Abstract
Meiotic recombination is essential for the segregation of chromosomes and the formation of normal haploid gametes, yet we know very little about the meiotic process in humans. We present the first (to our knowledge) recombination maps for every autosome in the human male obtained by new immunofluorescence techniques followed by centromere-specific multicolor fluorescence in situ hybridization in human spermatocytes. The mean frequency of autosomal recombination foci was 49.8±4.3, corresponding to a genetic length of 2,490 cM. All autosomal bivalents had at least one recombination focus. In contrast, the XY bivalent had a recombination focus in 73% of nuclei, suggesting that a relatively large proportion of spermatocytes may be at risk for nondisjunction of the XY bivalent or elimination by meiotic arrest. There was a very strong correlation between mean length of the synaptonemal complex (SC) and the number of recombination foci per SC. Each bivalent presented a distinct distribution of recombination foci, but in general, foci were near the distal parts of the chromosome, with repression of foci near the centromere. The position of recombination foci demonstrated positive interference, but, in rare instances, foci were very close to one another.
Introduction
Pairing of homologous chromosomes and meiotic recombination are essential for the segregation of chromosomes and the formation of normal haploid gametes, yet surprisingly little is known about meiotic processes in higher organisms, including humans. It is fortunate that the last few years have seen many exciting developments that have revolutionized research in human meiosis. A number of mammalian orthologues of recombination proteins of lower organisms have been identified, and novel immunocytological techniques have opened up new avenues of research.
There are two basic methods of analyzing meiotic recombination in human germ cells: standard genetic linkage analysis of human pedigrees and cytological analysis of chiasmata in human gametes (reviewed by Hultén and Tease [2003]). The first approach is an indirect method that uses genetic linkage (coinheritance of markers in families) to produce recombination maps for chromosome segments. These segments can then be linked to provide estimates of recombination frequencies for specific chromosomes. The second approach is a direct cytological analysis of recombination in human spermatocytes or oocytes.
The cytological approach has been restricted to very few laboratories. M. Hultén and her associates have studied diakinesis preparations in human spermatocytes over the past 3 decades (Hultén and Lindsten 1973; Hultén 1974; Laurie and Hultén 1985a, 1985b). This approach provided the first information on the frequency and distribution of chiasmata in individual chromosomes in human males. However, diakinesis preparations are laborious to analyze, and there are limited numbers of cells at this stage of spermatogenesis. Thus, analysis is slow, and there have been reports on chiasmata frequency in individual chromosomes in only five men (Hultén 1974; Laurie and Hultén 1985a) and detailed analysis of chiasmata locations in only two men (Hultén 1974; Laurie and Hultén 1985b).
It is fortunate that new immunofluorescence techniques provide an alternative approach that allows analysis of sites of recombination in meiocytes with greater precision in the location of the exchange. Various important meiotic structures can be identified by the use of immunofluorescence. Antibodies against SCP1 (transverse elements) or SCP3 (lateral elements) can be used to visualize the synaptonemal complexes (SCs, the proteinaceous structure linking homologous chromosomes in prophase of meiosis I). The centromere can be localized with CREST (calcinosis, Raynaud's phenomenon, esophageal dysfunction, sclerodactyly, telangiectasia) antisera. Most importantly, recent studies have demonstrated that antibodies against the DNA mismatch repair protein MLH1 identify the sites of meiotic exchange on SCs in both mouse (Baker et al. 1996; Anderson et al. 1999) and human spermatocytes and oocytes (Barlow and Hultén 1998; Lynn et al. 2002; Tease et al. 2002). The number and location of the MLH1 foci conform to that expected of a molecule that marks the site of recombination, and, recently, Marcon and Moens (2003) have demonstrated that MLH1 localizes to chiasmata precociously induced by okadaic acid.
These new immunofluorescence studies provide a great deal of information on the genomewide distribution of recombination. Froenicke et al. (2002) have used this technique, combined with multicolor FISH of chromosome-specific DNA libraries, to provide male mouse recombination maps for each autosome. In humans, specific recombination maps have been reported for only four chromosomes (1, 16, 21, and 22) in the male (Lynn et al. 2002) and four chromosomes (X, 18, 21, and 22) in the female (Tease et al. 2002). In this article, we present the first (to our knowledge) recombination maps for every autosome in the human male through use of the combined techniques of immunofluorescence in spermatocytes followed by centromere-specific multicolor FISH (cenM-FISH) (Nietzel et al. 2001; Oliver-Bonet et al. 2003).
Material and Methods
This study was approved by the University of Calgary institutional review board. A testicular sample with normal spermatogenesis was obtained from a 47-year-old man with a differentiated liposarcoma. The testicular tissue was processed using a modification of the technique of Barlow and Hulten (1998). Testicular tissue was shredded with two pairs of forceps, and the released pachytene cells were spread evenly over microscope slides layered with paraformaldehyde (Fisher Scientific)/Triton-X (Sigma) solution at pH 9.2. Slides were dried for ∼24 h at room temperature in a humid chamber, and then drying was completed on the bench for ∼30 min. Dried slides were washed, with agitation, for 4 min in 0.04% Photo-Flo (Kodak), air dried for 10 min, and soaked for 30 min in antibody dilution buffer (ADB) with agitation every 5 minutes. Four primary antibodies (human CREST [a gift from M. Fritzler, University of Calgary], rabbit MLH1 [Oncogene], goat synaptonemal complex protein 3 [SCP3] [a gift from T. Ashley, Yale University], and mouse synaptonemal complex protein 1 [SYN1] [a gift from P. Moens, York University]) were diluted in ADB; this cocktail was applied to each slide, covered with a glass cover slip, and sealed with rubber cement, and the slides were incubated for ∼24 h at 37°C in a humid chamber. On the following day, cover slips were removed by soaking in ADB for 20 min, and then slides were washed in ADB for 48 h at 4°C. Subsequent to the wash, a cocktail of secondary antibodies was prepared (1-amino-4-methylcoumarin-3-acetic acid [AMCA] donkey anti-human [Jackson Immunoresearch], Alexa 488 donkey anti-rabbit [Molecular Probes], Alexa 555 donkey anti-goat [Molecular Probes], and Cy3 donkey anti-mouse [Jackson Immunoresearch]) and applied to the slides under a plastic cover slip. Slides were incubated at 37°C in a humid chamber for 90 min and were washed three times in PBS solution for 10, 20, and 30 min, agitating every 5 minutes; a glass cover slip was then applied and sealed with rubber cement. Slides were scanned with a Zeiss Axiophot microscope, locations of the spreads were determined using a gridded finder slide, and images of the SCs, MLH1 sites, and CREST locations were captured using an Applied Imaging Cytovision 2.81 work station. Prints of the captured images were analyzed to determine the number of MLH1 sites on each individual SC and in the whole cell.
After analysis was complete, cenM-FISH was performed on the same cells. The techniques of Nietzel et al. (2001) and Oliver-Bonet et al. (2003) were modified to make use of the microwave-decondensed/codenatured FISH technique (Ko et al. 2001). Cells were decondensed for only 5 s in dithiothreitol (DTT) and 30 s in 3,5-diiodosalicylic acid, lithium salt (LIS)/DTT on medium power (550 watts). Hybridization buffer (10% dextran sulfate, 2× standard sodium citrate [SSC], and 55% formamide) was prewarmed to 50°C, added to the cenM-FISH probe, and warmed at 50°C until all probe was dissolved. Probe was applied to the slide, a glass cover slip was sealed in place with rubber cement, the probe and cells were microwave codenatured for 75 s at 1,100 watts, and the slide was incubated in a humid chamber at 37°C for ∼24 h. A posthybridization wash (200 μl of 0.4× SSC, flat on a slide warmer at 70°C) was performed, streptavidin-Alexa 647 (Molecular Probes) solution was applied under a plastic cover slip, and the slide was incubated at 37°C for 40 min in a humid chamber. The slide was washed, with constant agitation, for 10 min in 4× SSC, air dried, and cover slipped in 4′,6-diamidino-2-phenylindole (DAPI). The six probe colors (blue, aqua, green, gold, red, and far red) were captured, and cenM-FISH images were analyzed in the same cells that had SCs analyzed previously. Determination of positions of recombination foci and centromeres plus measurements of the length of the SCs were performed using MicroMeasure 3.3 (available from the MicroMeasure Web site).
Results and Discussion
In 100 pachytene stage cells analyzed, the mean frequency of recombination (MLH1) foci for autosomes was 49.8±4.3, with a range of 38–62 foci per cell (fig. 1). There were no autosomal bivalents without an MLH1 focus. This is very similar to other data obtained by immunocytological techniques (Barlow and Hultén 1998; Lynn et al. 2002) and by chiasma counts at diakinesis (Hultén 1974; Laurie and Hultén 1985a). The 49.8 recombination events marked by MLH1 foci correspond to a genetic length of 2,490 cM in human males, which is also very similar to the 2,590 cM obtained from linkage data (Kong et al. 2002). The XY bivalent was found to have an MLH1 focus in 73% of nuclei. This did not appear to vary with the stage of pachytene, since 71.1% (27/38) of cells in early pachytene (stages 1 and 2) had an MLH1 focus, compared with 74.2% (46/62) in late pachytene (stages 3, 4, and 5). No XY bivalent had two MLH1 foci. Barlow and Hultén (1998) have also found that the sex bivalent MLH1 focus is present at all pachytene stages. However, a significant proportion of cells do not have an XY focus. Other studies have suggested a susceptibility of the XY bivalent to nondisjunction and aneuploid sperm because of a lack of recombination in the pseudoautosomal region (Hassold et al. 1991; Martin et al. 1996; Shi et al. 2001). An unpaired XY bivalent could also lead to meiotic arrest (Miklos 1974; Hale 1994).
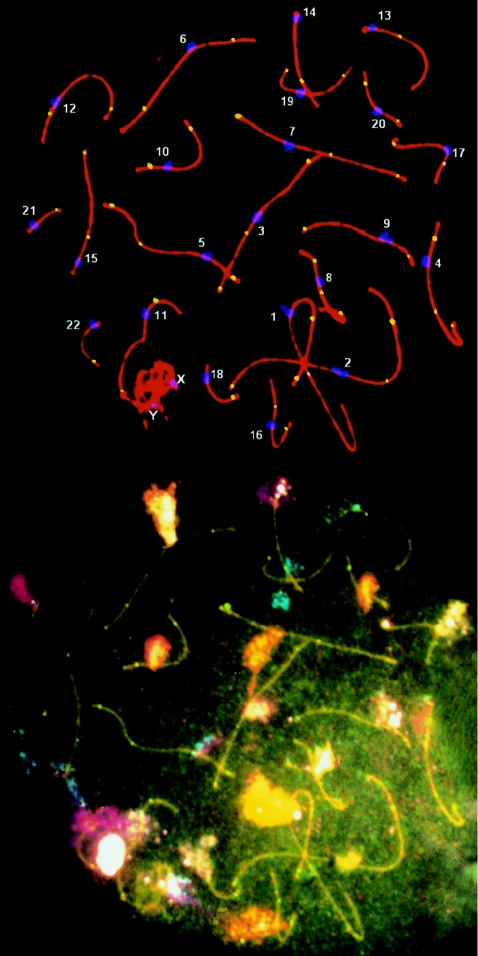
Figure 1.
Human pachytene spermatocyte with SCs shown in red, centromeres in blue, and MLH1 foci in yellow. Subsequent cenM-FISH analysis allows identification of individual chromosomes, so that recombination (MLH1) foci can be analyzed for each SC.
After synaptonemal complex spreads were prepared, individual autosomes were identified by cenM-FISH in 50 spreads. This technique combines centromeric probes for all chromosomes in a single assay. Thus, the number and distribution of MLH1 foci were established for individual chromosomes (fig. 1). The mean numbers of MLH1 foci for the short arm (p), long arm (q) and entire bivalent for individual chromosomes are presented in table 1. This ranged from a low of 1.00±0, for the smallest chromosome (21), to 3.90±.59, for the largest chromosome (1). Lynn et al. (2002) analyzed MLH1 foci in four individual chromosomes (1, 16, 21, and 22) and found results very similar to ours. Our results are also very similar to diakinesis chiasma counts performed by Laurie and Hultén (1985a), who found a mean of 1.07 chiasmata for chromosome 21 and 3.86 for chromosome 1. Finally, the genetic length (in cM) for each chromosome had a very close correspondence to genetic length determined by genotyping of >5,000 microsatellite markers in 146 families (Pearson correlation .97; P<.0001) (table 1) (Kong et al. 2002).
Table 1.
MLH1 Foci for Individual Autosomal SCs
|
No. of Foci |
|||||||||||
| p Arm |
q Arm |
Bivalent |
Map Units(cM) |
||||||||
| Chromosome | Mean | SD | Range | Mean | SD | Range | Mean | SD | Range | From SC Data | From Linkage Dataa |
| 1 | 1.97 | .48 | 1–3 | 1.93 | .44 | 1–3 | 3.90 | .60 | 3–5 | 195.0 | 195.1 |
| 2 | 1.47 | .62 | 0–3 | 2.03 | .66 | 1–4 | 3.50 | .76 | 2–5 | 175.0 | 189.6 |
| 3 | 1.67 | .47 | 1–2 | 1.47 | .50 | 1–2 | 3.13 | .67 | 2–4 | 156.5 | 160.7 |
| 4 | .93 | .25 | 0–1 | 1.62 | .61 | 1–3 | 2.57 | .62 | 2–4 | 128.5 | 146.5 |
| 5 | 1.03 | .18 | 1–2 | 1.67 | .54 | 1–3 | 2.70 | .53 | 2–4 | 135.0 | 151.2 |
| 6 | 1.17 | .37 | 1–2 | 1.47 | .50 | 1–2 | 2.63 | .55 | 2–4 | 131.5 | 137.6 |
| 7 | 1.20 | .40 | 1–2 | 1.47 | .50 | 1–2 | 2.67 | .65 | 2–4 | 133.5 | 128.4 |
| 8 | 1.00 | .00 | 1 | 1.24 | .43 | 1–2 | 2.20 | .48 | 1–3 | 110.0 | 107.9 |
| 9 | 1.03 | .18 | 1–2 | 1.43 | .50 | 1–2 | 2.43 | .56 | 1–3 | 121.5 | 117.3 |
| 10 | .97 | .18 | 0–1 | 1.33 | .47 | 1–2 | 2.30 | .53 | 1–3 | 115.0 | 133.9 |
| 11 | 1.00 | .37 | 0–2 | 1.27 | .44 | 1–2 | 2.27 | .44 | 2–3 | 113.5 | 109.4 |
| 12 | .90 | .30 | 0–1 | 1.73 | .57 | 1–3 | 2.63 | .61 | 1–4 | 131.5 | 135.5 |
| 13 | .05 | .23 | 0–1 | 1.90 | .47 | 1–3 | 2.00 | .37 | 1–3 | 100.0 | 101.3 |
| 14 | .02 | .13 | 0–1 | 2.03 | .18 | 2–3 | 2.07 | .25 | 2–3 | 103.5 | 94.6 |
| 15 | .08 | .26 | 0–1 | 1.77 | .42 | 1–2 | 1.90 | .40 | 1–3 | 95.0 | 102.6 |
| 16 | 1.00 | .00 | 1 | 1.13 | .56 | 0–3 | 2.13 | .56 | 1–4 | 106.5 | 108.1 |
| 17 | .87 | .34 | 0–1 | 1.31 | .53 | 0–2 | 2.13 | .43 | 1–3 | 106.5 | 108.6 |
| 18 | .77 | .42 | 0–1 | 1.17 | .45 | 0–2 | 1.83 | .45 | 1–3 | 91.5 | 98.6 |
| 19 | 1.03 | .18 | 1–2 | 1.00 | .00 | 1 | 2.03 | .18 | 2–3 | 101.5 | 92.6 |
| 20 | .83 | .45 | 0–2 | 1.00 | .00 | 1 | 1.83 | .45 | 1–3 | 91.5 | 74.7 |
| 21 | .00 | .00 | 0 | 1.00 | .00 | 1 | 1.00 | .00 | 1 | 50.0 | 47.3 |
| 22 | .03 | .18 | 0–1 | 1.07 | .25 | 1–2 | 1.10 |
.30 | 1–2 | 55.0 |
49.0 |
| Total | 50.95 | 2,547.5 | 2,590.5 | ||||||||
From Kong et al. (2002).
In general, we found a very good correspondence between the average relative length of SCs compared with mitotic chromosomes (Pearson correlation .97; P<.0001) (table 2). However, a few SCs are noticeably shorter or longer than would be expected on the basis of their relative mitotic lengths. For example, chromosome 8 has a shorter SC than chromosomes 9, 10, 11, or 12, and chromosome 17 has a longer SC than chromosomes 13, 14, 15, or 16. Thus, the relative physical length in mitosis does not seem to correlate exactly with the genetic length at meiosis. Kong et al. (2002) reported that the intensity of G-band staining is inversely related to the recombination frequency in humans. This would predict that chromosomes with the highest proportions of G bands should have shorter SCs and decreased levels of recombination than what would be expected from their mitotic chromosome length. Our results confirm this prediction: chromosomes 8 and 13 have very high proportions of G bands, and their relative SC lengths are much shorter than their mitotic lengths; their map units are similarly decreased. For example, for chromosome 13, the length rank is number 18, the mean absolute length of the SC is 9.8±1.45 μm (compared with 10.9–11.7 for chromosomes 14–17). In contrast, chromosomes 17 and 19 have very low proportions of G bands, and their SCs are longer than their mitotic lengths; the genetic lengths are also increased. For example, chromosome 19 has an SC length (10.6 μm) considerably longer than chromosome 18 (7.8 μm) or 20 (7.3 μm), commensurate with a genetic length of 101.5 cM, compared with 91.5 cM for chromosome 18 and 91.5 cM for chromosome 20. Thus, our cytological results are in agreement with recombination maps based on family genotyping. G-band regions generally have a decreased number of genes. Therefore, our results and those of others in various organisms (Nicolas 1998; Froenicke et al. 2002) suggest that recombination is generally initiated in gene-rich regions; this could account for some of the variability in recombination frequency in different chromosomes, as well as for the different recombination patterns observed in different bivalents.
Table 2.
Comparison of Autosomal SCs with Mitotic Chromosomes
|
Length of SC |
|||||||
| Absolute(in μm) |
Relativec |
Relative Length ofMitoticChromosomesa | |||||
| Chromosome | Mean | SD | Mean | SD | Mean | SD | % Difference from Expected Relative Length Ratiob |
| 1 | 26.56 | 4.27 | 8.92 | 1.43 | 9.11 | .53 | −2 |
| 2 | 23.33 | 3.73 | 7.83 | 1.25 | 8.61 | .41 | −9 |
| 3 | 20.29 | 3.89 | 6.81 | 1.31 | 6.97 | .36 | −2 |
| 4 | 16.91 | 3.63 | 5.68 | 1.22 | 6.49 | .32 | −13 |
| 5 | 16.81 | 3.14 | 5.64 | 1.05 | 6.21 | .50 | −9 |
| 6 | 16.66 | 3.63 | 5.59 | 1.22 | 6.07 | .44 | −8 |
| 7 | 16.58 | 2.75 | 5.57 | .92 | 5.43 | .47 | 3 |
| 8 | 13.64 | 2.20 | 4.58 | .74 | 4.94 | .28 | −7 |
| 9 | 14.23 | 2.90 | 4.78 | .97 | 4.78 | .39 | 0 |
| 10 | 14.17 | 1.92 | 4.76 | .64 | 4.80 | .58 | −1 |
| 11 | 13.92 | 2.23 | 4.67 | .75 | 4.82 | .30 | −3 |
| 12 | 13.98 | 1.85 | 4.69 | .62 | 4.50 | .26 | 4 |
| 13 | 9.80 | 1.45 | 3.29 | .49 | 3.87 | .26 | −15 |
| 14 | 10.89 | 1.76 | 3.66 | .59 | 3.74 | .23 | −2 |
| 15 | 11.45 | 1.93 | 3.84 | .65 | 3.30 | .25 | 16 |
| 16 | 10.93 | 2.00 | 3.67 | .67 | 3.14 | .35 | 17 |
| 17 | 11.70 | 2.10 | 3.93 | .71 | 2.97 | .30 | 32 |
| 18 | 7.79 | 2.01 | 2.62 | .67 | 2.78 | .18 | −6 |
| 19 | 10.60 | 2.13 | 3.56 | .72 | 2.46 | .31 | 45 |
| 20 | 7.34 | 1.30 | 2.46 | .44 | 2.25 | .24 | 9 |
| 21 | 4.28 | .70 | 1.44 | .24 | 1.70 | .32 | −15 |
| 22 | 5.99 |
.84 | 2.01 | .28 | 1.80 | .26 | 12 |
| Total | 297.85 | ||||||
% of total autosomal mitotic length (International System for Human Cytogenetic Nomenclature 1985).
%difference=[(relativeSClength/relativemitoticchromosomelength)-1]×100.
% of total autosomal SC length.
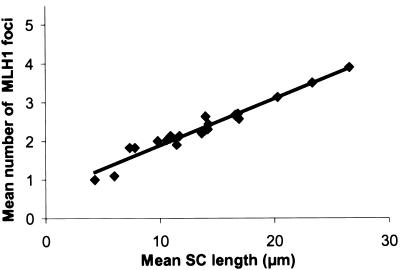
To determine whether SC length equates to genetic or physical length, we measured individual SC lengths through use of the computer application MicroMeasure 3.3 (MicroMeasure Web site). We found a very strong correlation between average SC length and average number of MLH1 foci per SC (Pearson correlation .98; P<.0001) (fig. 2). This relationship has previously been observed in mouse spermatocytes (Heng et al. 2001; Froenicke et al. 2002). It also holds true for chiasmata at diakinesis (Hultén 1974; Laurie and Hultén 1985b). Thus, the SC measures genetic distance.
Figure 2.
Relationship between the mean SC length (μm) and the mean number of MLH1 foci on human SCs. (Pearson correlation .98; P<.0001).
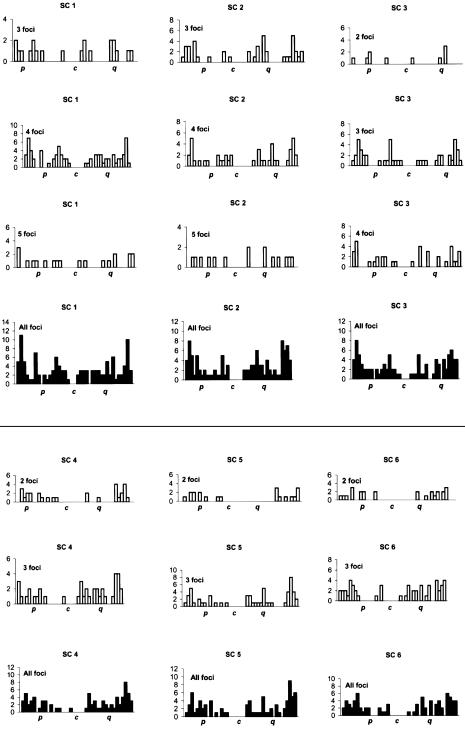
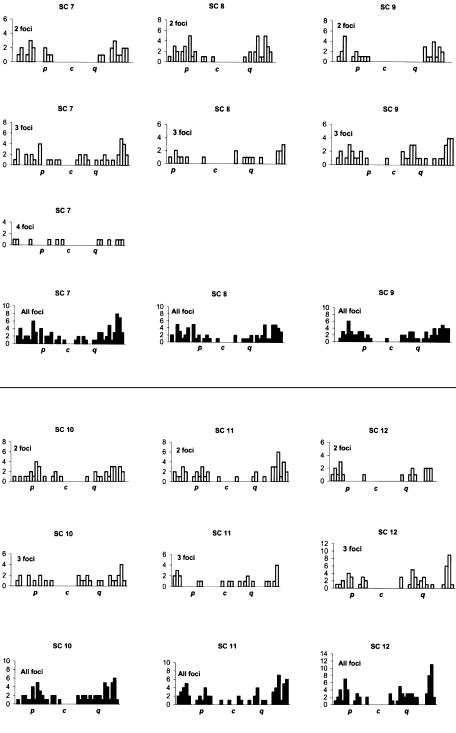
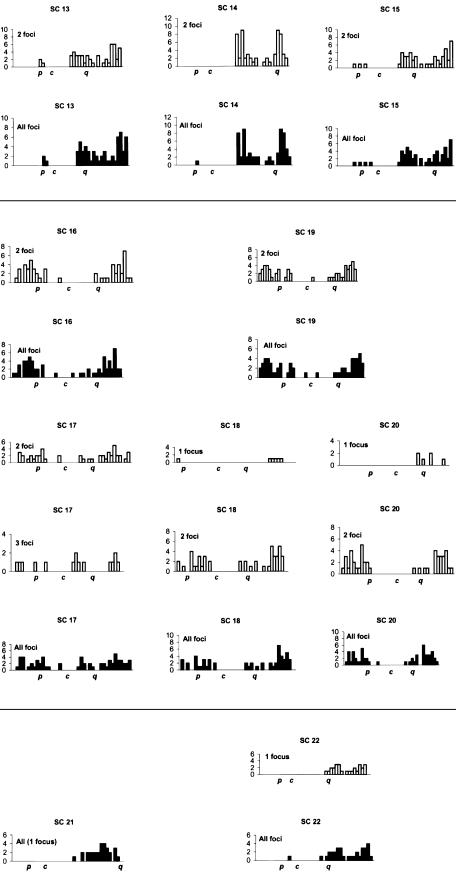
The distributions of MLH1 foci for individual chromosomes are presented in figure 3. For each SC, different distributions for one to five MLH1 foci are presented, as well as a final cumulative distribution. In general, many MLH1 foci were near the medial and terminal parts of the chromosome, with repression of foci near the centromeres. Every bivalent had at least one recombination focus per chromosome. Moreover, each chromosome arm generally had at least one MLH1 focus, with the exception of the short arms of acrocentric chromosomes, where exchanges seldom occur. The arms of chromosome 4, 5, 8, 9, 10, 12, 16, and 19 almost invariably had only one MLH1 focus, with mean arm frequencies of 0.9–1.03 (table 1). The distribution of recombination foci varied according to the number of crossovers present and the position of the centromere (fig. 3; table 3). For example, for chromosome 2, two foci were separated, on average, by 70.7% of the chromosome length, three foci by 39.8%, four foci by 29.6%, and five foci by 21.2%. For chromosome 12, two foci were separated, on average, by 70.9%, three foci by 40.9%, and four foci by 25.3% (table 3). For chromosome 21, only one MLH1 focus was present for each SC, always on the long arm and generally near the telomere. For chromosome 22, the distribution was bimodal, with medial and distal foci and some SCs with two sites of recombination. Tapper et al. (2002) have determined that recombination is associated with CT/CA repeats for chromosomes 21 and 22. These repeats are largely subtelomeric in distribution on chromosome 21, whereas they are widely distributed on chromosome 22, perhaps offering greater opportunities for double recombinants. “Hot” cytological regions (encompassing several megabases) were generally observed in the subtelomeric region, particularly for chromosomes 1p and 1q and the q arms of chromosomes 5, 12, 13, and 14 (fig. 3). Hotspots of recombination defined by molecular analysis appear to correspond to hot regions defined by cytological studies (Wintle et al. 1997).
Figure 3.
Distribution of MLH1 foci for individual SCs. The X-axis represents the position of the foci from the p telomere (left) to the q telomere (right). The centromere is labeled “c”. Each chromosome is divided into 5% intervals. From top to bottom, the histograms show the positions of foci for bivalents with one to five foci. Data were not displayed for bivalents with <3 cells with a specific number of MLH1 foci. The blackened histograms display overall frequencies.
Table 3.
Interfocal Distances on the Same SC with Different Numbers of Foci
|
Interfocal Distancec |
|||||
| Chromosomeand No. of Focia | MeanInterfocal Distanceb | 1 to 2 | 2 to 3 | 3 to 4 | 4 to 5 |
| Chromosome 1: | |||||
| 3 | 36.7 ± 1.7 | 38.1 ± 13.8 | 35.2 ± 17.3 | … | … |
| 4 | 28.5 ± 1.4 | 26.2 ± 8.1 | 34.5 ± 8.8 | 24.9 ± 5.6 | … |
| 5 | 23.1 ± 2.3 | 19.4 ± 5.9 | 35.8 ± 10.6 | 20.9 ± 7.3 | 16.2 ± 4.4 |
| Chromosome 2: | |||||
| 2 | 70.7 ± 5.4 | 70.7 ± 5.4 |
… | … | … |
| 3 | 39.8 ± .5 | 38.7 ± 10.9 | 40.9 ± 11.9 | … | … |
| 4 | 29.6 ± 1.5 | 26.6 ± 7.6 | 35.0 ± 11.2 | 27.1 ± 9.4 | … |
| 5 | 21.2 ± 2.8 | 22.5 ± 7.6 | 19.5 ± 10.9 | 22.5 ± 9.4 | 20.2 ± 15.2 |
| Chromosome 3: | |||||
| 2 | 61.3 ± 7.5 | 61.3 ± 7.5 |
… | … | … |
| 3 | 39.8 ± 1.2 | 46.5 ± 15.0 | 33.0 ± 12.5 | … | … |
| 4 | 30.3 ± 2.5 | 29.0 ± 8.2 | 37.8 ± 11.7 |
24.0 ± 5.7 | … |
| Chromosome 4: | |||||
| 2 | 68.1 ± 12.5 | 68.1 ± 12.5 | … | … | … |
| 3 | 40.0 ± 1.2 | 34.8 ± 11.0 | 45.2 ± 13.5 |
… | … |
| 4 | 29.2 ± 5.1 | 18.4 ± 6.3 | 22.5 ± 12.5 | 46.7 ± .1 |
… |
| Chromosome 5: | |||||
| 2 | 75.0 ± 13.4 | 75.0 ± 13.4 |
… | … | … |
| 3 | 42.3 ± 1.1 | 46.7 ± 10.5 | 37.9 ± 12.7 | … | … |
| 4 | 29.0 ± 0 | 28.3 ± 0 | 28.4 ± 0 | 30.2 ± 0 |
… |
| Chromosome 6: | |||||
| 2 | 69.1 ± 10.6 | 69.1 ± 10.6 |
… | … | … |
| 3 | 41.1 ± 1.9 | 34.2 ± 10.9 | 48.0 ± 14.7 | … | … |
| 4 | 30.6 ± 0 | 31.6 ± 0 | 32.0 ± 0 |
28.1 ± 0 | … |
| Chromosome 7: | |||||
| 2 | 70.5 ± 9.3 | 70.5 ± 9.3 |
… | … | … |
| 3 | 40.2 ± .4 | 40.6 ± 16.2 | 39.9 ± 17.1 | … | … |
| 4 | 30.0 ± 1.0 | 21.0 ± 6.6 | 48.9 ± 4.6 |
20.3 ± 4.1 | … |
| Chromosome 8: | |||||
| 2 | 70.1 ± 9.9 | 70.1 ± 9.9 |
… | … | … |
| 3 | 43.7 ± 2.5 | 41.4 ± 11.3 | 46.0 ± 6.2 |
… | … |
| Chromosome 9: | |||||
| 2 | 71.2 ± 9.0 | 71.2 ± 9.0 |
… | … | … |
| 3 | 41.4 ± .1 | 41.3 ± 8.0 | 41.5 ± 7.9 | … | … |
| Chromosome 10: | |||||
| 2 | 63.7 ± 14.5 | 63.7 ± 14.5 |
… | … | … |
| 3 | 39.5 ± .7 | 35.8 ± 12.6 | 43.3 ± 11.1 |
… | … |
| Chromosome 11: | |||||
| 2 | 70.0 ± 17.0 | 70.0 ± 17.0 | … | … | … |
| 3 | 41.2 ± 4.0 | 38.1 ± 13.8 | 44.4 ± 21.7 | … | … |
| Chromosome 12: | |||||
| 2 | 70.9 ± 13.5 | 70.9 ± 13.5 | … | … | … |
| 3 | 40.9 ± 1.7 | 41.9 ± 9.4 | 39.9 ± 12.8 | … | … |
| 4 | 25.3 ± 0 | 28.7 ± 0 | 29.2 ± 0 | 18.1 ± 0 |
… |
| Chromosome 13: | |||||
| 2 | 57.6 ± 15.0 | 57.6 ± 15.0 | … | … | … |
| 3 | 34.9 ± 2.5 | 33.2 ± 7.1 | 36.5 ± 2.0 | … | … |
| Chromosome 14: | |||||
| 2 | 58.4 ± 11.5 | 58.4 ± 11.5 | … | … | … |
| 3 | 45.3 ± 0 | 53.1 ± 0 | 37.5 ± 0 |
… | … |
| Chromosome 15: | |||||
| 2 | 60.9 ± 15.4 | 60.9 ± 15.4 | … | … | … |
| 3 | 34.7 ± 0 | 48.9 ± 0 | 20.5 ± 0 |
… | … |
| Chromosome 16: | |||||
| 2 | 70.9 ± 8.2 | 70.9 ± 8.2 |
… | … | … |
| 3 | 38.2 ± 0 | 18.5 ± 0 | 57.9 ± 0 |
… | … |
| 4 | 27.8 ± 4.2 | 33.0 ± .9 | 16.9 ± 6.3 | 33.4 ± 11.3 |
… |
| Chromosome 17: | |||||
| 2 | 61.6 ± 15.7 | 61.6 ± 15.7 | … | … | … |
| 3 | 40.8 ± .6 | 49.9 ± 5.3 | 31.8 ± 4.1 |
… | … |
| Chromosome 18: | |||||
| 2 | 65.5 ± 16.0 | 65.5 ± 16.0 | … | … | … |
| 3 | 38.9 ± 0 | 28.7 ± 0 | 49.1 ± 0 |
… | … |
| Chromosome 19: | |||||
| 2 | 73.9 ± 12.9 | 73.9 ± 12.9 |
… | … | … |
| 3 | 38.1 ± 0 | 46.1 ± 0 |
30.1 ± 0 | … | … |
| Chromosome 20: | |||||
| 2 | 73.0 ± 12.6 | 73.0 ± 12.6 |
… | … | … |
| 3 | 34.4 ± 0 | 44.0 ± 0 |
24.8 ± 0 | … | … |
| Chromosome 22: | |||||
| 2 | 76.4 ± 15.4 | 76.4 ± 15.4 | … | … | … |
Since every chromosome 21 SC had a single MLH1 focus, no distance between foci can be reported.
“Mean” refers to the mean distance between two foci (% of chromosome) in any category—for example, for chromosome 1 with three MLH1 foci, there was a mean interfocal distance of 36.7%.
Mean ± SD for a given interfocal interval (% of chromosome). Foci are numbered from the q-arm telomere, across the centromere, to the p-arm telomere. The transcentromere distance is underlined; intervals with a mixture of intra- and interarm distances are boldface italic.
The position of recombination foci was not random but exhibited positive interference (i.e., the presence of one exchange inhibits the formation of a second exchange in close proximity). For chromosomes with two exchanges, random placement of two foci on the SC leads to an expected average separation distance of ∼33% (Carpenter 1988). We found that the average separation distance between foci on SC with two foci was 67.6%±5.5%, more than twice this distance, indicating that MLH1 foci do demonstrate positive interference. This has previously been demonstrated for many organisms, including birds (Pigozzi and Solari 1999), mice (Froenicke et al. 2002), and humans (Barlow and Hultén 1998; Lynn et al. 2002; Tease et al. 2002). However, there were rare instances (∼3%–4% of cells) in which the MLH1 foci were very close, demonstrating that it is physically possible to accommodate neighboring exchange events.
In summary, we have presented immunocytological recombination maps for every autosome in the human male for the first time. Our maps demonstrate a preference for distal exchanges with repression of foci over the centromere, crossover interference, and specific patterns of recombination observed for each bivalent. The genetic length of individual bivalents is very similar to that determined by linkage studies. The SC length predicts the number of exchanges and, therefore, the genetic length, since we have demonstrated a very tight relationship between average SC length and average number of MLH1 foci per SC.
Acknowledgments
We thank T. Ashley, M. Fritzler, and P. Moens, for the generous gift of antibodies, and T. Hassold, for help in establishing the technique for analysis of synaptonemal complexes. R.H.M. holds the Canada Research Chair in Genetics, and the research was funded by Canadian Institutes of Health Research (CIHR) grant MA7961. F.S. is the recipient of a CIHR Strategic Training Fellowship in Genetics, Child Development and Health.
Electronic-Database Information
The URL for data presented herein is as follows:
- MicroMeasure Web site, http://www.colostate.edu/Depts/Biology/MicroMeasure (for MicroMeasure for Windows, version 3.3)
References
- Anderson LK, Reeves A, Webb LM, Ashley T (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151:1569–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 13:336–342 [DOI] [PubMed] [Google Scholar]
- Barlow AL, Hultén MA (1998) Crossing over analysis at pachytene in man. Eur J Hum Genet 6:350–358 10.1038/sj.ejhg.5200200 [DOI] [PubMed] [Google Scholar]
- Carpenter A (1988) Thoughts on recombination nodules, meiotic recombination, and chiasmata. In: Kucherlapati R, Smith GR (eds) Genetic recombination. American Society of Microbiology, Washington, DC, pp 529–548 [Google Scholar]
- Froenicke L, Anderson LK, Wienberg J, Ashley T (2002) Male mouse recombination maps for each autosome identified by chromosome painting. Am J Hum Genet 71:1353–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale D (1994) Is X-Y recombination necessary for spermatocyte survival during mammalian spermatogenesis? Cytogenet Cell Genet 65:278–282 [DOI] [PubMed] [Google Scholar]
- Hassold T, Sherman S, Pettay D, Page D, Jacobs P (1991) XY chromosome nondisjunction in man is associated with diminished recombination in the pseudoautosomal region. Am J Hum Genet 49:253–260 [PMC free article] [PubMed] [Google Scholar]
- Heng H, Liu G, Lu W, Bremer S, Ye G, Hughes M, Moens P (2001) Spectral karyotyping of mouse meiotic chromosomes. Genome 44:293–298 10.1139/gen-44-2-293 [DOI] [PubMed] [Google Scholar]
- Hultén M (1974) Chiasma distribution at diakinesis in the normal human male. Hereditas 76:55–78 [DOI] [PubMed] [Google Scholar]
- Hultén M, Lindsten J (1973) Cytogenetic aspects of human male meiosis. In: Harris H, Hirschhorn K (eds) Advances in human genetics. Vol 4. Plenum Press, New York, pp 327–387 [DOI] [PubMed] [Google Scholar]
- Hultén M, Tease C (2003) Genetic mapping: comparison of direct and indirect approaches. In: Cooper DN (ed) Nature encyclopedia of the human genome. Vol 2. Nature Publishing Group, London, pp 876–881 [Google Scholar]
- International System for Human Cytogenetic Nomenclature (1985) An international system for human cytogenetic nomenclature. In: Harnden DG, Klinger HP (eds) Birth defects original article series, vol 21. S. Karger, New York, pp 115 [PubMed] [Google Scholar]
- Ko E, Rademaker A, Martin R (2001) Microwave decondensation and codenaturation: a new methodology to maximize FISH data from donors with very low concentrations of sperm. Cytogenet Cell Genet 95:143–145 [DOI] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Laurie D, Hultén M (1985a) Further studies on bivalent chiasma frequency in human males with normal karyotypes. Ann Hum Genet 49:189–201 [DOI] [PubMed] [Google Scholar]
- ——— (1985b) Further studies on chiasma distribution and interference in the human male. Ann Hum Genet 49:203–214 [DOI] [PubMed] [Google Scholar]
- Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt P, Hassold TJ (2002) Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296:2222–2225 10.1126/science.1071220 [DOI] [PubMed] [Google Scholar]
- Marcon E, Moens P (2003) MLH1p and MLH3p localize to precociously induced chiasmata of okadaic acid treated mouse spermatocytes. Genetics 165:2283–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Spriggs E, Rademaker A (1996) Multicolor fluorescence in situ hybridization analysis of aneuploidy and diploidy frequencies in 225,846 sperm from 10 normal men. Biol Reprod 54:394–398 [DOI] [PubMed] [Google Scholar]
- Miklos GL (1974) Sex-chromosome pairing and male fertility. Cytogenet Cell Genet 13:558–577 [DOI] [PubMed] [Google Scholar]
- Nicolas A (1998) Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc Natl Acad Sci USA 95:87–89 10.1073/pnas.95.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietzel A, Rocchi M, Starke H, Heller A, Fiedler W, Wlodarska I, Longarevic IF, Beensen V, Claussen U, Liehr T (2001) A new multicolor-FISH approach for the characterization of marker chromosomes: centromere-specific multicolour-FISH (cenM-FISH). Hum Genet 108:199–204 10.1007/s004390100459 [DOI] [PubMed] [Google Scholar]
- Oliver-Bonet M, Liehr T, Nietzel A, Heller A, Starke H, Claussen U, Codina-Pascual M, Pujol A, Abad C, Egozcue J, Navarro J, Benet J (2003) Karyotyping of human synaptonemal complexes by cenM-FISH. Eur J Hum Genet 11:879–883 10.1038/sj.ejhg.5201067 [DOI] [PubMed] [Google Scholar]
- Pigozzi MI, Solari AJ (1999) Recombination nodule mapping and chiasma distribution in spermatocytes of the pigeon, Columba livia. Genome 42:308–314 10.1139/gen-42-2-308 [DOI] [PubMed] [Google Scholar]
- Shi Q, Spriggs E, Field L, Ko E, Barclay L, Martin R (2001) Single sperm typing demonstrates that reduced recombination is associated with the production of aneuploid 24,XY human sperm. Am J Med Genet 99:34–38 [DOI] [PubMed] [Google Scholar]
- Tapper WJ, Ke X, Morton NE, Collins A (2002) Recombination, interference and sequence: comparison of chromosomes 21 and 22. Ann Hum Genet 66:75–86 10.1017/S0003480001008946 [DOI] [PubMed] [Google Scholar]
- Tease C, Hartshorne GM, Hultén M (2002) Patterns of meiotic recombination in human fetal oocytes. Am J Hum Genet 70:1469–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintle RF, Nygaard TG, Herbrick JA, Kvaloy K, Cox DW (1997) Genetic polymorphism and recombination in the subtelomeric region of chromosome 14q. Genomics 40:409–414 10.1006/geno.1996.4572 [DOI] [PubMed] [Google Scholar]