Abstract
Oculocutaneous albinism (OCA) is a complex genetic disease with great clinical heterogeneity. Four different types of OCA have been reported to date (OCA1, OCA2, OCA3, and OCA4). MATP was recently reported in a single Turkish OCA patient as the fourth pathological gene, but no other patients with OCA4 have been reported. Here, we report the mutational profile of OCA4, determined by genetic analysis of the MATP gene in a large Japanese population with OCA. Of 75 unrelated patients that were screened, 18 individuals (24%) were identified as having OCA4; they harbored seven novel mutations, including four missense mutations (P58S, D157N, G188V, and V507L) and three frameshift mutations (S90CGGCCA→GC, V144insAAGT, and V469delG), showing that MATP is the most frequent locus for tyrosinase-positive OCA in Japanese patients. We discuss the functional melanogenic activity of each mutant allele, judging from the relationship between the phenotypes and genotypes of the patients. This is the first report on a large group of patients with OCA4.
Introduction
Oculocutaneous albinism (OCA) is a group of autosomal recessive disorders caused by mutations of melanogenic genes, including tyrosinase (for OCA1 [MIM 203100]) (Tomita et al. 1989), P protein (for OCA2 [MIM 203200]) (Rinchik et al. 1993), and tyrosinase-related protein 1 (TYRP1) (for OCA3 [MIM 203290]) (Boissy et al. 1996). Individuals with OCA1A are born with a complete absence of pigment in the hair, eyes, and skin. In patients with OCA1B and OCA2, the phenotypes are typically somewhat less severe than in those associated with OCA1A, and the two disorders display considerable clinical overlap (Lee et al. 1994). The OCA3 phenotype is characterized by reddish skin and hair color in African blacks and is caused by a mutation in TYRP1. Rufous OCA is now classified as OCA3 (Manga et al. 1997). Recently, a novel melanogenic gene located in chromosome segment 5p and named “MATP” (membrane-associated transporter protein) or “AIM-1” (antigen in melanoma-1) was identified as the fourth pathological OCA gene. The human MATP gene encodes a 530–amino-acid polypeptide that contains 12 putative transmembrane domains, exhibits structural homology to plant sucrose-proton symporters, and is expressed in a high percentage of melanoma cell lines (Harada et al. 2001). Its homolog in medaka fish, b, encodes a transporter that mediates melanin synthesis (Fukamachi et al. 2001). In a single Turkish patient with OCA, a homozygous G-to-A transition in the splice-acceptor sequence of exon 2 of the MATP gene was identified, and that type of OCA was termed “OCA4” (MIM 606574) (Newton et al. 2001). No other patient with OCA4 was subsequently reported, suggesting that OCA4 might be a very minor type of OCA in the worldwide OCA population. Little is known about the clinical phenotype of OCA4, although the Turkish patient with generalized hypopigmentation and ocular abnormalities was reported to be within the phenotypic range commonly associated with OCA2 (Newton et al. 2001).
We examined 75 Japanese patients with OCA, in which 35 (47%) and 6 (8%) were classified as having OCA1 and OCA2, respectively, according to the sequence analyses of the tyrosinase gene and the P gene (Tomita et al. 2000; Suzuki et al. 2003). The remaining 34 patients were further examined to see whether any of them had MATP mutations. In this study, we report 18 individuals who have been identified with OCA4.
Subjects and Methods
Patients
A total of 34 Japanese patients with OCA (20 females and 14 males), who were unrelated and in whom no mutation in either the tyrosinase gene or the P gene was detected, were included in this study. The degree of hypopigmentation in each patient varied from mild to severe, similar to tyrosinase-related OCA (OCA1A and OCA1B). No patient had a family history of inbreeding.
This study was approved by the ethics committee of the Nagoya University School of Medicine. Informed consent was obtained from each patient, or from the patient’s parents, in the case of children.
Identification of the Genomic Organization and Mutation Screening of the MATP Gene
The GenBank database was screened with the MATP complementary DNA sequence (AF172849). From one human genomic contig (NT_023085), the intron/exon boundaries of the MATP gene were analyzed, and primer sequences were designed for the mutation screening of the MATP gene (table 1). Genomic DNA was isolated from the peripheral blood of each patient by use of a genomic DNA purification kit (Qiagen). The human MATP gene spans 7 exons. In exon 1, three primer sets were designed—for the 5′ side, the middle part, and the 3′ side—because DNA fragments >350 bp were not suitable to detect mutations precisely in our system. In exon 3, two primer sets were designed, for the same reason as in exon 1. The amplified fragments were then screened for mutations by simultaneous analyses of SSCPs and the heteroduplex method (Spritz et al. 1992). Three kinds of SSCP gels, with glycerol concentrations of 0%, 7%, and 10%, were used to elevate the sensitivity of our mutation screening system. Standard PCR amplification procedures were employed, with an annealing temperature of 58°C for all primers, except the primer sets for exons 3 and 5, which were done at 61°C. PCR products showing aberrant patterns were reamplified and sequenced directly. In patients with one or no mutations detected by the SSCP method, all of their PCR products were directly sequenced to identify any mutations.
Table 1.
Primer Pairs Used to Amplify the MATP Exon Segments
|
PCR Primer |
|||
| Exon No. | Forward | Reverse | PCR Product Size(bp) |
| 1 (5′ side) | 5′-AGGCTCCACGTCAAATCCAG-3′ | 5′-GGTCACATACGCTGCCTCCA-3′ | 260 |
| 1 (middle) | 5′-CAGACTCATCATGCACAGCA-3′ | 5′-ATGCCCACGAGCATCATGAC-3′ | 252 |
| 1 (3′ side) | 5′-CAGCATTGTGTGGTTCCTCA-3′ | 5′-GGTCAAACACATGAACATCCTC-3′ | 261 |
| 2 | 5′-AACGTGGATGATTCTAAAACAGGA-3′ | 5′-CTCATTGTCTGGGGAGCTGA-3′ | 280 |
| 3 (5′ side) | 5′-GGGAGTGTCTATGCATGAGG-3′ | 5′-GATAGAACCATACTCGTACATTCC-3′ | 324 |
| 3 (3′ side) | 5′-GCCCCACTTACAGAGGTTGC-3′ | 5′-CAACAAAGAGCAAGAATATTTTCCCTTG-3′ | 224 |
| 4 | 5′-AGCTGGCTGAGTTTCTGCAG-3′ | 5′-CCTCAACAGGTGTTAATGGAGG-3′ | 265 |
| 5 | 5′-AGAGGTGGAGAAGCAGAGTG-3′ | 5′-GAAGACATCCTTAGGAGAGAG-3′ | 236 |
| 6 | 5′-ATGAGGCACTGCCAGCTGTA-3′ | 5′-CCCAAGGCAGAGGTTCAATG-3′ | 286 |
| 7 | 5′-GCCCTAAATGACAGTTCCTTG-3′ | 5′-TGTGCTTCACTGTCTCTGAG-3′ | 326 |
Results and Discussion
Mutations of the MATP Gene
Among the 75 Japanese patients with OCA in our study, 34 patients lacked any mutation in the coding or adjacent noncoding sequences of the tyrosinase gene or the P gene. The diagnoses of OCA1 or OCA2 were therefore essentially eliminated. We then screened for mutations in the MATP gene of those 34 patients with OCA. PCR-SSCP/heteroduplex screening and direct sequencing of the MATP gene finally detected seven novel mutations in 18 individuals (3 males and 15 females) (table 2). These novel mutations included four missense substitutions (P58S, D157N, G188V, and V507L), two deletion mutations (S90CGGCCA→GC and V469delG), and one insertion mutation (V144insAAGT). All of the four missense mutations were found at amino acid residues conserved among medaka fish, mouse, and human.
Table 2.
Mutations of the MATP Gene in 18 Japanese Patients with OCA4[Note]
|
Mutation |
Clinical Phenotype |
Parents |
|||||||
| Patient | Age | Sex | 1st | 2nd | Hair Color | Iris Color | Nystagmus | Mother | Father |
| 3 | 23 years | F | D157N | G188V | Light yellow | Blue | Positive | G188V | D157N |
| 4 | 1 year | M | G188V | NI | Brown | Red-brown | Negative | NM | G188V |
| 6 | 13 years | F | D157N | V469delG | Light yellow | Blue | Positive | V469delG | D157N |
| 8 | 71 years | F | D157N | D157N | White | Blue | Positive | ND | ND |
| 9 | 25 years | F | S90CGGCCA→GC | V144insAAGT | White | Blue | Positive | ND | ND |
| 10 | 1 year | F | G188V | S90CGGCCA→GC | Blond | Gray-blue | Negative | G188V | S90CGGCCA→GC |
| 11 | 25 years | F | D157N | NI | Blond | Brown | Negative | ND | NM |
| 12 | 5 years | M | G188V | G188V | Pale blond | Red-brown | Negative | G188V | G188V |
| 15 | 11 mo | F | G188V | NI | Light yellow | Brown | Negative | G188V | NM |
| 17 | 1 year | F | D157N | G188V | Yellow | Blue | Positive | G188V | D157N |
| 19 | 10 years | F | D157N | NI | Brown-black | Brown | Negative | ND | ND |
| 20 | 2 mo | F | D157N | NI | Light yellow | Gray-blue | Negative | D157N | NM |
| 25 | 6 years | F | D157N | NI | Blond | Gray | Positive | D157N | NM |
| 36 | 1 year | F | D157N | V507L | Blond | Gray-blue | Negative | V507L | D157N |
| 38 | 1 mo | M | D157N | V144insAAGT | Light yellow | Gray | Positive | D157N | V144insAAGT |
| 42 | 6 mo | F | P58S | S90CGGCCA→GC | Light yellow | Blue | Negative | P58S | S90CGGCCA→GC |
| 50 | 24 years | F | D157N | D157N | Light yellow | Blue | Positive | D157N | ND |
| 51 | 1 year | F | D157N | S90CGGCCA→GC | Light yellow | Blue | Positive | D157N | S90CGGCCA→GC |
Note.— ND = an examination was not done; NM = no mutation was found in our examination; NI = mutation not identified.
We examined the frequency of the seven mutant alleles in the MATP gene of 104 unrelated normally pigmented Japanese subjects (208 alleles), and no mutant allele was detected (table 3). This indicates that those seven alleles might be very rare in the general Japanese population and could be defined statistically as pathological alleles.
Table 3.
Positions of MATP Gene Mutations Identified in This Study and Its Frequencies in Japanese Patients with OCA4 and Normally Pigmented Japanese Subjects[Note]
|
No. of Alleles Identified in |
|||
| PathogenicVariant | Position in the MATP Protein | Japanese Patientswith OCA4(n=36) | Normally PigmentedJapanese Subjects(n=208) |
| P58S | 1st transmembrane domain | 1 (.03) | 0 |
| D157N | 2nd cytoplasmic loop | 14 (.39) | 0 |
| G188V | 5th transmembrane domain | 7 (.19) | 0 |
| V507L | 12th transmembrane domain | 1 (.03) | 0 |
| S90CGGCCA→GC | Frameshift in 1st cytoplasmic loop | 4 (.11) | 0 |
| V144insAAGT | Frameshift in 4th transmembrane domain | 2 (.06) | 0 |
| V469delG | Frameshift in 5th cytoplasmic loop | 1 (.03) | 0 |
Note.— n = number of alleles.
In six patients (4, 11, 15, 19, 20, and 25 [table 2]), only one heterozygous mutation was found, although we directly sequenced all PCR products amplified from the DNA of these six patients to find a second mutation. It seems likely that the other MATP allele contains an occult mutation that either was not detected by our SSCP/heteroduplex screening procedure or occurs in a gene region not sampled in the PCR products.
The frequency of OCA4 in the Japanese albino population was 24% (18/75), indicating that mutations in the MATP gene are one of the most common causes of tyrosinase-positive OCA in Japan. Although all PCR products amplified from their DNA were directly sequenced to find any mutations in the MATP gene, 16 patients still remained as an unclassified type of OCA.
Of the four missense mutations identified, three were within transmembrane domains and one was the first amino acid in the second cytoplasmic loop (table 3). This is in complete contrast to mutations of the P protein associated with OCA2, because most missense substitutions in patients with OCA2 occurred within the loops between the transmembrane domains (Spritz 1994). Both the MATP and the P proteins have 12 transmembrane domains arranged similarly to various transporters and appear to be integral membrane proteins of melanosomes.
Identification of Polymorphism
SSCP revealed several polymorphisms in the exonic or the nearby intronic sequences in the MATP gene, and the frequencies were determined for the 34 patients with OCA and the 104 unrelated normally pigmented Japanese subjects (table 4). Two of the exonic polymorphisms resulted in amino acid changes (E272K and T500P), whereas the remaining ones were silent. E272K is a common polymorphism (0.37), whereas T500P is rare (0.01) in the Japanese population. Three of the polymorphisms (IVS3+14A→G [MATP1-1; dbSNP accession number ss16339967], IVS4-6T→C [MATP1-2; dbSNP accession number ss16339968], and A1498C [MATP1-3; dbSNP accession number ss16339969]) were novel.
Table 4.
Polymorphisms Detected in the MATP Gene and Its Frequencies in Japanese Patients with OCA and Normally Pigmented Japanese Subjects
|
Allele Frequency in |
||||
| NucleotideChangea | Amino Acid Change | Exon | Japanese Patientswith OCA | Normally PigmentedJapanese Subjects |
| G814A | E272K | EX3 | 30/68 (.44) | 76/208 (.37) |
| IVS3+14A→G | None | IVS3 | 1/68 (.01) | 0/208 (.00) |
| G987A | T329T | EX4 | 18/68 (.26) | 57/208 (.27) |
| IVS4-6T→C | None | IVS4 | 1/68 (.01) | 0/208 (.00) |
| IVS4-44A→C | None | IVS4 | 34/68 (.50) | 129/208 (.62) |
| A1498C | T500P | EX7 | 0/68 (.00) | 2/208 (.01) |
| G1594A | None | EX7 | 0/68 (.00) | 1/208 (.005) |
Nucleotide 1 begins at the first nucleotide of codon 1.
The Relationship between the Genotype and the Phenotype in Patients with OCA4
The Pro(CCA)58Ser(TCA) mutation in exon 1 occurred within the first transmembrane domain (table 3) and was identified only in patient 42. The other mutation in that patient was a S90CGGCCA→GC frameshift mutation, which creates a stop codon at 111 and thus produces no functional protein. Patient 42 was a 6-mo-old girl with light yellow hair and blue irides.
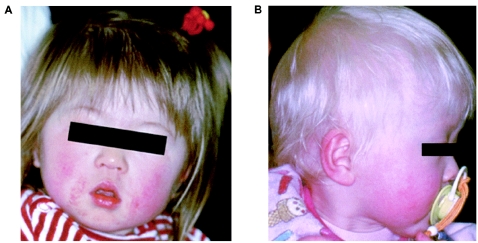
The Asp(GAC)157Asn(AAC) mutation in exon 2 causes a substitution from an acidic to a neutral amino acid at the first residue in the second cytoplasmic loop. This mutant allele was found in 12 patients, and 2 of them (patients 8 and 50) were homozygous for this allele. The allele frequency of D157N in all patients with OCA4 was 0.39 (14/36), indicating that D157N is the most common mutant allele in Japanese patients with OCA4. Patient 8 was 71 years old with nystagmus and had no pigmentation in her hair or eyes. Patient 50, who was 24 years old, also showed light yellow hair and blue irides with nystagmus. Patients 6, 38, and 51 (fig. 1B) had the D157N allele plus another mutant allele—V469delG, V144insAAGT, or S90CGGCCA→GC, respectively—which may have no functional activity in melanogenesis because the mutations caused frameshifts. All of those patients presented with light yellow hair, blue eyes, and nystagmus, as did patients 8 and 50. These results indicate that the D157N mutant allele might have a very low functional activity in melanogenesis.
Figure 1.
The diverse clinical phenotypes of patients with OCA4. Both patients are 1-year-old Japanese girls. Patient 36, with D157N and V507L mutations, has blond hair and gray-blue irides (A). Patient 51, with D157N and S90CGGCCA→GC mutations, has light yellow hair and blue irides (B). Note the milder hypopigmentation of patient 36 compared with that of patient 51.
The Gly(GGT)188Val(GTT) mutation resulted from a single-nucleotide mutation from G to T at the first nucleotide of exon 3, which caused an amino acid substitution within the fifth transmembrane domain (table 3). This mutant allele was found in six patients, including one homozygote for this allele (patient 12). The allele frequency was 0.19 (7/36). The clinical phenotype of patient 12 presented with some pigmentation—that is, the hair was pale blond, and the irides were red-brown without nystagmus. Patient 10, who had the G188V allele and the S90CGGCCA→GC allele, presented with blond hair and gray-blue eyes. All other patients who had the G188V allele also presented with some generalized pigmentation (patients 3, 4, 15, and 17), although the degree of hypopigmentation in each patient varied. These results indicate that the G188V allele might have some functional activity in melanogenesis.
The Val(GTG)507Leu(CTG) mutation in exon 7 occurred within the 12th transmembrane domain, which is nearest to the C-terminal of the MATP protein. A single individual (patient 36 [fig. 1A]) had this mutant allele and presented with blond hair and gray-blue irides. Her other mutant allele was D157N, which appears to have a very low functional activity in melanogenesis, as mentioned above.
Three different frameshift mutations were identified in this study. The first one was found in four patients (9, 10, 42, and 51) who exhibited clear heteroduplexes on SSCP gels with the primer set for exon 1; the consequent sequencing revealed a two-plus-two nucleotide deletion (CGGCCA→GC) from Ser90 to Ser92. This deletion results in a change after amino acid 90 with a consequent truncation of the distal MATP nonsense polypeptide, so that 21 new amino acids were translated before a premature stop occurred at codon 111. The second frameshift mutation was observed in two patients (9 and 38) who have a 4-bp insertion in exon 2 (V144insAAGT), which results in a 10–amino-acid frameshift followed by a premature stop at codon 154. A single nucleotide deletion of G in exon 7 (V469delG) was identified as the third frameshift mutation in one individual (patient 6). This deletion results in a frameshift with a truncation of the MATP nonsense polypeptide at codon 469 that lacks the C-terminal 2 transmembrane domains of the predicted protein. Patient 9 had two different frameshift mutation alleles, S90CGGCCA→GC and V144insAAGT, which are both predicted to have no functional activity in melanogenesis. This patient was 25 years old and had no generalized pigmentation, a phenotype similar to OCA1A (tyrosinase-negative OCA). Therefore, it was impossible to distinguish her or patient 8 from patients with OCA1A on the basis of their clinical phenotypes. However, we could have identified her as non-OCA1A with a positive DOPA test by use of electron microscopy, because her specimen showed stage IV melanosomes when treated with DOPA, indicating that the tyrosinase activity in her melanosomes was fairly high (data not shown).
Finally, we speculate that the D157N allele might suppress the functional activity of the G188V allele on melanogenesis, similar to the amino acid substitution of D153N in the mouse underwhite gene of Uwdbr, which is known as a dominant allele that reduces melanogenesis when heterozygous (Sweet et al. 1998; Newton et al. 2001). Patient 10, whose clinical phenotype included blond hair and gray-blue irides without nystagmus, had both the G188V and the S90CGGCCA→GC mutant alleles. On the other hand, patients 3 and 17, who had the D157N and the G188V mutation alleles, respectively, presented with yellow hair and blue irides with nystagmus, showing somewhat more severe hypopigmentation than patient 10. This may suggest that D157N might have a dominant negative effect, because the MATP mutant protein derived from the G188V mutant allele appeared to have some functional activity in melanogenesis by itself, as mentioned above. More advanced approaches for functional assays will be required to confirm the dominant negative effect of the D157N substitution on melanogenesis, because other differing background genes in these patients may contribute to their more severe hypopigmentation.
In conclusion, this study demonstrates that seven novel mutations in the MATP gene were found in 18 Japanese patients with OCA, indicating that OCA4 is one of the major types in Japan. The study also establishes that the clinical phenotype of the patients varied, depending on their mutant genotypes. Further accumulation of data correlating phenotypes with mutant genotypes is expected to give new insights into investigations on melanogenesis.
Acknowledgments
The authors are grateful to the volunteers for donating blood samples. This work was supported by grants 14570805, 14770403, 15659260, 15790574, and 15790575 from the Ministry of Education, Science, and Culture, Japan, and by the Japanese Society for Investigative Dermatology Fellowship SHISEIDO Award 2003.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for MATP1-1 [accession number ss16339967], MATP1-2 [accession number ss16339968], and MATP1-3 [accession number ss16339969])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for MATP genomic DNA [accession number NT 023085], MATP cDNA [accession number AF172849])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for OCA1, OCA2, OCA3, and OCA4)
References
- Boissy RE, Zhao H, Oetting WS, Austin LM, Wildenberg SC, Boissy YL, Zhao Y, Sturm RA, Hearing VJ, King RA, Nordlund JJ (1996) Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as “OCA3.” Am J Hum Genet 58:1145–1156 [PMC free article] [PubMed] [Google Scholar]
- Fukamachi S, Shimada A, Shima A (2001) Mutations in the gene encoding B, a novel transporter protein, reduce melanin content in medaka. Nat Genet 28:381–385 10.1038/ng584 [DOI] [PubMed] [Google Scholar]
- Harada M, Li YF, El-Gamil M, Rosenberg SA, Robbins PF (2001) Use of an in vitro immunoselected tumor line to identify shared melanoma antigens recognized by HLA-A*0201-restricted T cells. Cancer Res 61:1089–1094 [PubMed] [Google Scholar]
- Lee ST, Nicholls RD, Bundey S, Laxova R, Musarella M, Spritz RA (1994) Mutations of the P gene in oculocutaneous albinism, ocular albinism, and Prader-Willi syndrome plus albinism. N Engl J Med 330:529–534 10.1056/NEJM199402243300803 [DOI] [PubMed] [Google Scholar]
- Manga P, Kromberg JG, Box NF, Sturm RA, Jenkins T, Ramsay M (1997) Rufous oculocutaneous albinism in southern African blacks is caused by mutations in the TYRP1 gene. Am J Hum Genet 61:1095–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, Brilliant MH (2001) Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet 69:981–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinchik EM, Bultman SJ, Horstemke B, Lee ST, Strunk KM, Spritz RA, Avidano KM, Jong MT, Nicholls RD (1993) A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 361:72–76 10.1038/361072a0 [DOI] [PubMed] [Google Scholar]
- Spritz RA, Holmes SA, Ramesar R, Greenberg J, Curtis D, Beighton P (1992) Mutations of the KIT (mast/stem cell growth factor receptor) proto-oncogene account for a continuous range of phenotypes in human piebaldism. Am J Hum Genet 51:1058–1065 [PMC free article] [PubMed] [Google Scholar]
- Spritz RA (1994) Molecular genetics of oculocutaneous albinism. Hum Mol Genet 3 Spec: 1469–1475 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miyamura Y, Matsunaga J, Shimizu H, Kawachi Y, Ohyama N, Ishikawa O, Ishikawa T, Terao H, Tomita Y (2003) Six novel P gene mutations and oculocutaneous albinism type 2 frequency in Japanese albino patients. J Invest Derm 120:781–783 [DOI] [PubMed] [Google Scholar]
- Sweet HO, Brilliant MH, Cook SA, Johnson KR, Davisson MT (1998) A new allelic series for the underwhite gene on mouse chromosome 15. J Hered 89:546–551 10.1093/jhered/89.6.546 [DOI] [PubMed] [Google Scholar]
- Tomita Y, Takeda A, Okinaga S, Tagami H, Shibahara S (1989) Human oculocutaneous albinism caused by single base insertion in the tyrosinase gene. Biochem Biophys Res Commun 164:990–996 [DOI] [PubMed] [Google Scholar]
- Tomita Y, Miyamura Y, Kono M, Nakamura E, Matsunaga J (2000) Molecular bases of congenital hypopigmentary disorders in humans and oculocutaneous albinism 1 in Japan. Pigment Cell Res 13 Suppl 8:130–134 [DOI] [PubMed] [Google Scholar]