Abstract
Rett syndrome (RTT), a neurodevelopmental disorder affecting mostly females, is caused by mutations in the X-linked gene encoding methyl-CpG–binding protein 2 (MeCP2). Although the majority of girls with classic RTT have a random pattern of X-chromosome inactivation (XCI), nonbalanced patterns have been observed in patients carrying mutant MECP2 and, in some cases, account for variability of phenotypic manifestations. We have generated an RTT mouse model that recapitulates all major aspects of the human disease, but we found that females exhibit a high degree of phenotypic variability beyond what is observed in human patients with similar mutations. To evaluate whether XCI influences the phenotypic outcome of Mecp2 mutation in the mouse, we studied the pattern of XCI at the single-cell level in brains of heterozygous females. We found that XCI patterns were unbalanced, favoring expression of the wild-type allele, in most mutant females. It is notable that none of the animals had nonrandom XCI favoring the mutant allele. To explore why the XCI patterns favored expression of the wild-type allele, we studied primary neuronal cultures from Mecp2-mutant mice and found selective survival of neurons in which the wild-type X chromosome was active. Quantitative analysis indicated that fewer phenotypes are observed when a large percentage of neurons have the mutant X chromosome inactivated. The study of neuronal XCI patterns in a large number of female mice carrying a mutant Mecp2 allele highlights the importance of MeCP2 for neuronal viability. These findings also raise the possibility that there are human females who carry mutant MECP2 alleles but are not recognized because their phenotypes are subdued owing to favorable XCI patterns.
Introduction
Rett syndrome (RTT [MIM 312750]) is a neurodevelopmental disorder that affects predominantly females (Rett 1966; Hagberg et al. 1983; Rett Syndrome Diagnostic Criteria Work Group 1988). RTT is caused by mutations in MECP2, a gene, located on Xq28, that encodes for methyl-CpG–binding protein 2 (MeCP2) (Amir et al. 1999). Girls with classic RTT appear normal from birth until ∼6–18 mo of age, but then they fail to acquire new milestones and enter a period of regression during which motor and language skills are lost. These girls also have poor coordination, tremor, impaired social interaction, and stereotypic hand movements. In addition to these classic phenotypes, a vast range of phenotypes has been observed in patients with MECP2 mutations, including milder and more-severe forms of the disease. Patients with more-severe phenotypes develop the disease shortly after birth, without the period of normal development, and often have congenital hypotonia and infantile spasms. Patients with a milder variant may retain some speech and motor functions and do not have seizures (Hagberg 1995). In addition, MECP2 mutations have been associated with non-RTT phenotypes, including neonatal-onset encephalopathy; Angelman syndrome phenotype (Watson et al. 2001); mild, nonspecific mental retardation (Orrico et al. 2000; Yntema et al. 2002); and autism (Lam et al. 2000; Carney et al. 2003).
Several factors could explain this phenotypic variability, including mutation type and location, cis- and trans-acting modifiers, and the degree of cellular mosaicism for the mutant allele. The latter could be in the form of somatic mosaicism in males or nonrandom X-chromosome inactivation (XCI) in females, given that MECP2 is subject to XCI (Adler et al. 1995). Although the majority of patients with classic RTT have balanced XCI patterns in brain tissue (Shahbazian et al. 2002b), nonrandom patterns of XCI were observed in several cases (Renieri et al. 2003; Weaving et al. 2003). In particular, nonrandom XCI patterns favoring expression of the wild-type allele can enable females who carry classic RTT–causing mutations to be asymptomatic (Sirianni et al. 1998). If the mutation is severe (e.g., if the protein is absent or largely truncated), skewed XCI patterns can also enable females to have a milder phenotype than the mutation would predict (Wan et al. 1999; Bienvenu et al. 2000; Ishii et al. 2001; Zappella et al. 2001; Huppke et al. 2002). Recently, a girl with a 47,XXX karyotype and an RTT-causing MECP2 mutation has been reported to have a relatively mild, atypical form of RTT owing to unbalanced XCI patterns favoring one or the other maternal alleles, thus illustrating an interesting example of potential effects of XCI skewing (Hammer et al. 2003).
Studies of XCI in humans rely mainly on methylation patterns of X-linked polymorphic markers to distinguish the active from the inactive chromosome in blood or epithelial cells. Discordances between these two determinations have been repeatedly reported, suggesting that XCI in the brain cannot always be predicted on the basis of the study of peripheral tissues (Temudo and Maciel 2002). This poses a limitation for the study of XCI in RTT, as the associated phenotype is predominantly, if not exclusively, of neurologic origin. In addition, the type of mutation and the quality of care could also affect the clinical manifestations of the patients (Sharp et al. 2000).
So far, genotype-phenotype studies in mice have been limited to only two mutations, comprising null and hypomorphic alleles of Mecp2 (Chen et al. 2001; Guy et al. 2001; Shahbazian et al. 2002a). The former resembles more severe cases of RTT than the latter, as evidenced by the phenotypes of the mutant males. It is interesting that, although the female mice develop a progressive neurological phenotype in both cases, the degree of variability and patterns of XCI have not been documented from either mutant. To this end, we set out to investigate the patterns of XCI and their effect on the phenotypic outcome in female mice carrying the hypomorphic Mecp2308 mutation (Mecp2308/X) that results in the protein truncated at amino acid 308, similar to mutations found in patients with RTT (Shahbazian et al. 2002a). The study of XCI patterns in female mice carrying a mutant Mecp2 allele has several advantages. It allows the study of an unselected large population of females that have an identical genetic background and the same Mecp2 mutation. In mice, one can also control for environmental factors—such as diet, rearing, and nurturing—that could affect the phenotypic manifestations.
We found that the XCI patterns in most Mecp2308/X–mutant females were unbalanced, favoring expression of the wild-type allele. It is interesting that none of the animals had nonrandom XCI favoring expression of the mutant allele. Quantitative genotype-phenotype studies indicated that the unbalanced XCI patterns mitigate several phenotypes in the Mecp2308/X mice.
Material and Methods
Mouse Breeding and Phenotyping
For all the experiments described in this article, 6- or 12-month–old mice of a pure 129/SvEv genetic background were used. Physical characteristics of mice, such as appearance of fur and periocular region, were noted. Presence of body tremor (Tr) and forepaw stereotypies were assessed by holding the mice in the palm and suspending them by their tails, respectively.
Immunofluorescence
Mouse brains were fixed by transcardial perfusion followed by overnight immersion in PBS-buffered 4% formaldehyde. The brains were embedded in O.C.T. (Tissue Tek) and cryosectioned (50 μm). Sections were blocked for 1 h in 2% normal goat serum and 0.3% triton X-100 in PBS. After blocking, the sections were incubated for 48 h at 4°C in blocking solution containing primary anti-MeCP2 antibody (Upstate) (1:100) in combination with antibodies to one of the following proteins: calbindin (1:1,000), tyrosine hydroxylase (1:1,000), or parvalbumin (1:2,000). The sections were washed four times in PBS and were incubated for 48 h in blocking solution containing secondary antibody. Anti-mouse– or anti-rabbit–conjugated Alexa Fluor 488 (Molecular Probes) was used at a dilution of 1:500, and anti-mouse– or anti-rabbit–conjugated Cy3 (Jackson Immunoresearch) was used at a dilution of 1:500.
Cells in culture were fixed for 30 min in PBS-buffered 4% formaldehyde, were incubated for 15 min in 0.1% triton X-100 in PBS, and were blocked for 30 min in 1% bovine serum albumin, 2% normal goat serum, and 0.05% triton X-100 in PBS. After blocking, the sections were incubated for 2 h at 4°C in blocking solution containing primary anti-MeCP2 antibody (Upstate) (1:150) in combination with antityrosine-tubulin (TUB-1A2) (1:1,000).
Sections were washed as described above and were mounted on microscope slides, with Prolong (Molecular Probes) as mounting media. Cell counting was done manually by going through image stacks of optical sections collected using a Zeiss 510 confocal microscope. Only cells visualized in their totality were counted. Toto-3–iodide (molecular probes) staining of nuclear DNA performed in a subset of sections was consistent with the results obtained using confocal analysis.
Culture of Primary Neurons
Hippocampal neurons were prepared from embryonic day 18 (E18) 129/SvEv embryos, derived from an Mecp2+/Y–Mecp2308/X mating. Embryos were sexed by microscopic examination of internal reproductive organs, and neurons from male hippocampi were paired in every possible combination and were plated in duplicate chambers. Neurons from female embryos were plated in duplicate chambers without pooling. Dissociated cells were plated on poly-D-lysine–coated Lab-TekII glass chambers in Neurobasal medium supplemented with 2% B27, 0.5 mM L-glutamine, and 25 μM glutamate. After 3 d, the medium was changed to Neurobasal/2% B27 without glutamate. Genotyping and sex confirmation of the embryos was performed by PCR, as described elsewhere (Kunieda et al. 1992; Shahbazian et al. 2002a). Neuronal cultures from Mecp2308/X female mice were fixed and stained for immunofluorescence after 4 d and 7 d in culture, whereas cocultures from male mice of different genotypes were fixed and stained for immunofluorescence after 7 d in culture.
Statistical Analysis
Logistic regression was used to determine the association between the absolute level of XCI and the presence of each phenotype. A χ2 test was used to determine the association among three groups on the basis of XCI patterns (group 1 54%–70%, group 2 71%–77%, and group 3 >78%) and the presence of each phenotype. The Kruskal-Wallis test was used to compare the three XCI-pattern groups, with respect to percentage of phenotypes manifested.
Results
High Degree of Phenotypic Variability in Mecp2308/X Female Mice
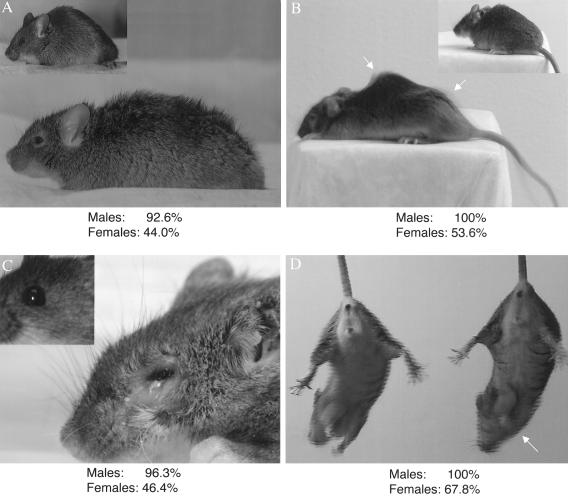
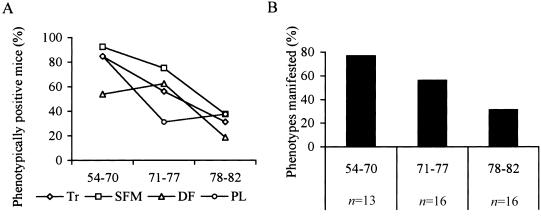
Initial phenotypic characterization of Mecp2308/X female mice indicated that there is variability in the phenotypic manifestations (Shahbazian et al. 2002a). To ascertain the extent and degree of phenotypic variability in a large number of animals, we focused on phenotypes that seemed to be fully penetrant in males and that are easily scored. Four phenotypes fulfilled the set criteria: (1) Tr, which typically appears at age 5 wk in males and is the first observable abnormality; (2) stereotypic forepaw movements (SFM), also evident early and a cardinal feature of classic RTT; (3) disheveled fur (DF), which could be a consequence of poor grooming related to the observed hypoactivity; and (4) periocular lesions (PL). Mutant mice develop periocular inflammation and bleeding (fig. 1C), usually accompanied by bacterial infections. The lesions appear as early as age 6 wk in males. The cause of this phenotype remains elusive, but one hypothesis posits that these lesions could result from excessive stereotypic face scratching, a behavior that has been observed in patients with RTT (Hammer et al. 2002).
Figure 1.
Penetrance of phenotypes caused by the Mecp2308 allele. Each of the four panels shows an image of a mutant female mouse and of a wild-type female litter mate (inset). The percentage of animals of each sex manifesting a particular phenotype is shown below each panel. A, Representative image of a female with DF. The inset shows a wild-type litter mate with shiny and groomed fur. B, A picture taken at a shutter speed of 1 s reveals the tremor as a shadow effect over the mouse’s silhouette (arrows). The sharp inset picture of a wild-type mouse was taken at the same shutter speed. C, PLs that sometimes extend to the ears. Note also the disarranged appearance of the fur. Inset shows a normal eye in a wild-type litter mate. D, SFMs, inferred by looking at this picture taken with a shutter speed of 1 s. The arrow points to the forepaws of the mutant female, which have a shadow because of the rapid movements (see video by Shahbazian et al. [2002a] at the Neuron Web site).
The presence or absence of these four phenotypes was noted in 108 males and in 84 females at age 6 mo and 12 mo, respectively. We did not score for the degree of severity, to avoid any ambiguity or subjectivity in the scoring. Last, we chose 12 mo as testing age for females, because the disease may occur later and progress more slowly in females than in males. All four phenotypes were ∼100% penetrant in males, whereas the penetrance varied from 44% to 68% in females (fig. 1).
XCI Is Predominantly Nonrandom in Favor of Expression of the Wild-Type Mecp2 Allele
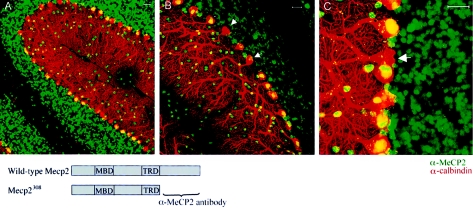
Variable patterns of XCI have been suggested as modifiers of clinical severity in human patients (Zoghbi et al. 1990; Amir et al. 2000; Bienvenu et al. 2000; Hoffbuhr et al. 2002). To evaluate whether XCI patterns influence the phenotypic outcome of the Mecp2308 mutation, we studied the pattern of XCI in the brains of Mecp2308/X females. To analyze the XCI patterns at the cellular level, we performed double immunofluorescence labeling, using an antibody directed against the C-terminus of MeCP2, which is deleted in the Mecp2308 allele and an antibody against calbindin; thus, the MeCP2 antibody will label only cells carrying the wild-type X chromosome as the active one, whereas the antibody against calbindin will label every Purkinje cell (PC) (fig. 2). The MeCP2 antibody labels all PCs in the cerebella of wild-type female mice (data not shown), whereas there is no immunoreactivity on cerebella of male mice carrying truncated MeCP2 (Shahbazian et al. 2002a).
Figure 2.
Determination of XCI patterns in the cerebellum of Mecp2308/X females by confocal laser scanning microscopy. Costaining with anti-calbindin antibody (red) and an antibody against the C-terminus of MeCP2 (green) (see lower diagram for antigen localization) allows us to label all PCs (calbindin) and identify neurons that express the wild-type Mecp2 allele (cells positive for calbindin and Mecp2 have yellow nuclei). Note that the majority of PCs are immunoreactive for the MeCP2 antibody. Arrows mark the few cells expressing mutant Mecp2 (nonimmunoreactive for α-MeCP2). Each panel is a representative example of immunofluorescence, visualized at increasingly higher magnifications. Scale bar = 20 μm.
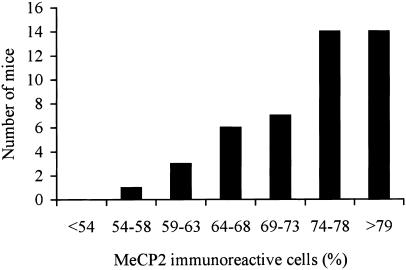
A quantitative analysis was done for 45 brains from 12-mo-old Mecp2308/X female mice by counting ⩾200 PCs per cerebellum. We observed variability in the patterns of XCI in PCs, with a range of 54%–82% (percentage of cells with the chromosome carrying the wild-type allele active). It is important to note that we observed that >60% of the females tested exhibited unbalanced XCI patterns (fig. 3). It is notable that the unbalanced pattern was always in favor of cells carrying the wild-type X chromosome as the active chromosome, as evidenced by the absence of females with <54% of cells with the wild-type X chromosome active. Immunostaining with an antibody directed against the N-terminus of MeCP2 (Shahbazian et al. 2002a), present in both the wild-type and the truncated MeCP2, labeled virtually every PC in the cerebella of Mecp2308/X female mice. This confirms that PCs that failed to label with the C-terminal anti-MeCP2 antibody were, in fact, expressing the truncated protein and carry an inactivated wild-type X chromosome.
Figure 3.
Distribution of XCI patterns in Mecp2308/X female mice. Values on the X-axis represent the percentage of cells that have the wild-type X chromosome active, as assayed by coimmunofluorescence. Among a total of 45 female mice, none had <54% of cells with the wild-type chromosome as the active one.
Variability in XCI Patterns in Different Brain Regions
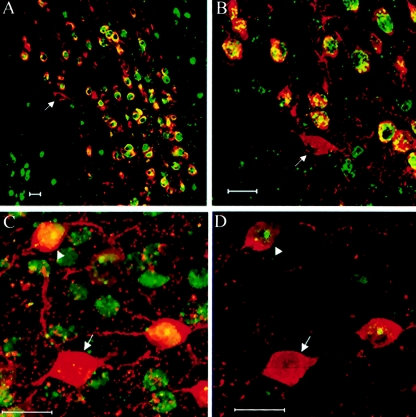
The observation that the majority of females exhibited nonrandom XCI patterns in PCs prompted us to investigate whether the same skewing is observed in different cell populations throughout the brain. For this purpose, we performed double immunofluorescence with the same anti-MeCP2 (C-terminus) combined with anti-calbindin, anti-TH (tyrosine hydroxylase), or antiparvalbumin antibodies on serial sections from seven Mecp2308/X female mouse brains. Because of the heterogeneity in MeCP2-expression levels (LaSalle et al. 2001), we focused our analysis on a specific subset of neurons: catecholaminergic neurons in the paraventricular hypothalamic nucleus (labeled by anti-TH) and on parvalbumin-positive cells in the cerebral cortex (layers II–VIa) (fig. 4). Two (28.6%) of seven mice analyzed showed variability across the different regions studied (table 1 [see mice number 4 and number 6]), whereas the remaining five had similar patterns of XCI in all the regions tested.
Figure 4.
XCI patterns in midbrain and cerebral cortex of Mecp2308/X female mice. A and B, Double immunofluorescence for the detection of wild-type MeCP2 (green) and TH (red) was used to determine the XCI pattern in catecholaminergic neurons in the midbrain. Arrows denote cells that have the X chromosome bearing the mutant Mecp2 as the active one. C, Visualization of coimmunolabeling of Mecp2 (green) and parvalbumin (red) in the cerebral cortex by confocal microscopy. D, Same as in panel C, but a single focal section is shown, to demonstrate the definitive identification of cells expressing the wild-type (arrowhead) versus the mutant (arrow) Mecp2 allele. Scale bar = 20 μm.
Table 1.
XCI Patterns in Neurons From Different Brain Regions of Mecp2308/X Female Mice
|
Percentage (No. of Neurons Analyzed)Expressing Wild-Type Allele |
Phenotype |
||||||
| Mouse | PCs | TH MidbrainNeurons | ParvalbuminCortical Neurons | Tr | SFM | DF | PL |
| 1 | 62.0 (200) | 61.2 (56) | 64.7 (153) | + | + | + | |
| 2 | 75.0 (200) | 70.9 (72) | 68.8 (125) | + | + | + | |
| 3 | 75.0 (200) | 71.9 (64) | 76.9 (130) | + | + | + | |
| 4 | 78.0 (200) | 85.9 (71) | 55.9 (177) | + | + | + | |
| 5 | 80.0 (200) | 77.5 (49) | 76.0 (121) | + | |||
| 6 | 80.0 (200) | 51.2 (82) | 78.0 (182) | + | + | ||
| 7 | 81.0 (200) | 80.9 (63) | 82.9 (94) | + | |||
Selective Survival of Cells Carrying the Wild-Type Mecp2 Allele Might Explain Favorable XCI Patterns
Nonrandom XCI may result from both selective and stochastic processes. In the mouse, the choice of which X chromosome to inactivate is under the control of the X-inactivation center and is made during embryonic development, sometime between implantation and completion of gastrulation (Plath et al. 2002). The absence of animals with XCI patterns favoring expression of the mutant allele suggests that stochastic events are not responsible for the observed skewing. However, we cannot rule out definitively the possibility that a dysfunctional MeCP2 has a primary effect on the X-inactivation process.
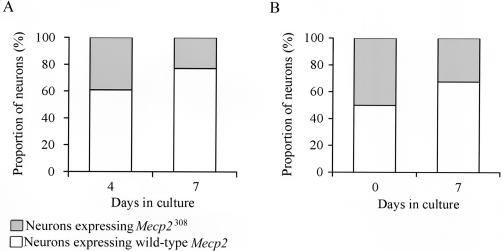
To determine the mechanism responsible for the unbalanced XCI patterns, we resorted to cell culture experiments. We cultured primary neurons derived from the hippocampus of E18 Mecp2308/X mice, a time when the virtually irreversible process of XCI has already taken place. Using the same strategy of coimmunolabeling of wild-type MeCP2 and a neuronal marker (tyrosine α-tubulin), we observed that the percentage of cells expressing the wild-type allele changed from 61% (74 wild type of 122 total) to 77% (155 wild type of 201 total) after 3 d in culture (days 4–7) (fig. 5A). Furthermore, mixing of cultures of cells derived from wild-type Mecp2+/Y and mutant Mecp2308/Y male mice and plating them at a ratio of ∼1:1 gave rise to a predominantly wild-type population of neurons after 7 d in culture (67.5%±3.5%; N=2) (fig. 5B). These data suggest that there is a selective advantage for cells expressing the wild-type Mecp2 allele and/or pressure against cells expressing the mutant allele, highlighting the importance of MeCP2 in neuronal survival.
Figure 5.
Selective growth advantage of neurons expressing the wild-type allele when grown in culture. The proportion of neurons expressing either wild-type or truncated Mecp2 allele was determined by indirect immunofluorescence in hippocampal neurons from an E18 female Mecp2308/X embryo cultured for 4 or 7 d (A) and in neurons from mixed cultures derived from Mecp2308/Y and Mecp2+/Y embryos (1:1 ratio) cultured for 7 d (B). White bars = neurons expressing wild-type MeCP2. Gray bars = neurons expressing truncated MeCP2.
Phenotypic Manifestations in Mecp2308/X Female Mice Mitigated by Unbalanced XCI
The high prevalence of skewing in Mecp2308/X female mice suggested that the pattern of XCI could be responsible for the variability in their phenotypes. To address this directly, we looked at how XCI patterns in PCs (by use of data from 45 brains) correlate with the four phenotypes systematically evaluated for the same set of mice. Logistic regression analysis indicated that the more the XCI patterns favored expression of the wild-type allele the less were the odds of exhibiting any of the phenotypes. The highest level of correlation was observed for Trs and SFMs, but a correlation was evident for every phenotype, suggesting that each phenotype is affected by XCI patterns (fig. 6A).
Figure 6.
Correlation between degree of XCI skewing and phenotype. We divided the population of 45 females into three groups, according to their XCI patterns, and plotted them against the percentage of mice expressing a particular phenotype for each range of XCI patterns (A). Logistic regression analysis indicated that a unit increase in XCI was associated with a 16%, a 21%, a 9%, and a 12% decrease in the odds of Tr (P<.01), SFM (P<.01), DF (P<.06), and PL (P<.02), respectively. B, The cumulative number of phenotypes presented by the animals in each group plotted against the XCI groups. A significant difference was detected among XCI pattern groups (P<.001), and further testing indicated differences between groups 1 (54%–70%) and 2 (71%–77%) (P<.05), between groups 1 and 3 (>78%) (P<.001), and between groups 2 and 3 (P<.02).
Another way to study the XCI-phenotype relationship is to group the mice according to their pattern of XCI, expressed as the percentage of cells having the wild-type allele active (54%–70%, 71%–77%, and >78%) and to plot every group against the cumulative number of phenotypes manifested by the mice that belong to that particular group. We found that significantly fewer phenotypes are observed when a large percentage of neurons has the mutant X chromosome silenced, confirming that the ratio of cells expressing a wild-type versus mutant MeCP2 is critical for the phenotypic manifestation in females (fig. 6B).
Discussion
The study of female mice harboring a late-truncating mutation in Mecp2 (Mecp2308) revealed that they exhibit large variability in phenotypic manifestations. Four phenotypes with full penetrance in males (Tr, repetitive forepaw movements, periocular injuries, and DF) exhibited high variability when scored for presence or absence in the Mecp2308/X female mice. This finding is surprising, since the range of phenotypes presented by human patients carrying similar MECP2 mutations is less variable. Although a small number of patients carrying comparable mutations (such as R294X and Q297X [Ishii et al. 2001]) have been reported to have nonclassic phenotypes, the majority of patients with similar mutations have classic RTT (RettBASE: IRSA MECP2 Variation Database). In a way, the observed high degree of variability in the Mecp2308/X female mice is counterintuitive, as the differential presence of genetic and environmental modifiers that could occur in a human population is most likely not present in our controlled conditions (pure genetic background and controlled husbandry and housing conditions).
We found that the majority of the mutant females had unbalanced XCI patterns. This was another surprising result, since most female patients with classic RTT have balanced XCI (Shahbazian et al. 2002b; Armstrong et al. 2003). The XCI patterns in Mecp2308/X female mice were determined by use of a simple, efficient, and sensitive method that allows for precise quantification of the proportion of cells that have inactivated the mutant or the wild-type X chromosome. Using this method, we demonstrate here that the phenotypic variability is accompanied by variable XCI patterns and that the majority of the female mice studied showed unbalanced XCI that favors the expression of the wild-type allele. Nonrandom XCI could be caused by a bias in the initial choice of which X chromosome to inactivate (primary cause) or by the result of a selection against cells having elected to inactivate a given X chromosome (secondary cause). Examples of the former include nonrandom XCI due to chance events or dysfunction of gene(s) that regulate XCI (Cattanach et al. 1969; Rastan 1982; Newall et al. 2001). Examples of secondary causes include many X/autosome translocations in which there is selection against cells that inactivate the translocated X chromosome (see, e.g., McMahon and Monk [1983]) and several X-linked disorders in which the mutated gene has deleterious effects in a cell-autonomous manner (Plenge et al. 2002). Biochemical studies indicate that XCI is complete in all cells in female mice by the onset of gastrulation (Monk 1992) and that the inactive state of the X chromosome is stable and inherited in female somatic cells throughout the lifetime of the mouse.
We did not find a single female mouse with nonrandom XCI patterns favoring expression of the mutant allele, suggesting that there is a selective advantage of cells expressing the wild-type Mecp2 allele (consistent with a secondary cause of nonrandom XCI). This hypothesis is supported by the fact that, although the unbalanced pattern of XCI was not linked to a particular region of the brain—in the majority of the cases, it was rather similar across the analyzed regions (cerebellum, midbrain, and cerebral cortex)—∼30% of the females exhibited differences in the patterns of XCI in the brain regions tested.
Additional support for this hypothesis comes from the finding that, over time, neurons expressing the wild-type protein show a relative increase in primary cultures of hippocampal cells from Mecp2308/X females, when compared with neurons expressing the mutant allele. Furthermore, the ratio of wild-type and mutant cells cultured together (obtained from Mecp2+/Y and Mecp2308/Y males, respectively) depart form the initial equality at the expense of the mutant cells. Although we recognize the limitations of evaluating the effects of mutations in in vitro culture conditions, we believe that these data together with the in vivo data (always favoring the wild-type allele) strongly support the hypothesis that a selective advantage of cells expressing the wild-type allele over the cells expressing the mutant allele accounts for unbalanced XCI patterns in this mouse model. Balmer et al. (2002) reported that MECP2 mutations cause a growth disadvantage in cultured clonal T cells, providing additional support for our hypothesis. Furthermore, our results also suggest that the selective advantage that results in preferential survival or proliferation of wild-type versus mutant cells does not depend on an anatomical or histological context. Examination of three diverse neuronal populations and finding nonrandom XCI patterns in all of them suggests that preferential survival of cells expressing the wild-type allele can occur in all brain regions, but we cannot exclude the possibility that some brain regions or peripheral tissues do not pose this selective advantage and, hence, have balanced XCI patterns. Whether the effect on XCI patterns observed for the Mecp2308 allele is shared by several or all forms of dysfunctional MeCP2 remains to be determined. To our knowledge, only one girl who has the 1157del32 mutation has been reported to show unfavorable XCI patterns, with ∼66% of mutated X active and only 34% of the normal paternal X active (Zappella et al. 2001). In that case, the unbalanced pattern is probably a result of chance, since it is hard to envision a nonfunctional MeCP2 providing a survival advantage.
Nonrandom XCI patterns can affect the phenotypes of females carrying mutant X-linked genes (Lyon 2002). The outcome depends on whether the skewing favors the normal or the mutant allele-harboring X chromosome.
The effect of XCI patterns on the RTT phenotype is illustrated by a number of reports of females who carry MECP2 mutations but who are apparently unaffected, owing to extreme (90%–100%) unbalanced XCI patterns favoring expression of the X chromosome carrying the wild-type allele (Sirianni et al. 1998; Wan et al. 1999; Amir et al. 2000; Bienvenu et al. 2000; Villard et al. 2000; Hoffbuhr et al. 2001). Other females with MECP2 mutations but with slightly lower levels of skewing (85%–95%) were reported to have atypical RTT, showing some but not all diagnostic features of RTT (Hoffbuhr et al. 2001; Zappella et al. 2001). However, the majority of patients with RTT have balanced XCI patterns (Shahbazian et al. 2002b; Armstrong et al. 2003). We hypothesized that this is due to ascertainment bias; if nonrandom XCI affects the phenotypic outcome of MECP2 mutations, there might be females who carry mutations in MECP2 who are asymptomatic or misdiagnosed. Some of them may be recognized only after having an affected child, whereas some may go unrecognized if they are subfertile. To test this hypothesis, we studied the association of nonrandom XCI with the phenotypic outcome of the Mecp2308 mutation. Our results indicate that XCI patterns affect the phenotypic manifestations. A clear correlation was observed between the pattern of XCI in PC of the cerebellum and the cumulative phenotype, so that those females with a higher degree of skewing toward the wild-type X chromosome are much less likely to exhibit the scored phenotypes than those with more balanced XCI. A high degree of skewing will even prevent the appearance of any of the four phenotypes and will cause the females carrying most of the cells that express the wild-type Mecp2 allele to be indistinguishable from their wild-type litter mates.
Our finding that the pattern of XCI correlates with the phenotypic outcome of a truncation in Mecp2 is relevant to the human disease in several ways. First, on the basis of the data presented here, we propose that there might be more females carrying mutations in MECP2 than are currently accounted for. These females are probably misdiagnosed or not even identified unless they have an affected child. Second, although the limited amount of information precludes a definitive ascertainment, the data suggest that the forepaw/hand stereoptypies might be associated with dysfunction of catecholaminergic neurons (mouse 6 in table 1) and that the Trs might have a cortical origin (mouse 4 in table 1). Thus, the observation that nonrandom XCI patterns mitigate the phenotypic manifestations and that these patterns might diverge in different regions of the brain could be exploited to identify which neurons might mediate a particular phenotype in this mouse model.
Acknowledgments
We thank Dong Cheul Kang, for excellent technical assistance; Richard Atkinson, for advice on confocal microscopy; E. O’Brian Smith, for help with the statistic analysis; and A. Bowman, for critical reading of the manuscript. These studies were supported by a Rett Syndrome Research Foundation grant (to J.I.Y.) and by National Institutes of Health grant P01HD40301 (to H.Y.Z.). H.Y.Z. is an investigator with the Howard Hughes Medical Institute.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Neuron, http://www.neuron.org/content/full/35/2/243/DC1 (for Shahbazian et al. video)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RTT) [PubMed]
- RettBASE: IRSA MECP2 Variation Database, http://mecp2.chw.edu.au/ [DOI] [PubMed]
References
- Adler DA, Quaderi NA, Brown SD, Chapman VM, Moore J, Tate P, Disteche CM (1995) The X-linked methylated DNA binding protein, Mecp2, is subject to X inactivation in the mouse. Mamm Genome 6:491–492 [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Schultz R, Malicki DM, Tran CQ, Dahle EJ, Philippi A, Timar L, Percy AK, Motil KJ, Lichtarge O, Smith EO, Glaze DG, Zoghbi HY (2000) Influence of mutation type and X chromosome inactivation on Rett syndrome phenotypes. Ann Neurol 47:670–679 [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl- CpG-binding protein 2. Nat Genet 23:185–188 10.1038/13810 [DOI] [PubMed] [Google Scholar]
- Armstrong DD, Deguchi K, Antallfy B (2003) Survey of MeCP2 in the Rett syndrome and the non-Rett syndrome brain. J Child Neurol 18:653–660 [DOI] [PubMed] [Google Scholar]
- Balmer D, Arredondo J, Samaco RC, LaSalle JM (2002) MECP2 mutations in Rett syndrome adversely affect lymphocyte growth, but do not affect imprinted gene expression in blood or brain. Hum Genet 110:545–552 10.1007/s00439-002-0724-4 [DOI] [PubMed] [Google Scholar]
- Bienvenu T, Carrie A, de Roux N, Vinet MC, Jonveaux P, Couvert P, Villard L, Arzimanoglou A, Beldjord C, Fontes M, Tardieu M, Chelly J (2000) MECP2 mutations account for most cases of typical forms of Rett syndrome. Hum Mol Genet 9:1377–1384 10.1093/hmg/9.9.1377 [DOI] [PubMed] [Google Scholar]
- Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, Vance JM, Pericak-Vance MA (2003) Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol 28:205–211 [DOI] [PubMed] [Google Scholar]
- Cattanach BM, Pollard CE, Perez JN (1969) Controlling elements in the mouse X-chromosome. I. Interaction with the X-linked genes. Genet Res 14:223–235 [DOI] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R (2001) Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet 27:327–331 10.1038/85906 [DOI] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A (2001) A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet 27:322–326 10.1038/85899 [DOI] [PubMed] [Google Scholar]
- Hagberg B (1995) Clinical delineation of Rett syndrome variants. Neuropediatrics 26:62 [DOI] [PubMed] [Google Scholar]
- Hagberg B, Aicardi J, Dias K, Ramos O (1983) A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett’s syndrome: report of 35 cases. Ann Neurol 14:471–479 [DOI] [PubMed] [Google Scholar]
- Hammer S, Dorrani N, Dragich J, Kudo S, Schanen C (2002) The phenotypic consequences of MECP2 mutations extend beyond Rett syndrome. Ment Retard Dev Disabil Res Rev 8:94–98 10.1002/mrdd.10023 [DOI] [PubMed] [Google Scholar]
- Hammer S, Dorrani N, Hartiala J, Stein S, Schanen NC (2003) Rett syndrome in a 47,XXX patient with a de novo MECP2 mutation. Am J Med Genet 122A:223–226 12966522 [DOI] [PubMed] [Google Scholar]
- Hoffbuhr K, Devaney JM, LaFleur B, Sirianni N, Scacheri C, Giron J, Schuette J, Innis J, Marino M, Philippart M, Narayanan V, Umansky R, Kronn D, Hoffman EP, Naidu S(2001) MeCP2 mutations in children with and without the phenotype of Rett syndrome. Neurology 56:1486–1495 [DOI] [PubMed] [Google Scholar]
- Hoffbuhr KC, Moses LM, Jerdonek MA, Naidu S, Hoffman EP (2002) Associations between MeCP2 mutations, X-chromosome inactivation, and phenotype. Ment Retard Dev Disabil Res Rev 8:99–105 10.1002/mrdd.10026 [DOI] [PubMed] [Google Scholar]
- Huppke P, Held M, Hanefeld F, Engel W, Laccone F (2002) Influence of mutation type and location on phenotype in 123 patients with Rett syndrome. Neuropediatrics 33:63–68 10.1055/s-2002-32365 [DOI] [PubMed] [Google Scholar]
- Ishii T, Makita Y, Ogawa A, Amamiya S, Yamamoto M, Miyamoto A, Oki J (2001) The role of different X-inactivation pattern on the variable clinical phenotype with Rett syndrome. Brain Dev Suppl 23:S161–S164 11738865 [DOI] [PubMed] [Google Scholar]
- Kunieda T, Xian M, Kobayashi E, Imamichi T, Moriwaki K, Toyoda Y (1992) Sexing of mouse preimplantation embryos by detection of Y chromosome-specific sequences using polymerase chain reaction. Biol Reprod 46:692–697 [DOI] [PubMed] [Google Scholar]
- Lam CW, Yeung WL, Ko CH, Poon PM, Tong SF, Chan KY, Lo IF, Chan LY, Hui J, Wong V, Pang CP, Lo YM, Fok TF (2000) Spectrum of mutations in the MECP2 gene in patients with infantile autism and Rett syndrome. J Med Genet 37:E41 10.1136/jmg.37.12.e41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, Goldstine J, Balmer D, Greco CM (2001) Quantitative localization of heterogeneous methyl-CpG-binding protein 2 (MeCP2) expression phenotypes in normal and Rett syndrome brain by laser scanning cytometry. Hum Mol Genet 10:1729–1740 10.1093/hmg/10.17.1729 [DOI] [PubMed] [Google Scholar]
- Lyon MF (2002) X-chromosome inactivation and human genetic disease. Acta Paediatr Suppl 91:107–112 10.1080/080352502762458030 [DOI] [PubMed] [Google Scholar]
- McMahon A, Monk M (1983) X-chromosome activity in female mouse embryos heterozygous for Pgk-1 and Searle’s translocation, T(X; 16) 16H. Genet Res 41:69–83 [DOI] [PubMed] [Google Scholar]
- Monk M (1992) The X chromosome in development in mouse and man. J Inherit Metab Dis 15:499–513 [DOI] [PubMed] [Google Scholar]
- Newall AE, Duthie S, Formstone E, Nesterova T, Alexiou M, Johnston C, Caparros ML, Brockdorff N (2001) Primary non-random X inactivation associated with disruption of Xist promoter regulation. Hum Mol Genet 10:581–589 10.1093/hmg/10.6.581 [DOI] [PubMed] [Google Scholar]
- Orrico A, Lam C, Galli L, Dotti MT, Hayek G, Tong SF, Poon PM, Zappella M, Federico A, Sorrentino V (2000) MECP2 mutation in male patients with non-specific X-linked mental retardation. FEBS Lett 481:285–288 10.1016/S0014-5793(00)01994-3 [DOI] [PubMed] [Google Scholar]
- Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B (2002) Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet 36:233–278 10.1146/annurev.genet.36.042902.092433 [DOI] [PubMed] [Google Scholar]
- Plenge RM, Stevenson RA, Lubs HA, Schwartz CE, Willard HF (2002) Skewed X-chromosome inactivation is a common feature of X-linked mental retardation disorders. Am J Hum Genet 71:168–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastan S (1982) Primary non-random X-inactivation caused by controlling elements in the mouse demonstrated at the cellular level. Genet Res 40:139–147 [DOI] [PubMed] [Google Scholar]
- Renieri A, Meloni I, Longo I, Ariani F, Mari F, Pescucci C, Cambi F (2003) Rett syndrome: the complex nature of a monogenic disease. J Mol Med 81:346–354 [DOI] [PubMed] [Google Scholar]
- Rett A (1966) On an unusual brain atrophy syndrome in hyperammonemia in childhood. Wien Med Wochenschr 116:723–726 [PubMed] [Google Scholar]
- Rett Syndrome Diagnostic Criteria Work Group (1988) Diagnostic criteria for Rett syndrome. Ann Neurol 23:425–428 [DOI] [PubMed] [Google Scholar]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong D, Paylor R, Zoghbi H (2002a) Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron 35:243–254 [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Sun Y, Zoghbi HY (2002b) Balanced X chromosome inactivation patterns in the Rett syndrome brain. Am J Med Genet 111:164–168 10.1002/ajmg.10557 [DOI] [PubMed] [Google Scholar]
- Sharp A, Robinson D, Jacobs P (2000) Age- and tissue-specific variation of X chromosome inactivation ratios in normal women. Hum Genet 107:343–349 10.1007/s004390000382 [DOI] [PubMed] [Google Scholar]
- Sirianni N, Naidu S, Pereira J, Pillotto RF, Hoffman EP (1998) Rett syndrome: confirmation of X-linked dominant inheritance, and localization of the gene to Xq28. Am J Hum Genet 63:1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temudo T, Maciel P (2002) Rett's syndrome: clinical features and advances in genetics. Rev Neurol Suppl 34:S54–S58 12447790 [DOI] [PubMed] [Google Scholar]
- Villard L, Cardoso AK, Chelly PJ, Tardieu PM, Fontes M (2000) Two affected boys in a Rett syndrome family: clinical and molecular findings. Neurology 55:1188–1193 [DOI] [PubMed] [Google Scholar]
- Wan M, Lee SSJ, Zhang X, Houwink-Manville I, Song H-R, Amir RE, Budden S, Naidu S, Pereira JLP, Lo IFM, Zoghbi HY, Schanen NC, Francke U (1999) Rett syndrome and beyond: recurrent spontaneous and familial MECP2 mutations at CpG hotspots. Am J Hum Genet 65:1520–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P, Black G, Ramsden S, Barrow M, Super M, Kerr B, Clayton-Smith J (2001) Angelman syndrome phenotype associated with mutations in MECP2, a gene encoding a methyl CpG binding protein. J Med Genet 38:224–228 10.1136/jmg.38.4.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaving LS, Williamson SL, Bennetts B, Davis M, Ellaway CJ, Leonard H, Thong MK, Delatycki M, Thompson EM, Laing N, Christodoulou J (2003) Effects of MECP2 mutation type, location and X-inactivation in modulating Rett syndrome phenotype. Am J Med Genet 118A:103–114 12655490 [DOI] [PubMed] [Google Scholar]
- Yntema HG, Kleefstra T, Oudakker AR, Romein T, de Vries BB, Nillesen W, Sistermans EA, Brunner HG, Hamel BC, van Bokhoven H (2002) Low frequency of MECP2 mutations in mentally retarded males. Eur J Hum Genet 10:487–490 10.1038/sj.ejhg.5200836 [DOI] [PubMed] [Google Scholar]
- Zappella M, Meloni I, Longo I, Hayek G, Renieri A (2001) Preserved speech variants of the Rett syndrome: molecular and clinical analysis. Am J Med Genet 104:14–22 10.1002/ajmg.10005 [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Percy AK, Schultz RJ, Fill C (1990) Patterns of X chromosome inactivation in the Rett syndrome. Brain Dev 12:131–135 [DOI] [PubMed] [Google Scholar]