Abstract
Age-related macular degeneration (AMD) is a complex multifactorial disease that affects the central region of the retina. AMD is clinically heterogeneous, leading to geographic atrophy (GA) and/or choroidal neovascularization (CNV) at advanced stages. Considerable data exists in support of a genetic predisposition for AMD. Recent linkage studies have provided evidence in favor of several AMD susceptibility loci. We have performed a high-resolution (5-cM) genome scan of 412 affected relative pairs that were enriched for late-stage disease (GA and/or CNV). Nonparametric linkage analysis was performed using two different diagnostic criteria and also by dividing the affected individuals according to GA or CNV phenotype. Our results demonstrate evidence of linkage in regions that were suggested in at least one previous study at chromosomes 1q (236–240 cM in the Marshfield genetic map), 5p (40–50 cM), and 9q (111 cM). Multipoint analysis of affected relatives with CNV provided evidence of additional susceptibility loci on chromosomes 2p (10 cM) and 22q (25 cM). A recently identified Gln5345Arg change in HEMICENTIN-1 on chromosome 1q25 was not detected in 274 affected members in the restricted group with AMD, 346 additional patients with AMD, and 237 unaffected controls. Our results consolidate the chromosomal locations of several AMD susceptibility loci and, together with previous reports, should facilitate the search for disease-associated sequence variants.
Introduction
Age-related macular degeneration (AMD [MIM 603075]) is a complex, multifactorial neurodegenerative disease that leads to visual impairment and results in half of all cases of legal blindness in Western populations >65 years of age (Klein et al. 1992; Vingerling et al. 1995; Bird 2003). In the United States, >1.6 million individuals are affected with late-stage AMD, and 30% of individuals >75 years of age show some signs of disease (Prevent Blindness America and National Eye Institute 2002). A dramatic increase in the size of the aging population makes AMD a significant public health problem and a major focus of research efforts (National Advisory Eye Council 1998). There is currently no cure, and the impact of specific interventions is limited.
AMD is a progressive disease encompassing a broad spectrum of clinical findings that primarily affect the central (macular) region of the retina (Bressler et al. 1988; Ambati et al. 2003). Early manifestations of the disease include the appearance of drusen, accumulations of acellular debris in the basement membrane of the retinal pigment epithelium (RPE), and pigmentary abnormalities in the macular region. Although small drusen are commonly observed in the aging macula, the presence of large drusen is considered diagnostic for AMD (Bird et al. 1995). Late stages of AMD are typically grouped in two forms: (1) a “dry” form associated with the loss of RPE and photoreceptors and diminished retinal function, known as “geographic atrophy” (GA); and (2) a “wet” exudative form, which is associated with the formation of choroidal neovascularization (CNV), leading to sub-RPE or subretinal hemorrhage, subretinal fluid, macular edema, and, eventually, fibrotic scarring. Although GA and CNV together account for only 10% of patients with AMD (Smith et al. 2001), the two are responsible for most cases of associated blindness. At present, it is unclear whether these forms represent different clinical entities with distinct genetic etiology or stages in a single disease continuum (Gorin et al. 1999). During the course of disease, ophthalmic findings in a single individual can include multiple clinical pathologies. Large drusen and pigmentary changes typically precede both GA and CNV, and a single affected individual may exhibit both forms of the late-stage disease (Green 1999; Ambati et al. 2003). Oxidative stress and immune-mediated processes are believed to contribute to AMD pathogenesis (Cai et al. 2000; Hageman et al. 2001; Crabb et al. 2002; Johnson et al. 2002; Ambati et al. 2003); however, precise molecular events have not yet been elucidated.
Epidemiological studies have implicated interaction of environmental and genetic factors in AMD pathogenesis. Although a number of lifestyle factors (e.g., alcohol consumption, light exposure, diet, smoking) may influence disease manifestation (Hirvela et al. 1996; Klein et al. 1997; Smith et al. 2001), the strongest risk factors identified to date are age and family history (Evans 2001). Aging itself appears to be a multifactorial and genetically determined process (Hekimi and Guarente 2003), and it influences expression of several genes in the retina (Yoshida et al. 2002; S. Yoshida, S. Zareparsi, and A. Swaroop, unpublished data). Population studies have suggested familial aggregation of AMD and genetic predisposition to disease pathogenesis (Seddon et al. 1997; Klaver et al. 1998; Gorin et al. 1999). The phenotypic similarities between monogenic early-onset macular disease and AMD prompted several association studies; however, the genes responsible for monogenic diseases do not appear to make a significant contribution to AMD (Stone et al. 2001). To date, only allelic variations in apolipoprotein-E (APOE) and possibly the retinal ATP-binding cassette transporter gene ABCR have been associated with susceptibility to AMD (Allikmets et al. 2000; Schmidt et al. 2002).
The first tentative localization of a genetic locus for AMD (ARMD1) on chromosome 1q25-31 was provided by linkage analysis of data from a family with a predominantly dry form of disease that exhibited a dominant mode of inheritance (Klein et al. 1998). Thereafter, a genome scan of affected sib pairs provided suggestive evidence for AMD susceptibility loci on chromosomes 5, 9, and 10 (Weeks et al. 2000). A follow-up genome scan, including some of the same samples, however, indicated linkage to 1q31 and 17q25 (Weeks et al. 2001). Recently, three independent genomewide scans have provided evidence for a number of suggestive AMD susceptibility loci, which include the locus at 1q (Majewski et al. 2003; Schick et al. 2003; Seddon et al. 2003). Recently, a Gln5345Arg change in the HEMICENTIN-1 gene was reported to be associated with the disease in the original AMD family that exhibited linkage to chromosome 1q25-31 (Schultz et al. 2003).
The power of affected relative pair studies depends on the increased risk of disease to individuals who share a particular locus identical by descent (IBD) with an affected relative (Risch 1990). As prevalence increases, the maximum possible increase in risk attributable to any one locus decreases. The late-stage AMD phenotypes (GA and CNV) are rarer and may occur in individuals who are genetically more susceptible. We hypothesized that the individuals with late-stage AMD would typically carry a heavier load of genetic susceptibility factors than other patients with AMD and thus would be well suited for genetic investigations. Consistent with this hypothesis, studies by Klaver et al. (1998) show that first-degree relatives of individuals with late-stage AMD have substantially increased risk and earlier onset of maculopathy than the general population. Here, we report the results of a 5-cM genomewide linkage study in families enriched for late-stage AMD, with the majority of individuals affected by GA and/or CNV. In addition to the ARMD1 locus, our results demonstrate evidence of linkage on chromosomes 2p (10 cM on the Marshfield genetic map), 5p (40–50 cM), 9q (111 cM), and 22q (25 cM).
Material and Methods
Family Recruitment and Selection
Families with AMD were primarily ascertained and recruited from the clinical practice at the Kellogg Eye Center, University of Michigan Hospitals. Since the Retina Clinic serves as a tertiary health care center for the State of Michigan and the surrounding Great Lakes region, the patient population is biased toward late-stage AMD (69% of the affected individuals in our study have GA and/or CNV). The patient population used for genotyping in this study is white and primarily of Western European ancestry, reflecting the genetic constitution of the Great Lakes region. Ophthalmic records for current and previous eye examinations, fundus photographs, and fluorescein angiograms were obtained for all probands and family members. All records and ophthalmic documentation were scored for the presence of AMD clinical findings in each eye and were updated every 1–2 years. The recruitment and research protocols were reviewed and approved by the University of Michigan institutional review board, and informed consent was obtained from all study participants.
Disease Classification
Fundus findings in each eye were classified on the basis of a standardized set of diagnostic criteria established by the International Age-Related Maculopathy Epidemiological Study (Bird et al. 1995). The following individuals were excluded from the study: patients with a history of severe macular disease or vision loss before 40 years of age; others with signs of a juvenile macular or retinal degeneration (such as Stargardt or Best disease) or macular damage resulting from ocular trauma, retinal detachment, high myopia, chorioretinal infective or inflammatory processes, or choroidal dystrophy; and those with inadequate documentation.
For the genetic studies reported here, macular findings were scored in each individual by use of two clinical grading systems: a restricted definition (R-AMD) and a broad description (B-AMD) of AMD. For an individual to be graded as affected in the R-AMD category, the clinical findings had to include: (1) CNV and/or GA and/or large macular drusen in at least one eye with clearly documented coarse RPE changes and/or macular drusen in the fellow eye or (2) at least 3 years of medical records from a collaborating ophthalmologist documenting a history of atrophic AMD bilaterally or small macular drusen and coarse RPE changes bilaterally. In the B-AMD analysis, the requirements for the quality, consistency, and length of the ophthalmic documentation were relaxed. This double-grading system minimized the ambiguities in distinguishing macular pathology caused by AMD or resulting from the natural aging process, accommodated variations in the quality of clinical documentation, which varied because of enrollment procedures, and allowed the inclusion of individuals who had not been directly evaluated by a collaborating ophthalmologist. Clinical documentation for 331 affected individuals (under both the broad and restricted models) spanned, on average, 5.3 years.
In total, our sample included >242 informative affected relative pairs (including 179 sibling pairs, 40 cousin pairs, 20 avuncular pairs, 3 half-sibling pairs, and additional more distant relatives) under the restricted phenotype definition (R-AMD), and it included >412 affected relative pairs (including 255 sibling pairs, 116 cousin pairs, 36 avuncular pairs, and 5 half-sibling pairs) under the broader classification scheme (B-AMD).
Sample Preparation
DNA from all study participants was extracted from 10 ml of EDTA-treated blood by use of the Wizard Genomic DNA Purification Kit (Promega) (Miller et al. 1988). In families with only two affected individuals, we collected DNA samples for genotyping from unaffected first-degree relatives (parents and siblings), whenever possible. Inclusion of three genotyped individuals per family improved our ability to detect genotyping errors and estimate allele sharing.
Genome Scan
A high-resolution 5-cM genomewide screen was performed at the Marshfield Clinic Research Foundation (Marshfield, WI). This higher marker density enhanced our ability to detect linkage and assisted in detecting misspecified relationships and genotyping errors in this population with AMD, in which parental genotypes were not available for most of the families. A total of 427 samples were screened using 734 autosomal and 39 X-linked microsatellite markers. The marker set was composed primarily of trinucleotide and tetranucleotide repeat markers and was based on the Marshfield Genetics screening sets 12 and 52. For autosomes, the average distance between consecutive markers was 4.71 cM (±2.53 cM). The maximum intermarker distance was 14.3 cM, and 29 pairs of consecutive markers were separated by ⩾10 cM. We collected a total of 306,134 genotypes, which corresponded to an overall genotyping success rate of 93%. Average heterozygosity was 72% for autosomal markers and 68% for markers on the X chromosome (in females).
Error Checking
In the absence of parental genotypes, deviations from Hardy-Weinberg equilibrium (HWE) can bias the evidence for linkage. We checked HWE for all markers by first selecting one individual from each pedigree, collapsing rare alleles so that expected counts were more than three for all genotype classes, and, finally, conducting a χ2 test (Weir 1990). For markers on the X chromosome, we considered only female individuals. We identified two markers (AAT035 and GAAA1C11) with significant (P < .0001) deviations from HWE and excluded these markers from further analysis. Familial relationships were verified using the GRR (Abecasis et al. 2001b) and RELPAIR (Epstein et al. 2000) programs. Nine families showed pedigree errors. In four of these families, individuals reported to be full siblings were almost certainly half siblings. In another family, an individual reported to be a full sibling appeared to be unrelated. Parents’ sex was misspecified in two families. Finally, in two other families we identified two pairs of identical twins, allowing us to estimate the error rate in our experiments. One twin pair was genotyped at 700 markers and another at 754 markers. In total, we observed one discrepancy between twin genotypes, for an overall discrepancy rate of <1/1,000. All these pedigree errors were corrected prior to linkage analysis. We also used the MERLIN program (Abecasis et al. 2002) to check for Mendelian inconsistencies and to identify genotypes associated with an excessive number of recombination events.
Genetic Map Validation
During the course of the analyses, we compared the genetic and physical maps for the microsatellite markers used in our study. We identified a few previously documented discrepancies (DeWan et al. 2002). For every set of five adjacent markers, we calculated the likelihood of the observed genotypes, assuming (1) the ordering implied by the genetic map; (2) the ordering implied by the physical map; and (3) that one of the markers was unlinked. The genetic map was well supported for nearly all markers, but we identified three markers (TAT024, GATA43F06, and GATA72A06) for which relatively large log-likelihood ratios supported a different order (155.637, 139.796, and 16.558, respectively). These markers were not considered in subsequent analyses.
Nonparametric Linkage Analysis
We performed genomewide, nonparametric linkage analyses using the SALL statistic (Whittemore and Halpern 1994), as recommended by Sengul et al. (2001). This statistic combines information from all affected individuals in a pedigree and can detect regions of excess IBD sharing. We conducted single-point and multipoint analyses with the MERLIN program (Abecasis et al. 2002). For X-chromosome analyses, we used MINX (Merlin-In-X), a modified version of MERLIN. Since we cannot specify an accurate genetic model for a complex disease such as AMD, we report only nonparametric LOD scores. It is our experience that, although parametric LOD scores for some models would be slightly higher for each of the identified loci, a similar set of susceptibility loci would be discovered.
Empirical Significance Levels
We calculated genomewide significance levels by use of 100 (for single-point analysis) or 25 (for multipoint analysis) gene-dropping simulations. Computational constraints precluded additional multipoint simulations: our largest pedigree included a total of 30 individuals (including 10 affected), and performing analysis at equally spaced 1-cM intervals throughout the genome required ∼4 d. In each simulation, we retained the original phenotypes and generated a new data set with the same allele frequencies, marker spacings, phenotypes, and missing-data patterns. Any evidence for linkage in this simulated data is due to chance. We analyzed each replicate and recorded the highest peak for each chromosome to evaluate the false-positive rate. These simulations allowed us to account for uneven marker spacing and informativeness, as well as the diversity of family structures in our study, and to calculate the probability of observing multiple linkage peaks of a certain height. (See the work of Kruglyak and Daly [1998] for a discussion of the utility of empirical significance levels in linkage analysis.)
Correlations to Age and between Different Linkage Signals
We calculated each family’s contribution to the overall nonparametric LOD score. To evaluate whether the loci we identified are associated with an earlier age at onset, we calculated Pearson’s correlation coefficient between the LOD score and the average age at diagnosis of affected individuals in each family. We used a one-tailed test to examine whether linkage was stronger in families with a lower mean age at diagnosis. To evaluate whether there was evidence for interaction between different loci, we calculated Pearson’s correlation coefficient between the LOD score contributions of each family to different pairs of loci. When testing for interaction, we used a two-sided test, similar to the analysis performed by Majewski et al. (2003).
Analysis of HEMICENTIN-1
We screened all affected members in the R-AMD group (n=274), 346 additional patients with AMD, and 237 unaffected controls for the Gln5345Arg mutation. A 225-bp region of exon 104 of the HEMICENTIN-1 gene was amplified by PCR by use of the following primers: F-5′-CATTAATGAATGTGAACAAGTGC-3′ and R-5′-TGTCTGTAATGCTGTTGAGGTTG-3′. The amplified product was sequenced at the University of Michigan DNA Sequencing Core, and the chromatograms were read using the Chromas software, version 1.45 (Griffith University, School of Health Science).
Results
The characteristics of the families with AMD that were genotyped in our study, under both the restricted and broad models of disease, are summarized in table 1. Note that all of the families in our study include at least one individual who satisfies the strict definition of AMD, and 82% of families (93/113) include two such affected individuals (table 1). In addition, 93% of all of the families in our study include one genotyped individual who was diagnosed with bilateral AMD and has the late-stage disease (GA or CNV). Broadening the definition of disease did not significantly alter the characteristics of the study population (table 1). Under both definitions, the average age, sex, and percentage of individuals with late-stage forms of disease were roughly equivalent. The overall family structures were also similar: in the R-AMD model, 84% of the families were composed of three or fewer affected individuals, whereas in the B-AMD model, the families were slightly larger, and 78% had three or fewer affected individuals (table 1). Under both models, >95% reported a family history that included at least one affected first-degree relative, and >55% reported a family history with at least three affected individuals in two generations of the family.
Table 1.
Sample Characteristics Using the Restricted and Broad Diagnostic Criteria (Genotyped Individuals Only)
|
Using Restricted Diagnostic Criteria (R-AMD) |
Using Broad Diagnostic Criteria (B-AMD) |
|||||||
| Characteristic | AMD | GA | CNV | GA or CNV | AMD | GA | CNV | GA or CNV |
| No. (%) of affected individuals | 274 (100) | 95 (35) | 133 (49) | 190 (69) | 331 (100) | 102 (31) | 134 (40) | 195 (59) |
| No. (%) of affected females | 184 (67) | 61 (64) | 91 (68) | 128 (67) | 221 (67) | 65 (64) | 91 (68) | 130 (67) |
| No. of families with: | ||||||||
| ⩾1 affected individual | 113 | 64 | 89 | 105 | 113 | 65 | 89 | 105 |
| ⩾2 affected individuals | 93 | 19 | 28 | 56 | 113 | 22 | 28 | 57 |
| ⩾3 affected individuals | 35 | 7 | 12 | 18 | 47 | 7 | 12 | 18 |
| Affected individuals per family with ⩾2 affected individuals | 2.73 | 2.63 | 2.57 | 2.52 | 2.9 | 2.7 | 2.6 | 2.6 |
| Average age at diagnosis: | ||||||||
| Mean (stdev) | 70.2 (9.2) | 68.4 (10.2) | 70.5 (9.2) | 69.8 (9.5) | 70.3 (9.1) | 68.7 (10.2) | 70.5 (9.2) | 69.8 (9.5) |
| Median (Q1−Q3) | 71.0 (64.0−76.0) | 70.0 (61.0−75.0) | 71.0 (65.0−76.0) | 71.0 (63.5−76.0) | 71.0 (64.0−76.0) | 70.0 (61.0−76.0) | 71.0 (65.0−76.0) | 71.0 (63.5−76.0) |
| Average age at last examination: | ||||||||
| Mean (stdev) | 79.0 (7.6) | 80.2 (7.1) | 79.4 (7.6) | 79.5 (7.5) | 78.2 (7.9) | 80.3 (7.2) | 79.5 (7.6) | 79.6 (7.5) |
| Median (Q1−Q3) | 79.2 (74.3−84.4) | 80.8 (74.8−86.1) | 79.8 (74.4−84.1) | 79.8 (74.8−84.7) | 78.6 (73.6−84.0) | 80.9 (74.9−86.1) | 79.8 (74.5−84.1) | 79.8 (74.8−84.8) |
In the R-AMD analysis, 274 individuals were analyzed; the probands and their family members were predominantly affected with late-stage forms of AMD (table 1), and 75% of the family pairs were composed of siblings. The B-AMD clinical model added 57 affected individuals and 22 informative families (table 1). Although the bias toward late-stage forms (GA and/or CNV) was maintained in this expanded study population (58%–70% of all affected individuals), the B-AMD model included nearly double the number of individuals with other forms of AMD.
Single-Point Results
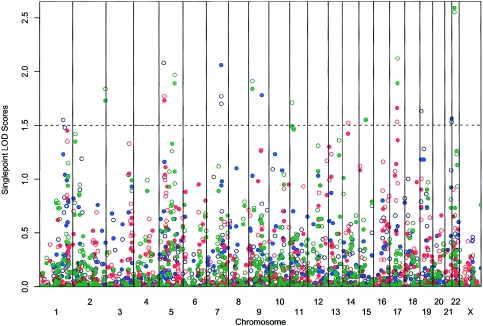
Our initial single-point analyses identified several linkage signals at a LOD score (Kong and Cox 1997) threshold of 1.5 (summarized in fig. 1 and in table 2). We observed the strongest evidence for linkage (LOD=2.59; asymptotic P=.0003) among patients who met the strict definition of CNV on chromosome 22 at marker AGAT055Z (25 cM on the Marshfield genetic map). This finding overlaps with a previous report of linkage (Seddon et al. 2003) and is near the TIMP3 gene (Weber et al. 1994), which, when mutated, can cause Sorsby fundus dystrophy. We also observed suggestive evidence for linkage with the CNV phenotype on chromosomes 9 and 19. The chromosome 9 signal overlaps with two previously reported findings (Weeks et al. 2001; Majewski et al. 2003). Using the broader AMD classification and GA phenotypes, we observed the strongest evidence for linkage (LOD>2.0) on chromosome 5. This peak is 25 cM telomeric to a recent linkage finding (Schick et al. 2003).
Figure 1.
Nonparametric linkage analysis at each marker by use of the NPLALL statistics (Whittemore and Halpern 1994). LOD scores (Kong and Cox 1997) for six disease subtypes were plotted for every marker location. The solid symbols denote the strict trait definition, and the empty ones denote the broad trait definition. The blue symbols indicate AMD, the red symbols indicate GA, and the green symbols indicate CNV.
Table 2.
Peak Single-Point LOD Scores[Note]
|
Peak LOD for Phenotype |
|||||||
| Chromosomeand Marker | Location(cM) | B-AMD | R-AMD | B-GA | R-GA | B-CNV | R-CNV |
| 1: | |||||||
| GATA51H01 | 205 | 1.55 (.004) | … | … | … | … | … |
| 2: | |||||||
| AGAT021 | 266 | … | … | … | … | 1.84 (.002) | 1.73 (.002) |
| 5: | |||||||
| GATA12A08 | 40 | 2.08 (.001) | … | … | … | … | … |
| GATA7C06 | 45 | … | … | 1.77 (.002) | 1.73 (.002) | … | … |
| AAT255 | 130 | … | … | … | … | 1.97 (.001) | 1.89 (.002) |
| 7: | |||||||
| TTTA001 | 125 | 1.77 (.002) | 2.06 (.001) | … | … | … | … |
| AGAT133 | 126 | 1.70 (.003) | … | … | … | … | … |
| 9: | |||||||
| AGAT142 | 34 | … | … | … | … | 1.91 (.0015) | 1.84 (.002) |
| GGAA22E01 | 11 | … | 1.78 (.002) | … | … | … | … |
| 11: | |||||||
| GATA48E02 | 21 | … | … | … | … | 1.71 (.003) | … |
| 14: | |||||||
| GATA90G11 | 51 | … | … | 1.52 (.004) | … | … | … |
| 15: | |||||||
| GATA151F03N | 60 | … | … | … | … | 1.55 (.004) | 1.55 (.004) |
| 17: | |||||||
| GATA25A04 | 62 | … | … | 1.53 (.004) | 1.66 (.003) | … | … |
| ATC6A06N | 67 | … | … | … | … | 2.12 (.0009) | 1.89 (.002) |
| 19: | |||||||
| GATA146H09 | 11 | 1.63 (.003) | … | … | … | … | … |
| 21: | |||||||
| GATA70B08 | 58 | 1.53 (.004) | 1.56 (.004) | … | … | … | … |
| 22: | |||||||
| AGAT055Z | 25 | … | … | … | … | 2.55 (.0003) | 2.59 (.0003) |
| X: | |||||||
| TTTA062 | 181 | … | … | … | … | 1.91 (.002) | 1.82 (.002) |
Note.— Nonparametric linkage analysis was performed using the SALL statistic (Whittemore and Halpern 1994). The table lists all markers with a LOD score (Kong and Cox 1997) >1.5 in single-point analyses, with the peak P value in parentheses. Marker locations are given according to the Marshfield genetic map.
Finally, our study also replicates another promising linkage finding. With the broad AMD phenotype, we observed suggestive evidence of linkage on chromosome 1, at marker GATA51H01 (LOD=1.55; P=.004; 205 cM). Linkage in this region has been reported in three other affected relative pair studies (Weeks et al. 2001; Majewski et al. 2003; Seddon et al. 2003) and in the study of a single large pedigree with AMD (Klein et al. 1998).
Multipoint Analyses
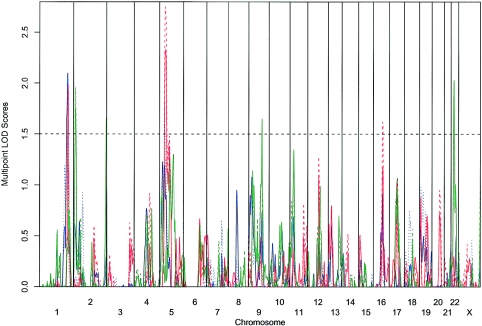
Overall, our multipoint analyses (figs. 2 and 3; table 3) identified a subset of the regions observed in the single-point analysis. With the AMD and GA traits, multipoint analysis modestly increased the evidence for linkage on chromosome 1 (LOD=2.13; P=.0009) near the peak that was reported in other affected relative pair studies (Weeks et al. 2001; Majewski et al. 2003; Seddon et al. 2003); it also increased the evidence for linkage on chromosome 5 (LOD=2.55; P=.0003) near the linkage finding of Schick et al. (2003). In contrast, evidence for linkage was weakened for the chromosome 9 and 22 findings. Although multipoint analysis is theoretically more powerful, it is also more sensitive to errors in genotyping and in genetic map specification (Abecasis et al. 2001a; Sullivan et al. 2003). Thus, although it is likely that some of the unreplicated single-point linkage signals may be false positives, it is possible that other signals were lost due to genotyping errors or genetic map inaccuracies.
Figure 2.
Multipoint nonparametric linkage analysis at equally spaced locations throughout the genome by use of the NPLALL statistics (Whittemore and Halpern 1994). LOD scores (Kong and Cox 1997) for six disease subtypes are plotted at 1-cM intervals. The solid lines denote the strict trait definition, and the dotted lines denote the broad trait definition. The blue line indicates AMD, the red line indicates GA, and the green line indicates CNV.
Figure 3.
Multipoint nonparametric linkage analysis at equally spaced locations throughout the genome by use of the NPL-ALL statistics (Whittemore and Halpern 1994). LOD scores (Kong and Cox 1997) for six disease subtypes are plotted at 1-cM intervals. The solid lines denote the strict trait definition, and the dotted lines denote the broad trait definition. The blue lines indicate AMD, the red lines indicate GA, and the green lines indicate CNV. The multipoint LOD score plots for four chromosomes of interest (1, 5, 9, and 22) are shown.
Table 3.
Peak Multipoint LOD Scores[Note]
| Chromosomeand Disease Trait | Nearest Marker | MarkerLocation (cM) | Position(cM) | LOD | Interval(cM) | Peak P Value |
| 1: | ||||||
| AMD BROAD | GAT004 | 236 | 235 | 1.51 | (208–239) | .004 |
| AMD RESTR | GAT004 | 236 | 236 | 2.13 | (223–244) | .0009 |
| GA BROAD | GATA4H09 | 240 | 238 | 1.95 | (233–243) | .0014 |
| GA RESTR | GATA4H09 | 240 | 240 | 1.98 | (234–245) | .0013 |
| 2: | ||||||
| CNV BROAD | GATA72G11N | 10 | 10 | 1.96 | (4–18) | .0013 |
| CNV RESTR | GATA72G11N | 10 | 10 | 1.76 | (4–17) | .002 |
| CNV RESTR | AGAT021 | 266 | 267 | 1.63 | (262–267) | .003 |
| 5: | ||||||
| AMD BROAD | GATA12A08 | 40 | 42 | 1.72 | (38–56) | .002 |
| GA BROAD | GATA63C02 | 50 | 50 | 2.55 | (39–91) | .0003 |
| GA RESTR | GATA63C02 | 50 | 50 | 2.13 | (40–89) | .0009 |
| 9: | ||||||
| CNV BROAD | GGAA22E01 | 111 | 111 | 1.64 | (99–112) | .003 |
| CNV RESTR | GGAA22E01 | 111 | 111 | 1.65 | (103–112) | .003 |
| 22: | ||||||
| CNV BROAD | AGAT055Z | 25 | 25 | 1.87 | (19–31) | .002 |
| CNV RESTR | AGAT055Z | 25 | 25 | 2.03 | (19–31) | .0011 |
Note.— Nonparametric linkage analysis was performed using the SALL statistic (Whittemore and Halpern 1994). The table lists all linkage peaks with a LOD score (Kong and Cox 1997) >1.5 in multipoint analyses. For each peak, the LOD score cM location and P values are listed (according to the Marshfield genetic map), as well as the interval in which the LOD score is >1.0.
Simulation Results (Single-Point Analysis)
Overall, none of our individual linkage signals reached genomewide significance. For example, our most significant finding, a LOD score of 2.55 for the broad CNV phenotype, was observed by chance in 13/100 simulated genomes (table 4). The result is similar when we consider the restricted CNV phenotype, for which a LOD score of 2.59 was observed in 9/100 replicates. However, our simulations suggest that at least some of our observed linkage signals are likely to be real. For example, whereas we observed 7 linkage signals for the CNV phenotype at a LOD score threshold of 1.55, only 2.5 such signals are expected to occur by chance and 0/100 (none) simulated data sets showed as many signals. This is consistent with an oligogenic basis for AMD. That is, multiple loci are likely to be involved, and, although the effect of each locus is small (so that larger samples are required to achieve genomewide significance), they result in an excess of modest linkage peaks in studies such as this one. The results were less striking for the B-AMD analysis (5 peaks observed, 2.70 expected) and GA phenotypes (3 peaks observed, 2.53 expected; table 4). When all three phenotype classifications are considered, we observed a total of 14 linkage signals at a LOD score threshold of 1.52, compared with 6.87 expected to occur by chance. Only one of our replicate data sets included ⩾14 signals by chance. Results were similar for the stricter disease definition (not shown).
Table 4.
Summary of Simulation Results (Single-Point Analyses) Using the Broad Phenotype Definition
| Trait andThreshold | ObservedPeak (x) | ExpectedFalse Positive | Proportion of ReplicatesWhere X⩾x | Observed Signal |
| AMD-B: | ||||
| 2.08 | 1 | .70 | 54/100 | GATA12A08, Chr 5, 40 cM |
| 1.77 | 2 | 1.51 | 47/100 | TTTA001, Chr 7, 125 cM |
| 1.63 | 3 | 2.21 | 42/100 | GATA146H09, Chr 19, 11 cM |
| 1.55 | 4 | 2.62 | 32/100 | GATA51H01, Chr 1, 205 cM |
| 1.53 | 5 | 2.70 | 17/100 | GATA70B08, Chr 21, 58 cM |
| GA-B: | ||||
| 1.77 | 1 | 1.11 | 65/100 | GATA7C06, Chr 5, 45 cM |
| 1.53 | 2 | 2.46 | 73/100 | GATA25A04, Chr 17, 62 cM |
| 1.52 | 3 | 2.53 | 45/100 | GATA90G11, Chr 14, 51 cM |
| CNV-B: | ||||
| 2.55 | 1 | .13 | 13/100 | AGAT055Z, Chr 22, 25 cM |
| 2.12 | 2 | .58 | 16/100 | ATC6A06N, Chr 17, 67 cM |
| 1.97 | 3 | .95 | 7/100 | AAT255, Chr 5, 130 cM |
| 1.91 | 5 | 1.13 | 0/100 | AGAT142, Chr 9, 34 cM |
| TTTA062, Chr X, 181 cM | ||||
| 1.84 | 6 | 1.40 | 0/100 | AGAT021, Chr 2, 266 cM |
| 1.71 | 7 | 1.96 | 0/100 | GATA48E02, Chr 11, 21 cM |
| 1.55 | 8 | 2.80 | 0/100 | GATA151F03N, Chr 15, 60 cM |
Simulation Results (Multipoint Analysis)
Because of computational constraints, we only generated 25 simulated genomes for multipoint analysis. As in the single-point analysis, no individual peak appears to reach genomewide significance, and peaks with similar or higher LOD scores were observed in some of the simulated genomes. For example, the most significant LOD score in our original multipoint analysis was a LOD=2.55 observed on chromosome 5 for the broad GA phenotype. For this phenotype, we observed a similar or higher LOD score on 3/25 simulated genomes. When we considered all six (three broad and three restricted) phenotype classifications, a similar or higher LOD score was observed on 11/25 simulated genomes. Nevertheless, for all the LOD score thresholds and phenotype classifications we considered, the original data set shows more linkage signals than the average simulated genome. For example, at a LOD score threshold of 1.64 we observed linkage signals on three chromosomes for the broad CNV phenotype—an event that occurred in only 2/25 simulated genomes. At the same LOD score threshold, we observed evidence for linkage in five autosomes when all phenotypes were considered—an event that occurred in 9/25 genomes.
Evidence for Interaction
We found only weak evidence for interaction between different linkage signals. The correlation between the signals on chromosomes 9 and 22 was r=.34 (P=.04, but not significant after adjusting for multiple testing). We also found that the mean age at diagnosis was typically lower in families showing evidence of linkage to chromosome 1 and 2 (correlation r=.52 and r=.34, respectively; P=.01 and P=.03), but again this result is not significant after adjustments for multiple testing.
HEMICENTIN-1
The Gln5345Arg change in HEMICENTIN-1 was not detected in any of the 620 (274 R-AMD and 346 other) affected individuals and 237 unaffected controls. Moreover, no sequence variation was identified in exon 104 in any of the DNA samples examined.
Discussion
We report here the results of a nonparametric linkage analysis of the first high-resolution genomewide scan for AMD susceptibility loci. Our AMD population included affected relative pairs that were enriched for late-stage disease forms (GA and/or CNV); this allowed us to search for susceptibility loci for the more severe AMD phenotypes. Our approach is biased toward disease phenotypes with higher genetic susceptibility load. It is unclear if GA and CNV are separate disease entities or part of a disease spectrum. Some individuals can be affected by only one form of late-stage disease, whereas others can be affected by both, suggesting that the two late-stage forms may be distinct yet share some common genetic predisposition. Our results provide stronger evidence of genetic linkage when analyses are focused on a subset of individuals with one of the late-stage phenotypes. Overall, we identified several modest linkage signals; both single- and multipoint analyses provided indications of linkage on chromosomes 1q, 2p, 5p, 9q, and 22q.
Four groups have recently published whole genome scan linkage results; however, so far only suggestive evidence has been obtained for AMD susceptibility loci (Weeks et al. 2001; Majewski et al. 2003; Schick et al. 2003; Seddon et al. 2003). Identification of numerous putative chromosomal locations in these studies is consistent with the underlying genetic complexity of AMD pathogenesis. Yet, it is most encouraging that, in spite of differences in patient population and diverse methods of analysis, many of our linkage signals were also detected in at least one of the previous reports. The validation of these loci by independent studies argues in favor of these loci harboring genes that contribute to the development of AMD.
One of the signals we identified on 1q31 (with a multipoint LOD score = 2.13, using the R-AMD classification) was originally described by linkage analysis in a large AMD family (Klein et al. 1998) and has now been reported in three (Weeks et al. 2001; Majewski et al. 2003; Seddon et al. 2003) of four published affected relative pair studies for AMD. The fourth study (Schick et al. 2003) used a population with both mild and severe forms of disease and observed weak evidence for linkage. Consistent with Klein et al. (1998), most of this linkage signal in our study appears to be contributed by families with GA (correlation between per-family LOD scores for GA and AMD at this position is 0.56; P=.006). Thus, it appears that the chromosome 1q locus is involved in AMD susceptibility; however, the extent of its involvement in disease pathogenesis is unclear at this stage. The estimates for the percentage of families linked to chromosome 1q varies from 7% to 40% (Weeks et al. 2001; Majewski et al. 2003). In our study, >49% of the families in each disease classification provide evidence for linkage of AMD susceptibility to chromosome 1q (LOD>0).
Recently, Schultz et al. (2003) suggested that a Gln5345Arg variant in exon 104 of HEMICENTIN-1 is responsible for ARMD1 locus. We did not detect this variant, or any other sequence change, in exon 104 of HEMICENTIN-1 either in 274 individuals in the R-AMD group, which exhibited the highest LOD score for this region, or in 346 additional patients with AMD. Our results suggest that variation(s) in another susceptibility gene at 1q, and not the Gln5345Arg change in HEMICENTIN-1, contribute to the disease in our AMD cohort. Our linkage peak is telomeric of HEMICENTIN-1, and, in our view, the results of Schultz et al. (2003) are compatible with the hypothesis that Gln5345Arg variant is a marker for the disease haplotype in their large pedigree but is not itself causal. We believe that further detailed studies of the candidate region on chromosome 1q are warranted. The region encompasses many other interesting candidate genes, including phosducin, two genes involved in the protein-ubiquitination pathway (ubiquitin carboxyl-terminal hydrolase L5 and HSPC150), the regulator of fas-induced apoptosis, and several genes for immune modulatory proteins (e.g., interleukin 10, interferon regulatory factor 6, complement related molecules, and TNF receptor-associated factor 5).
In multipoint analyses, our most striking linkage finding in families with GA is on chromosome 5p (50 cM on the Marshfield genetic map), close to another previously reported linkage signal (Schick et al. 2003). Interesting candidate genes in this region include caspase recruitment domain family member 6 (CARD6) and genes for several immune-related molecules (gp130-like monocyte receptor, interleukin 6 signal transducer, complement-c1q tumor necrosis factor-related protein 3, and leukemia inhibitory factor receptor).
Our data support the linkage peak obtained by Majewski et al. (2003) on chromosome 9q, which was their most robust peak, observed in both their initial and expanded sample and by both parametric and nonparametric linkage analysis. It is interesting to note that Weeks et al. (2001) reported a possible locus at a location ∼40 cM away; but their evidence was weakened after the number of affected sib pairs analyzed was increased. Our evidence for linkage to chromosome 9 was strengthened when the analysis was performed using only CNV sib pairs. This region contains genes for forkhead box E1 (FOXE1) and ATP-binding cassette, subfamily A (ABC1); the latter is associated with Tangier disease and familial high-density lipoprotein deficiency.
In addition to the three linkage signals (chromosome 1q, 5p, and 9q) that we consider significant, we found evidence for a susceptibility locus on chromosome 22 near TIMP3, a gene mutated in Sorsby fundus dystrophy (Weber et al. 1994), when we focused our analysis on the subset of patients with CNV. Most patients with Sorsby fundus dystrophy develop night blindness in their 20s and subfoveal choroidal neovascularization by 40 years of age. In contrast, the average age at diagnosis for our CNV patients is >70 years (see table 1). Although no TIMP3 association was evident by the analysis of 188 patients with AMD (Stone et al. 2001), we speculate that sequence variations in TIMP3 may lead to a later-onset neovascularization phenotype, possibly in association with other AMD susceptibility loci and environmental factors. It will therefore be of interest to perform TIMP3 association study in patients with CNV. Other potentially interesting candidates on chromosome 22 include the genes for heat shock 27 protein 3, leukemia inhibitory factor (LIF), calcineurin, ubiquinol-cytochrome c reductase complex, complement-c1q tumor necrosis factor-related protein 6, caspase recruitment domain family member 10, and several APOB-editing enzymes. Recently, both mRNA and protein for APOB were found to be expressed in normal adult RPE, and APOB was detected in drusen and basal deposits, suggesting a link between APOB and development of cholesterol-enriched lesions in AMD (Malek et al. 2003).
We did not find significant evidence for other previously suggested AMD loci in our analysis (Weeks et al. 2001; Majewski et al. 2003; Schick et al. 2003; Seddon et al. 2003). Prominent among these is the putative locus on chromosome 10q26, which was reported in three studies; although the evidence was weak in two reports (Weeks et al. 2001; Seddon et al. 2003), Majewski et al. (2003) obtained a HLOD score of 3 in nuclear families when parametric analysis with high penetrance was performed. Our nonparametric analysis did not demonstrate any evidence for an AMD susceptibility locus on chromosome 10.
In summary, this nonparametric linkage analysis of a high-resolution (5-cM) whole genome scan provides support for several previously reported chromosomal locations of AMD loci. In addition, we suggest new genetic loci (e.g., on chromosome 2) that may predispose individuals to late-stage AMD (GA or CNV). We have also found weak but interesting evidence for interaction between different susceptibility loci. When searching for genetic loci involved in complex diseases, crossvalidation of genetic data by different groups is important, since it is not easy for one group to obtain statistically significant evidence of linkage. The consistency between our results and previous studies suggests that genetic studies should be successful in identifying the molecular basis of AMD, thereby assisting in diagnosis, as well as in the design of novel prevention and therapeutic strategies.
Acknowledgments
We are grateful to numerous individuals and families who generously donated their time and resources. We thank Drs. Paul R. Lichter, Ron Kurtz, Don Puro, Michael Smith Wheelock, David Zacks, and Stephen Saxe; Ms. Kathryn B. Moore and Laurie Jessup, for their efforts in patient collection; many retina physicians in the State of Michigan and the Great Lakes region, for their support of this project; and the clinical staff at the Kellogg Eye Center Retina Clinic, whose assistance made this work possible. We acknowledge Dr. Michael Boehnke and Heather Stringham, for their suggestions and input during the early stages of the project, and Ms. Sharyn Ferrara, for administrative assistance. The genomewide scan was performed by the NHLBI Mammalian Genotyping Service (http://research.marshfieldclinic.org/genetics/), under the direction of Dr. James Weber. This research was supported by grants from the Macula Vision Research Foundation (West Conshohocken, PA), the Foundation Fighting Blindness (Owings Mills, MD), the Elmer and Sylvia Sramek Charitable Foundation (Chicago, IL), the National Institutes of Health (F32-EY014085), the American Federation for Aging Research (New York, NY), Research to Prevent Blindness (New York, NY), and the Harold F. Falls Collegiate Professorship.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Abecasis Lab Web Site, http://www.sph.umich.edu/csg/abecasis/ (for MERLIN and MINX)
- Marshfield Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/ (for Marshfield genetic map)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AMD) [PubMed]
References
- Abecasis GR, Cherny SS, Cardon LR (2001a) The impact of genotyping error on linkage and association analysis of quantitative traits. Eur J Hum Genet 9:130–134 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001b) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 10.1093/bioinformatics/17.8.742 [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin: rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Allikmets R, International ABCR Screening Consortium (2000) Further evidence for an association of ABCR alleles with age-related macular degeneration. The International ABCR Screening Consortium. Am J Hum Genet 67:487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP (2003) Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv Ophthalmol 48:257–293 10.1016/S0039-6257(03)00030-4 [DOI] [PubMed] [Google Scholar]
- Bird AC (2003) Towards an understanding of age-related macular disease. Eye 17:457–466 10.1038/sj.eye.6700562 [DOI] [PubMed] [Google Scholar]
- Bird AC, Bressler NM, Bressler SB, Chisholm IH, Coscas G, Davis MD, de Jong PT, et al (1995) An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol 39:367–374 [DOI] [PubMed] [Google Scholar]
- Bressler NM, Bressler SB, Fine SL (1988) Age-related macular degeneration. Surv Ophthalmol 32:375–413 [DOI] [PubMed] [Google Scholar]
- Cai J, Nelson KC, Wu M, Sternberg P Jr, Jones DP (2000) Oxidative damage and protection of the RPE. Prog Retin Eye Res 19:205–221 10.1016/S1350-9462(99)00009-9 [DOI] [PubMed] [Google Scholar]
- Crabb JW, Miyagi M, Gu, Xiaorong, Shadrach K, West KA, Sakahuchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Galomon RG, Hollyfield JG (2002) Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci USA 99:14682–14687 10.1073/pnas.222551899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWan AT, Parrado AR, Matise TC, Leal SM (2002) The map problem: a comparison of genetic and sequence-based physical maps. Am J Hum Genet 70:101–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M (2000) Improved inference of relationship for pairs of individuals. Am J Hum Genet 67:1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR (2001) Risk factors for age-related macular degeneration. Prog Retin Eye Res 20:227–253 10.1016/S1350-9462(00)00023-9 [DOI] [PubMed] [Google Scholar]
- Gorin MB, Breitner JC, De Jong PT, Hageman GS, Klaver CC, Kuehn MH, Seddon JM (1999) The genetics of age-related macular degeneration. Mol Vis 5:29 [PubMed] [Google Scholar]
- Green WR (1999) Histopathology of age-related macular degeneration. Mol Vis 5:27 [PubMed] [Google Scholar]
- Hageman GS, Luthert PJ, Chong NHV, Johnson LV, Anderson DH, Mullins RF (2001) An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res 20:705–732 10.1016/S1350-9462(01)00010-6 [DOI] [PubMed] [Google Scholar]
- Hekimi S, Guarente L (2003) Genetics and the specificity of the aging process. Science 299:1351–1354 10.1126/science.1082358 [DOI] [PubMed] [Google Scholar]
- Hirvela H, Luukinen H, Laara E, Sc L, Laatikainen L (1996) Risk factors of age-related maculopathy in a population 70 years of age or older. Ophthalmology 103:871–877 [DOI] [PubMed] [Google Scholar]
- Johnson LV, Leitner WP, Rivest AJ, Staples MK, Radeke MJ, Anderson DH (2002) The Alzheimer’s Aβ-peptide is deposited at sites of complement activation in pathologic deposits associated with aging and age-related macular degeneration. Proc Natl Acad Sci USA 99:11830–11835 10.1073/pnas.192203399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaver CCW, Wolfs RCW, Assink JJM, van Duijn CM, Hofman, A, de Jong TVM (1998) Genetic risk of age-related maculopathy. Arch Ophthalmol 116:1646–1651 [DOI] [PubMed] [Google Scholar]
- Klein ML, Schultz DW, Edwards A, Matise TC, Rust K, Berselli CB, Trzupek K, Weleber RG, Ott J, Wirtz MK, Acott TS (1998) Age-related macular degeneration. Clinical features in a large family and linkage to chromosome 1q. Arch Ophthalmol 116:1082–1088 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Jensen SC, Meuer SM (1997) The five-year incidence and progression of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology 104:7–21 [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Linton KL (1992) Prevalence of age-related maculopathy. The Beaver Dam Eye Study. Ophthalmology 99:933–943 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ (1998) Linkage thresholds for two-stage genome scans. Am J Hum Genet 62:994–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Schultz DW, Weleber RG, Schain MB, Edwards AO, Matise TC, Acott TS, Ott J, Klein ML (2003) Age-related macular degeneration—a genome scan in extended families. Am J Hum Genet 73:540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek G, Li C-M, Guidry C, Medeiros NE, Curcio CA (2003) Apolipoprotein B in cholesterol-containing drusen and basal deposits of human eyes with age-related maculopathy. Am J Path 162:413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Advisory Eye Council (1998) Vision research: a national plan. National Institutes of Health (publication 98-4120). http://www.nei.nih.gov/resources/strategicplans/plan.htm (accessed February 9, 2004)
- Prevent Blindness America and National Eye Institute (2002) Vision problems in the United States: prevalence of adult vision impairment and age-related eye disease in America. http://www.nei.nih.gov/eyedata/pdf/VPUS.pdf (accessed February 9, 2004)
- Risch N (1990) Linkage strategies for genetically complex traits II: the power of affected relative pairs. Am J Hum Genet 46:229–241 [PMC free article] [PubMed] [Google Scholar]
- Schick JH, Iyengar SK, Klein BE, Klein R, Reading K, Liptak R, Millard C, Lee KE, Tomany SC, Moore EL, Fijal BA, Elston RC (2003) A whole-genome screen of a quantitative trait of age-related maculopathy in sibships from the Beaver Dam Eye Study. Am J Hum Genet 72:1412–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Klaver C, Saunders A, Postel E, De La Paz M, Agarwal A, Small K, Udar N, Ong J, Chalukya M, Nesburn A, Kenney C, Domurath R, Hogan M, Mah T, Conley Y, Ferrell R, Weeks D, de Jong PT, van Duijn C, Haines J, Pericak-Vance M, Gorin M (2002) A pooled case-control study of the apolipoprotein E (APOE) gene in age-related maculopathy. Ophthalmic Genet 23:209–223 10.1076/opge.23.4.209.13883 [DOI] [PubMed] [Google Scholar]
- Schultz DW, Klein ML, Humpert AJ, Luzier CW, Persun V, Schain M, Mahan A, Runckel C, Cassera M, Vittal V, Doyle TM, Martin TM, Weleber RG, Francis PJ, Acott TS (2003) Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum Mol Genet 12:3315–3323 10.1093/hmg/ddg348 [DOI] [PubMed] [Google Scholar]
- Seddon JM, Ajani UA, Mitchell BD (1997) Familial aggregation of age-related maculopathy. Am J Ophthalmol 123:199–206 [DOI] [PubMed] [Google Scholar]
- Seddon JM, Santangelo SL, Book K, Chong S, Cote J (2003) A genomewide scan for age-related macular degeneration provides evidence for linkage to several chromosomal regions. Am J Hum Genet 73:780–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengul H, Weeks DE, Feingold E (2001) A survey of affected-sibship statistics for nonparametric linkage analysis. Am J Hum Genet 69:179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W, Assink J, Klein R, Mitchell P, Klaver CC, Klein BE, Hofman A, Jensen S, Wang JJ, de Jong PT (2001) Risk factors for age-related macular degeneration: pooled findings from three continents. Ophthalmology 108:697–704 10.1016/S0161-6420(00)00580-7 [DOI] [PubMed] [Google Scholar]
- Stone EM, Sheffield VC, Hageman GS (2001) Molecular genetics of age-related macular degeneration. Hum Mol Genet 10:2285–2292 10.1093/hmg/10.20.2285 [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale BM, Neale MC, van den Oord E, Kendler KS (2003) Multipoint and single point non-parametric linkage analysis with imperfect data. Am J Med Genet 121B:89–94 [DOI] [PubMed] [Google Scholar]
- Vingerling JR, Dielemans I, Hofman A, Grobbee DE, Hijmering M, Kramer CF, De Jong PT (1995) The prevalence of age-related maculopathy in the Rotterdam study. Ophthalmology 102:205–210 [DOI] [PubMed] [Google Scholar]
- Weber BH, Vogt G, Pruett RC, Stohr H, Felbor U (1994) Mutations in the tissue inhibitor of metalloproteinases-3 (TIMP3) in patients with Sorsby’s fundus dystrophy. Nat Genet 8:352–356 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Mah TS, Paul TO, Morse L, Ngo-Chang J, Dailey JP, Ferrell RE, Gorin MB (2000) A full genome scan for age-related maculopathy. Hum Mol Genet 9:1329–1349 10.1093/hmg/9.9.1329 [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai HJ, Mah TS, Rosenfeld PJ, Paul TO, Eller AW, Morse LS, Dailey JP, Ferrell RE, Gorin MB (2001) Age-related maculopathy: an expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions. Am J Ophthalmol 132:682–692 10.1016/S0002-9394(01)01214-4 [DOI] [PubMed] [Google Scholar]
- Weir BS (1990) Genetic data analysis. Sinauer Associates, Sunderland, MA [Google Scholar]
- Whittemore AS, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Yoshida S, Yashar BM, Hiriyanna S, Swaroop A (2002) Microarray analysis of gene expression in the aging human retina. Invest Ophthalmol Vis Sci 43:2554–2560 [PubMed] [Google Scholar]