, Herman Pretorius, Brian Robertson, Sean Kaliski, and Stephen Lay; and the nurses Ria van Wyk, Cheryl Botha, and Glenda Jansen. We would also like to thank Yu Zhao for help with figure preparation. Genotyping services were provided by CIDR. CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number N01-HG-65403. This research was supported by National Institutes of Health grant MH61399 and by the EJLB Foundation (support to M.K.).
Electronic-Database Information
The URLs for data presented herein are as follows:
- Authors' Web site, http://www.sph.umich.edu/csg/abecasis/public/ (for results of parametric linkage analyses)
- Center for Medical Genetics, Marshfield Clinic, http://research.marshfieldclinic.org/genetics/ (for the Marshfield genetic map)
- CIDR Human Marker Set, http://www.cidr.jhmi.edu/markerset.html (for marker screening set)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for schizophrenia)
References
- Abecasis GR, Cherny SS, Cardon LR (2001a) The impact of genotyping error on family-based analysis of quantitative traits. Eur J Hum Genet 9:130–134 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001b) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 10.1093/bioinformatics/17.8.742 [DOI] [PubMed] [Google Scholar]
- ——— (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association (1994) DSM-IV: diagnostic and statistical manual of mental disorders, 4th ed. American Psychiatric Association, Washington, DC [Google Scholar]
- Bailer U, Leisch F, Meszaros K, Lenzinger E, Willinger U, Strobl R, Gebhardt C, Gerhard E, Fuchs K, Sieghart W, Kasper S, Hornik K, Aschauer HN (2000) Genome scan for susceptibility loci for schizophrenia. Neuropsychobiology 42:175–182 10.1159/000026690 [DOI] [PubMed] [Google Scholar]
- Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, et al (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 20:70–73 10.1038/1734 [DOI] [PubMed] [Google Scholar]
- Brink PA, Ferreira A, Moolman JC, Weymar HW, van der Merwe PL, Corfield VA (1995) Gene for progressive familial heart block type I maps to chromosome 19q13. Circulation 91:1633–1640 [DOI] [PubMed] [Google Scholar]
- Brink PA, Steyn LT, Coetzee GA, Van der Westhuyzen DR (1987) Familial hypercholesterolemia in South African Afrikaners. PvuII and StuI DNA polymorphisms in the LDL-receptor gene consistent with a predominating founder gene effect. Hum Genet 77:32–35 [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS (2000) Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science 288:678–682 10.1126/science.288.5466.678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp NJ, Neuhausen SL, Tiobech J, Polloi A, Coon H, Myles-Worsley M (2001) Genomewide multipoint linkage analysis of seven extended Palauan pedigrees with schizophrenia, by a Markov-chain Monte Carlo method. Am J Hum Genet 69:1278–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Gottesman II (2000) Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet 97:12–17 [DOI] [PubMed] [Google Scholar]
- Cardon LR, Abecasis GR (2003) Using haplotype blocks to map human complex trait loci. Trends Genet 19:135–140 10.1016/S0168-9525(03)00022-2 [DOI] [PubMed] [Google Scholar]
- Chen H, Young R, Mu X, Nandi K, Miao S, Prouty L, Ursin S, Gonzalez J, Yanamandra K (1999) Uniparental isodisomy resulting from 46,XX,i(1p),i(1q) in a woman with short stature, ptosis, micro/retrognathia, myopathy, deafness, and sterility. Am J Med Genet 82:215–218 [DOI] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, et al (2002) Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Nat Acad Sci USA 99:13675–13680 10.1073/pnas.182412499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciulla TA, Sklar RM, Hauser SL (1988) A simple method for DNA purification from peripheral blood. Analyt Biochem 174:485–488 [DOI] [PubMed] [Google Scholar]
- Cloninger CR (1989) Schizophrenia: genetic etiological factors. In: Kaplan HI, Saddock BJ (eds) Comprehensive textbook of psychiatry, 5th ed. William & Wilkins, Baltimore, pp 732–744 [Google Scholar]
- Collier DA, Sham PC (1997) Catch me if you can: are catechol- and indoleamine genes pleiotropic QTLs for common mental disorders? Mol Psychiatry 2:181–183 10.1038/sj.mp.4000274 [DOI] [PubMed] [Google Scholar]
- Cookson WO, Ubhi B, Lawrence R, Abecasis GR, Walley AJ, Cox HE, Coleman R, Leaves NI, Trembath RC, Moffatt MF, Harper JI (2001) Genetic linkage of childhood atopic dermatitis to psoriasis susceptibility loci. Nat Genet 27:372–373 10.1038/86867 [DOI] [PubMed] [Google Scholar]
- Coon, H, Holik J, Hoff M, Reimherr F, Wender P, Myles-Worsley M, Waldo M, Freedman R, Byerley W (1994) Analysis of chromosome 22 markers in nine schizophrenia pedigrees. Am J Med Genet 54:72–79 [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Wright FA (1998) Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc Natl Acad Sci USA 95:12416–12423 10.1073/pnas.95.21.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisi LE, Mesen A, Rodriguez C, Bertheau A, LaPrade B, Llach M, Riondet S, Razi K, Relja M, Byerley W, Sherrington R (2002a) Genome-wide scan for linkage to schizophrenia in a Spanish-origin cohort from Costa Rica. Am J Med Genet 114:497–508 10.1002/ajmg.10538 [DOI] [PubMed] [Google Scholar]
- DeLisi LE, Shaw SH, Crow TJ, Shields G, Smith AB, Larach VW, Wellman N, Loftus J, Nanthakumar B, Razi K, Stewart J, Comazzi M, Vita A, Heffner T, Sherrington R (2002b) A genome-wide scan for linkage to chromosomal regions in 382 sibling pairs with schizophrenia or schizoaffective disorder. Am J Psychiatry 159:803–812 10.1176/appi.ajp.159.5.803 [DOI] [PubMed] [Google Scholar]
- Devlin B, Bacanu SA, Roeder K, Reimherr F, Wender P, Galke B, Novasad D, Chu A, TCuenco K, Tiobek S, Otto C, Byerley W (2002) Genome-wide multipoint linkage analyses of multiplex schizophrenia pedigrees from the oceanic nation of Palau. Mol Psychiatry 7:689–694 10.1038/sj.mp.4001056 [DOI] [PubMed] [Google Scholar]
- Dufourcq-Lagelouse R, Lambert N, Duval M, Viot G, Vilmer E, Fischer A, Prieur M, de Saint Basile G (1999) Chediak-Higashi syndrome associated with maternal uniparental isodisomy of chromosome 1. Eur J Hum Genet 7:633–637 10.1038/sj.ejhg.5200355 [DOI] [PubMed] [Google Scholar]
- Dyer TD, Blangero J, Williams JT, Goring HH, Mahaney MC (2001) The effect of pedigree complexity on quantitative trait linkage analysis. Genet Epidemiol 21 Suppl 1:S236–S243 [DOI] [PubMed] [Google Scholar]
- Ekelund J, Lichtermann D, Hovatta I, Ellonen P, Suvisaari J, Terwilliger JD, Juvonen H, Varilo T, Arajarvi R, Kokko-Sahin ML, Lonnqvist J, Peltonen L (2000) Genome-wide scan for schizophrenia in the Finnish population: evidence for a locus on chromosome 7q22. Hum Mol Genet 9:1049–1057 10.1093/hmg/9.7.1049 [DOI] [PubMed] [Google Scholar]
- Engel E (1980) A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet 6:137–143 [DOI] [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M (2000) Improved inference of relationship for pairs of individuals. Am J Hum Genet 67:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone SV, Matise T, Svrakic D, Pepple J, Malaspina D, Suarez B, Hampe C, Zambuto CT, Schmitt K, Meyer J, Markel P, Lee H, Harkavy Friedman J, Kaufmann C, Cloninger CR, Tsuang MT (1998) Genome scan of European-American schizophrenia pedigrees: results of the NIMH Genetics Initiative and Millennium Consortium. Am J Med Genet 81:290–295 [DOI] [PubMed] [Google Scholar]
- Field LL, Tobias R, Robinson WP, Paisey R, Bain S (1998) Maternal uniparental disomy of chromosome 1 with no apparent phenotypic effects. Am J Hum Genet 63:1216–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb BD, Willner JP, Dunn TM, Kardon NB, Verloes A, Poncin J, Desnick RJ (1998) Paternal uniparental disomy for chromosome 1 revealed by molecular analysis of a patient with pycnodysostosis. Am J Hum Genet 62:848–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E, Clerget-Darpoux F (1996) Consanguinity and the sib-pair method, an approach using identity by descent between and within individuals. Am J Hum Genet 59:1149–1162 [PMC free article] [PubMed] [Google Scholar]
- Goldman A, Krause A, Ramsay M, Jenkins T (1996) Founder effect and prevalence of myotonic dystrophy in South Africans: molecular studies. Am J Hum Genet 59:445–452 [PMC free article] [PubMed] [Google Scholar]
- Göring HH, Terwilliger JD (2000) Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet 66:1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Shields J (1982) Schizophrenia: the epigenetic puzzle. Cambridge University Press, Cambridge [Google Scholar]
- Groenewald JZ, Liebenberg J, Groenewald IM, Warnich L (1998) Linkage disequilibrium analysis in a recently founded population: evaluation of the variegate porphyria founder in South African Afrikaners. Am J Hum Genet 62:1254–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 10.1038/75514 [DOI] [PubMed] [Google Scholar]
- Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D (2001) Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet 68:661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastbacka J, de la Chapelle A, Mahtani MM, Clines G, Reeve-Daly MP, Daly M, Hamilton BA, Kusumi K, Trivedi B, Weaver A, Coloma A, Lovett M, Buckler A, Kaitila I, Lander ES (1994) The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell 78:1073–1087 [DOI] [PubMed] [Google Scholar]
- Hattori E, Liu C, Badner JA, Bonner TI, Christian SL, Maheshwari M, Detera-Wadleigh SD, Gibbs RA, Gershon ES (2003) Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet 72:1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MR, Hopkins HC, Macrea M, Beighton PH (1980) The origin of Huntington’s chorea in the Afrikaner population of South Africa. S Afr Med J 58:197–200 [PubMed] [Google Scholar]
- Hovatta I, Varilo T, Suvisaari J, Terwilliger JD, Ollikainen V, Arajärvi R, Juvonen H, Kokko-Sahin ML, Väisänen L, Mannila H, Lönnqvist J, Peltonen L (1999) A genomewide screen for schizophrenia genes in an isolated Finnish subpopulation, suggesting multiple susceptibility loci. Am J Hum Genet 65:1114–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaydjieva L, Hallmayer J, Chandler D, Savov A, Nikolova A, Angelicheva D, King RH, Ishpekova B, Honeyman K, Calafell F, Shmarov A, Petrova J, Turnev I, Hristova A, Moskov M, Stancheva S, Petkova I, Bittles AH, Georgieva V, Middleton L, Thomas PK (1996) Gene mapping in Gypsies identifies a novel demyelinating neuropathy on chromosome 8q24. Nat Genet 14:214–217 [DOI] [PubMed] [Google Scholar]
- Karayiorgou M, Torrington M, Abecasis GR, Pretorius H, Robertson B, Kaliski S, Lay S, Sobin C, Moller N, Lundy LS, Blundell ML, Gogos JA, Roos JL (2004) Phenotypic characterization and genealogical tracing in an Afrikaner schizophrenia database. Am J Med Genet 124B:20–28 [DOI] [PubMed] [Google Scholar]
- Kendler KS, O’Neill FA, Burke J, Murphy B, Duke F, Straub RE, Shinkwin R, Ni Nuallain M, MacLean CJ, Walsh D (1996) Irish study on high-density schizophrenia families: field methods and power to detect linkage. Am J Med Genet 67:179–190 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ (1998) Linkage thresholds for two-stage genome scans. Am J Hum Genet 62:994–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter DH, Engel E (1995) Uniparental disomy in humans: development of an imprinting map and its implications for prenatal diagnosis. Hum Mol Genet 4:1757–1764 [DOI] [PubMed] [Google Scholar]
- Leitersdorf E, Van der Westhuyzen DR, Coetzee GA, Hobbs HH (1989) Two common low density lipoprotein receptor gene mutations cause familial hypercholesterolemia in Afrikaners. J Clin Invest 84:954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Mahtani MM, Nancarrow DJ, Brown DM, Kruglyak L, Kirby A, Hayward NK, Crowe RR, Andreasen NC, Black DW, Silverman JM, Endicott J, Sharpe L, Mohs RC, Siever LJ, Walters MK, Lennon DP, Jones HL, Nertney DA, Daly MJ, Gladis M, Mowry BJ (1998) Genome scan of schizophrenia. Am J Psychiatry 155:741–750 [DOI] [PubMed] [Google Scholar]
- Lindholm E, Ekholm B, Shaw S, Jalonen P, Johansson G, Pettersson U, Sherrington R, Adolfsson R, Jazin E (2001) A schizophrenia-susceptibility locus at 6q25, in one of the world’s largest reported pedigrees. Am J Hum Genet 69:96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziade M, Fournier A, Phaneuf D, Cliche D, Fournier JP, Roy MA, Merette C (2002) Chromosome 1q12-q22 linkage results in eastern Quebec families affected by schizophrenia. Am J Med Genet 114:51–55 10.1002/ajmg.1616 [DOI] [PubMed] [Google Scholar]
- Miura Y, Hiura M, Torigoe K, Numata O, Kuwahara A, Matsunaga M, Hasegawa S, Boku N, Ino H, Mardy S, Endo F, Matsuda I, Indo Y (2000) Complete paternal uniparental isodisomy for chromosome 1 revealed by mutation analyses of the TRKA (NTRK1) gene encoding a receptor tyrosine kinase for nerve growth factor in a patient with congenital insensitivity to pain with anhidrosis. Hum Genet 107:205–209 10.1007/s004390000369 [DOI] [PubMed] [Google Scholar]
- Moises HW, Yang L, Kristbjarnarson H, Wiese C, Byerley W, Macciardi F, Arolt V, et al (1995) An international two-stage genome-wide search for schizophrenia susceptibility genes. Nat Genet 11:321–324 [DOI] [PubMed] [Google Scholar]
- Nurnberger JI Jr, Blehar MC, Kaufmann CA, York-Cooler C, Simpson SG, Harkavy-Friedman J, Severe JB, Malaspina D, Reich T (1994) Diagnostic interview for genetic studies. Rationale, unique features, and training. NIMH Genetics Initiative. Arch Gen Psychiatry 51:849–859 [DOI] [PubMed] [Google Scholar]
- Nystuen A, Benke PJ, Merren J, Stone EM, Sheffield VC (1996) A cerebellar ataxia locus identified by DNA pooling to search for linkage disequilibrium in an isolated population from the Cayman Islands. Hum Mol Genet 5:525–531 10.1093/hmg/5.4.525 [DOI] [PubMed] [Google Scholar]
- Ober C, Cox NJ (1998) The genetics of asthma: mapping genes for complex traits in founder populations. Clin Exp Allergy 28 Suppl 1:101–105 [DOI] [PubMed] [Google Scholar]
- Paunio T, Ekelund J, Varilo T, Parker A, Hovatta I, Turunen JA, Rinard K, Foti A, Terwilliger JD, Juvonen H, Suvisaari J, Arajarvi R, Suokas J, Partonen T, Lonnqvist J, Meyer J, Peltonen L (2001) Genome-wide scan in a nationwide study sample of schizophrenia families in Finland reveals susceptibility loci on chromosomes 2q and 5q. Hum Mol Genet 10:3037–3048 10.1093/hmg/10.26.3037 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 10.1038/35042049 [DOI] [PubMed] [Google Scholar]
- Pronk JC, Gibson RA, Savoia A, Wijker M, Morgan NV, Melchionda S, Ford D, Temtamy S, Ortega JJ, Jansen S, Havenga C, Cohn RJ, de Ravel TJ, Roberts I, Westerveld A, Easton DF, Joenje H, Mathew CG, Arwert F (1995) Localisation of the Fanconi anaemia complementation group A gene to chromosome 16q24.3. Nat Genet 11:338–340 [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Bullrich F, Czarnecki P, Weiss L, Uitto J (1997) Maternal uniparental disomy of chromosome 1 with reduction to homozygosity of the LAMB3 locus in a patient with Herlitz junctional epidermolysis bullosa. Am J Hum Genet 61:611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees MI, Fenton I, Williams NM, Holmans P, Norton N, Cardno A, Asherson P, Spurlock G, Roberts E, Parfitt E, Mant R, Vallada H, Dawson E, Li MW, Collier DA, Powell JF, Nanko S, Gill M, McGuffin P, Owen MJ (1999) Autosome search for schizophrenia susceptibility genes in multiply affected families. Mol Psychiatry 4:353–359 10.1038/sj.mp.4000521 [DOI] [PubMed] [Google Scholar]
- Roby P, Eyre S, Worthington J, Ramesar R, Cilliers H, Beighton P, Grant M, Wallis G (1999) Autosomal dominant (Beukes) premature degenerative osteoarthropathy of the hip joint maps to an 11-cM region on chromosome 4q35. Am J Hum Genet 64:904–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendorff J, Bernstein R, Macdougall L, Jenkins T (1987) Fanconi anemia: another disease of unusually high prevalence in the Afrikaans population of South Africa. Am J Med Genet 27:793–797 [DOI] [PubMed] [Google Scholar]
- Sengul H, Weeks DE, Feingold E (2001) A survey of affected-sibship statistics for nonparametric linkage analysis. Am J Hum Genet 69:179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, et al (2002) Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet 71:877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, O’Neill FA, Walsh D, Kendler KS (2002) Genome-wide scans of three independent sets of 90 Irish multiplex schizophrenia families and follow-up of selected regions in all families provides evidence for multiple susceptibility genes. Mol Psychiatry 7:542–559 10.1038/sj.mp.4001051 [DOI] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, O’Neill FA, Burke J, Murphy B, Duke F, Shinkwin R, Webb BT, Zhang J, Walsh D, Kendler KS (1995) A potential vulnerability locus for schizophrenia on chromosome 6p24-22: evidence for genetic heterogeneity. Nat Genet 11:287–293 [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale BM, Neale MC, van den Oord E, Kendler KS (2003) Multipoint and single point non-parametric linkage analysis with imperfect data. Am J Med Genet 121B:89–94 [DOI] [PubMed] [Google Scholar]
- Takizawa Y, Pulkkinen L, Chao SC, Nakajima H, Nakano Y, Shimizu H, Uitto J (2000) Mutation report: complete paternal uniparental isodisomy of chromosome 1: a novel mechanism for Herlitz junctional epidermolysis bullosa. J Invest Dermatol 115:307–311 10.1046/j.1523-1747.2000.00052.x [DOI] [PubMed] [Google Scholar]
- Thompson DA, McHenry CL, Li Y, Richards JE, Othman MI, Schwinger E, Vollrath D, Jacobson SG, Gal A (2002) Retinal dystrophy due to paternal isodisomy for chromosome 1 or chromosome 2, with homoallelism for mutations in RPE65 or MERTK, respectively. Am J Hum Genet 70:224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrington M, Viljoen DL (1991) Founder effect in 20 Afrikaner kindreds with pseudoxanthoma elasticum. S Afr Med J 79:7–11 [PubMed] [Google Scholar]
- Tsuang MT, Faraone SV (1994) The genetic epidemiology of schizophrenia. Compr Ther 20:130–135 [PubMed] [Google Scholar]
- Visapaa I, Fellman V, Vesa J, Dasvarma A, Hutton JL, Kumar V, Payne GS, Makarow M, Van Coster R, Taylor RW, Turnbull DM, Suomalainen A, Peltonen L (2002) GRACILE syndrome, a lethal metabolic disorder with iron overload, is caused by a point mutation in BCS1L. Am J Hum Genet 71:863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnich L, Kotze MJ, Groenewald IM, Groenewald JZ, van Brakel MG, van Heerden CJ, de Villiers JN, van de Ven WJ, Schoenmakers EF, Taketani S, Retief AE (1996) Identification of three mutations and associated haplotypes in the protoporphyrinogen oxidase gene in South African families with variegate porphyria. Hum Mol Genet 5:981–984 10.1093/hmg/5.7.981 [DOI] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Wijsman EM, Amos CI (1997) Genetic analysis of simulated oligogenic traits in nuclear and extended pedigrees: summary of GAW10 contributions. Genet Epidemiol 14:719–735 [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Rosenthal EA, Hall D, Blundell ML, Sobin C, Heath SC, Williams R, Brownstein MJ, Gogos JA, Karayiorgou M (2003) Genome-wide scan in a large complex pedigree with predominantly male schizophrenics from the island of Kosrae: evidence for linkage to chromosome 2q. Mol Psychiatry 8:695–705 10.1038/sj.mp.4001356 [DOI] [PubMed] [Google Scholar]
- Williams NM, Rees MI, Holmans P, Norton N, Cardno AG, Jones LA, Murphy KC, Sanders RD, McCarthy G, Gray MY, Fenton I, McGuffin P, Owen MJ (1999) A two-stage genome scan for schizophrenia susceptibility genes in 196 affected sibling pairs. Hum Mol Genet 8:1729–1739 10.1093/hmg/8.9.1729 [DOI] [PubMed] [Google Scholar]