Abstract
In the yeast Saccharomyces cerevisiae, septins form a scaffold in the shape of a ring at the future budding site that rearranges into a collar at the mother-bud neck. Many proteins bind asymmetrically to the septin collar. We found that the protein Bni4-CFP was located on the exterior of the septin ring before budding and on the mother side of the collar after budding, whereas the protein kinase Kcc4-YFP was located on the interior of the septin ring before budding and moved into the bud during the formation of the septin collar. Unbudded cells treated with the actin inhibitor latrunculin-A assembled cortical caps of septins on which Bni4-CFP and Kcc4-YFP colocalized. Bni4-CFP and Kcc4-YFP also colocalized on cortical caps of septins found in strains deleted for the genes encoding the GTPase activating proteins of Cdc42 (RGA1, RGA2, and BEM3). However, Bni4-CFP and Kcc4-YFP were still partially separated in mutants (gin4, elm1, cla4, and cdc3-1) in which septin morphology was severely disrupted in other ways. These observations provide clues to the mechanisms for the asymmetric localization of septin-associated proteins.
INTRODUCTION
Septins are conserved GTPases that were first discovered in Saccharomyces cerevisiae by analysis of mutants defective in cytokinesis (Hartwell, 1971; Longtine et al., 1996) and were subsequently found to be crucial for cell division in some, but not all, fungal and animal cells (Longtine et al., 1996; Field and Kellogg, 1999; Trimble, 1999; Nguyen et al., 2000; Westfall and Momany, 2002). Septins seem to play roles in vesicle transport and exocytosis and have been implicated in neurodegenerative diseases and oncogenesis (Trimble, 1999; Dent et al., 2002; Hall and Russell, 2004). However, the ways in which they contribute to these processes are still poorly understood. The yeast genome encodes seven septin proteins, five of which (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1) are expressed in vegetative cells and two of which (Spr3 and Spr28) act exclusively during sporulation (De Virgilio et al., 1996; Fares et al., 1996; Longtine et al., 1996; Longtine and Bi, 2003). All five vegetative septins form a ring adjacent to the plasma membrane at the incipient bud site ∼15 min before bud emergence, at which time the septin ring rearranges into an hourglass-shaped collar that spans both the mother and the daughter sides of the mother-bud neck. This collar persists until just before cytokinesis, when it splits into two rings that occupy opposite sides of the neck.
The molecular architecture of septin complexes and the mechanisms that govern septin organization and remodeling are not well understood. Septins purified from yeast (Frazier et al., 1998; Versele et al., 2004; Versele and Thorner, 2004), Drosophila (Field et al., 1996), rat (Hsu et al., 1998), and Xenopus (Mendoza et al., 2002) form filaments in vitro. In mouse NIH3T3 fibroblasts, the septins Sept2 and Sept6 colocalize with actin in the form of bundles and rearrange into ∼0.7-μm-diameter rings when actin organization is disturbed (Kinoshita et al., 2002). Similar septin rings reconstituted in vitro consist of individual septin filaments that run circumferentially (Kinoshita et al., 2002). Electron microscopic examination of the yeast bud neck revealed 10-nm striations that run perpendicular to the mother-daughter axis (Byers and Goetsch, 1976). These striations are absent in temperature-sensitive septin mutants at restrictive temperature, which suggests that septins form filaments that encircle the neck perpendicular to the mother-daughter axis. However, in strains deleted for genes GIN4, NAP1, or CLA4, the septins often look like bars that run parallel to the mother-daughter axis (Longtine et al., 1998, 2000), suggesting that septins can form filaments that run along the mother-daughter axis.
The septins at the bud neck act as a scaffold for proteins involved in such fundamental and diverse processes as the establishment of cell polarity (Sheu et al., 2000), cell wall synthesis (DeMarini et al., 1997), spindle positioning (Adames and Cooper, 2000; Kusch et al., 2002), regulation of the mitotic checkpoint (Barral et al., 1999; McMillan et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000; Hanrahan and Snyder, 2003; Theesfeld et al., 2003), and cytokinesis (Bi et al., 1998; Lippincott and Li, 1998; Vallen et al., 2000). Many septin-dependent proteins that occur in the neck region are asymmetrically distributed across the mother-daughter axis (reviewed by Gladfelter et al., 2001). Such proteins associate with the mother side of the neck, with the daughter side, or with the middle zone between the mother and the daughter cell. Some of these proteins localize exclusively to one side of the neck, whereas others change their locations during the cell cycle. These asymmetries are likely to have important roles in growth, cell cycle regulation, and morphogenesis. For example, the protein Bni4 coordinates the synthesis of chitin in a ring on the mother side of the neck (DeMarini et al., 1997). Loss or delocalization of this complex from the mother side of the neck results in a defect in morphology in a wild-type background and inviability in cells that also lack the formin protein Bni1 (Tong et al., 2001). In contrast, the protein kinase Hsl1 and its binding partner Hsl7 localize exclusively on the daughter side of the neck, where they act to regulate the stability of the morphogenesis checkpoint kinase Swe1 (Barral et al., 1999; McMillan et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000; Cid et al., 2001). The Kcc4 protein kinase also associates exclusively with the daughter side (Barral et al., 1999; Okuzaki and Nojima, 2001), where it may act with the related protein kinases Gin4 and Hsl1 to regulate septins and the cell cycle (Barral et al., 1999).
Several models have been proposed to explain the asymmetric localization of proteins at the bud neck (Gladfelter et al., 2001). One possibility, first proposed by DeMarini et al. (1997) for the asymmetry of Bni4, is that Bni4 binds irreversibly to the initial septin ring in unbudded cells. If septins are added asymmetrically to form the collar at bud emergence, this process would leave Bni4 on the mother side of the neck. This model does not explain the asymmetric localization of proteins such as Kcc4, which occur at the site of bud emergence and remain on the daughter site of the neck during budding. On the other hand, there is no evidence that a single mechanism accounts for the asymmetry of all bud neck-associated proteins, and individual proteins may localize by distinct mechanisms. A second possibility is that the asymmetry is generated because of the diffusion barrier formed by the septin collar (Barral et al., 2000; Takizawa et al., 2000; Dobbelaere and Barral, 2004). This barrier limits the diffusion of the membrane-associated protein Ist2, whose mRNA is delivered to the bud tip by an actomyosin-based process. A hypothetical asymmetry-initiating factor also could be transported to and retained in the membrane compartment of the bud by a similar mechanism. This factor could then direct the asymmetric localization of other proteins at the neck. A third hypothesis posits a specific septin organization within the septin ring and/or collar as the basis for the asymmetry. It is possible that septins are arranged into polar filaments running parallel to the mother-daughter axis in the neck. Such an organization of septins would create different environments on the two sides of the septin collar.
To investigate the mechanisms responsible for the asymmetry of septin-binding proteins, we have focused on Bni4 and Kcc4, which localize to the mother and daughter sides of the neck, respectively. These proteins have several attractive attributes for this study. A green fluorescent protein (GFP) fusion of each protein localizes to the neck, and the Bni4-GFP fusion, at least, seems to have normal activity. Both proteins localize to the incipient bud site early in the cell cycle before bud emergence, yet neither is necessary for the normal localization of the other. Using these two proteins as probes, we have discovered that the spatial separation between Bni4 and Kcc4 occurs before bud emergence. Furthermore, our data indicate that the actin cytoskeleton is involved in normal septin ring formation or maintenance in unbudded cells and for the concomitant asymmetric distribution of Bni4 and Kcc4.
MATERIALS AND METHODS
Yeast Strains, Media, and Reagents
The yeast strains used (Table 1) are all congenic to JC482 (Cannon and Tatchell, 1987) except for strains YLK281 and YLK282, which were derived by sporulation of KWY264 and KWY295 (Wertman et al., 1992; Ayscough et al., 1997), and the deletion panel strains (Winzeler et al., 1999), which were obtained from Research Genetics (Open Biosystems, Huntsville, AL). Mutant alleles were introduced into the JC482 background by at least seven serial backcrosses or by transformation. The cdc3-1 allele in YLK271 was derived from SLTD3-6B (Johnson et al., 1987). The cla4::TRP1 allele in JRL55 was derived from strain TRY-1D (Richman et al., 1999). The gin4::LEU2 allele in YLK158 and YLK202 was derived from a strain obtained from Douglas Kellogg (University of California, Santa Cruz, Santa Cruz, CA) (Altman and Kellogg, 1997). The construction of BNI4-CFP::kanMX6, CDC10-GFP::kanMX6, and CDC10-YFP::His3MX6 alleles was described previously (Kozubowski et al., 2003).
Table 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| KT1357 | MATaleu2 ura3 his3 trp1 | Frederick and Tatchell (1996) |
| KT1358 | MATα leu2 ura3 his3 trp1 | Venturi et al. (2000) |
| YLK66 | MATα leu2 ura3 his3 trp1 CDC10-GFP::kanMX6 | Kozubowski et al. (2003) |
| YLK158 | MATα leu2 ura3 his3 trp1 gin4::LEU2 CDC10-GFP::kanMX6 | See text |
| YLK189 | MATα leu2 ura3 his3 trp1 BNI4-CFP::kanMX6 KCC4-YFP::His3MX6 | See text |
| YLK202 | MATα leu2 ura3 his3 trp1 gin4::LEU2 BNI4-CFP::kanMX6 KCC4-YFP:: His3MX6 | See text |
| YLK214 | MATα leu2 ura3 his3 trp1 KCC4-CFP::kanMX6 CDC10-YFP::His3MX6 | See text |
| YLK215 | MATα leu2 ura3 his3 trp1 BNI4-CFP::kanMX6 CDC10-YFP::His3MX6 | See text |
| YLK217 | MATa/MATα leu2/leu2 ura3/ura3 his3/his3 trp1/trp1 KCC4-CFP::kanMX6/KCC4-CFP::kanMX6 CDC10-YFP::His3MX6/CDC10 | See text |
| YLK218 | MATa/MATα leu2/leu2 ura3/ura3 his3/his3 trp1/trp1 BNI4-CFP::kanMX6/BNI4-CFP::kanMX6 CDC10-YFP::His3MX6/CDC10 | See text |
| YLK271 | MATα leu2 ura3 his3 trp1cdc3-1 BNI4-CFP::kanMX6 KCC4-YFP::His3MX6 | See text |
| YLK281 | MATα leu2-3112 ura3-52 his3Δ200 ACT1:HIS3 tub2-201 CAN1? CRY1? | See text |
| YLK282 | MATaleu2-3112 ura3-52 his3Δ200 act1-117:HIS3 tub2-201 CAN1? CRY1? | See text |
| JRL18 | MATα leu2 ura3 his3 trp1shs1Δ::kanMX4 KCC4-CFP::kanMX6 BNI4-YFP::His3MX6 | See text |
| JRL49 | MATα leu2 ura3 his3 trp1 elm1Δ::kanMX6 BNI4-CFP::kanMX6 KCC4-YFP::His3MX6 | See text |
| JRL55 | MATα leu2 ura3 his3 trp1 cla4::TRP1 BNI4-CFP::kanMX6 KCC4-YFP::His3MX6 | See text |
| JRL159 | MATα leu2 ura3 his3 trp1 bem3Δ::kanMX6 rga1Δ::natMX6 rga2Δ::natMX6 BNI4-CFP::kanMX6 KCC4-YFP::His3MX6 | See text |
To generate gene-deletion and GFP-fusion strains by the PCR method, each amplified cassette was transformed into a KT1357 × KT1358 diploid strain, drug-resistant or amino acid-prototrophic transformants were sporulated, and haploid meiotic segregants were isolated by tetrad analysis. To generate the shs1Δ::kanMX4 deletion in strain JRL18, the pFA6a-kanMX4 cassette (Guldener et al., 1996) was amplified using primers SEP7F (CCAAAGATCTGCTTATAATTGCTAGAAAAATATATTATTAGCATAGGCCACTAGTGGATCTG) and SEP7R (GCTCAGCTTTGGATTTTGTACAGATACAACTCAATCTCTACAGCTGAAGCTTCGTACGC). To generate an elm1Δ::kanMX6 deletion, a deletion cassette was amplified using DNA from the elm1Δ strain from the Research Genetics panel and primers SP15-F (CCGGAATTCATAGTGCTTCGAGGA) and SP16-R (CGCGACACAGTGGGATCAAGATAA). To generate the rga1Δ rga2Δ bem3Δ mutant strain JRL159, deletion cassettes were amplified using DNA from the rga1Δ, rga2Δ, and bem3Δ strains from the Research Genetics panel and primers SP25-F (GCGCGCTTTATGCCGTATTAGGAA) and SP26-R (CAGACTGCTTGGCGCTTATGATTC) (rga1Δ), SP21-F (CCCAGTTAATTAGCCACAGTG) and SP22-R (GACGCAATACCAAGAGCATG) (rga2Δ), and SP23-F (GAAAGTTATATGGCGGCGGTGATG) and SP24-R (CCTTCTTTATCTCAGCTCTTCGACC) (bem3Δ). The cassettes were then individually transformed into a KT1357 × KT1358 diploid strain. Nat-resistant strains were then created by restriction digesting pAG25 (Goldstein and McCusker, 1999) with EcoRI and transforming into the Kan-resistant haploids. Crosses among these strains and YLK189 then yielded strain JRL159.
All integrated GFP, cyan fluorescent protein (CFP), and yellow fluorescent protein (YFP) fusions were constructed according to the method described by the Yeast Resource Center (University of Washington, Seattle, WA) (http://depts.washington.edu/∼yeastrc/fm_home3.htm), which is based on the method described by Wach et al. (1997). BNI4-YFP in strain JRL18 was generated using the primer pairs described in Kozubowski et al. (2003). For the generation of GFP, CFP, and YFP fusions to Kcc4, primers KCC4F (AATCCAAATTATTTTACAAAAAGAAGGTGTTTTGGACAAAGGTCGACGGATCCCCGGG) and KCC4R (CGTATTGTCCATTTGGGGATCGATTATCCCTCCCTTTTTTATCGATGAATTCGAGCTCG) were used. Strains were grown at 30°C on YPD medium (2% bacto peptone, 1% yeast extract, and 2% glucose) or synthetic complete medium that was made as described by Sherman et al. (1986) unless stated otherwise.
To generate pLK10, a BNI4-GFP LEU2 CEN plasmid, plasmid p366 (DeMarini et al., 1997), was transformed into a bni4Δ1::TRP1 deletion strain, which was then transformed with a PCR-generated EGFP-kanMX6 cassette amplified from plasmid pLK3 by using primer pairs BGFP1 and BGFP2 (Kozubowski et al., 2003). The resulting BNI4-GFP–containing plasmid was recovered from yeast by transformation into Escherichia coli. pTD150-CDC3 is a URA3 CEN plasmid containing CDC3-GFP expressed from the ACT1 promoter and was a gift from Brian Haarer (SUNY Upstate Medical University, Syracuse, NY).
Microscopy
Indirect immunofluorescence was done as described previously (Kozubowski et al., 2003). The primary antibodies were anti-Bni4 (DeMarini et al., 1997), anti-Cdc11 (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-Cdc12 (kindly provided by M. Versele and J. Thorner, University of California, Berkeley, Berkeley, CA), and the secondary antibody was Alexa Fluor 488 anti-rabbit-IgG (Molecular Probes, Eugene, OR). GFP, CFP, and YFP fusion proteins were imaged as described by Kozubowski et al. (2003). The CFP and YFP signals were pseudocolored green and red, respectively, and superimposed on the differential interference contrast (DIC) images by using IPlab Spectrum software (Scanalytics, Fairfax, VA). Confocal images were collected with a Bio-Rad Radiance 2000 confocal system (Nikon TE300 stand; 25-mW Ar laser; 100×, 1.4 numerical aperture, Plan Apo objective). The three-dimensional (3D) image projections were made using either the Radiance 2000 software (Bio-Rad, Hercules, CA) or the IPlab Spectrum software.
Cell synchronization and treatment with latrunculin A (Lat-A) (Sigma-Aldrich, St. Louis, MO) were carried out as described by Ayscough et al. (1997). Briefly, a 200-μl aliquot of overnight culture was spread onto YPD medium and incubated at 30°C overnight; the cells were then scraped into 20 ml of 50% YPD + 1 M sorbitol, and budded cells were eliminated by successive centrifugations at 500 × g. The unbudded cells (mostly daughter cells) remaining in the supernatant were pelleted and resuspended in YPD. Typically, aliquots of 200 μl of unbudded cells (at ∼5 × 107cells/ml) were incubated at 24°C in the presence of either Lat-A added from a 20 mM stock solution in dimethyl sulfoxide (DMSO) or the equivalent amount of DMSO. To evaluate the data and determine whether polarized fluorescence was in the shape of a ring or a cap, two or three images at different focal planes were acquired. The actin cytoskeleton was visualized using rhodamine-phalloidin (Molecular Probes) as described by Yang et al. (1997).
RESULTS
Bni4 and Kcc4 Are Spatially Separated on Septin Rings of Unbudded Cells
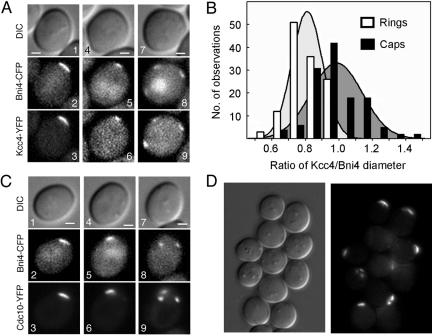
We constructed strains in which the blue-shifted and red-shifted variants of GFP were fused to Bni4 and Kcc4, respectively. Consistent with previous reports (DeMarini et al., 1997; Barral et al., 1999; Okuzaki and Nojima, 2001; Kozubowski et al., 2003), in small-budded cells, Bni4-CFP and Kcc4-YFP bound exclusively to the mother and daughter sides of the septin collar, respectively (Figure 1A). In unbudded cells of the same strain, Bni4-CFP and Kcc4-YFP occurred essentially simultaneously at the presumptive bud site: 95% (n = 92) of unbudded cells with Bni4-CFP at the incipient bud site also had Kcc4-YFP at that site, whereas 98% (n = 89) of unbudded cells with Kcc4-YFP at the incipient bud site also had Bni4-CFP there. If bud emergence were necessary for the Bni4-Kcc4 asymmetry, as originally proposed for the asymmetric localization of Bni4 (DeMarini et al., 1997), then there should be no separation between Bni4 and Kcc4 in unbudded cells. However, Bni4-CFP and Kcc4-YFP did not colocalize in unbudded cells. In cross-section (Figure 1B, 1–4), Kcc4-YFP was detected as either a single patch or two patches positioned inside two patches of Bni4-CFP. Examination of these signals in different focal planes revealed that both Bni4-CFP and Kcc4-YFP form rings at the future budding site. In cells for which the entire ring of Bni4-CFP was imaged (Figure 1B, 5–8), Kcc4-YFP occurred as either a smaller ring or a patch positioned inside the Bni4-CFP ring. To quantify these results, we measured the diameters of the Bni4-CFP and Kcc4-YFP rings in 128 unbudded cells. The measured diameters varied between 0.4 and 1.5 μm for Kcc4 and between 0.6 and 2.0 μm for Bni4. The ratio of the Kcc4 and Bni4 diameters was less variable and was significantly less than one (0.81 ± 0.09, p < 0.01).
Figure 1.
Bni4 and Kcc4 do not colocalize in unbudded cells. DIC and fluorescence images are shown. For merged images, Bni4-CFP and Kcc4-YFP are pseudocolored green and red, respectively. (A) Asymmetric localization of Bni4-CFP and Kcc4-YFP in small- and medium-budded cells of strain YLK189. (B) Spatial separation of Bni4-CFP and Kcc4-YFP in unbudded cells of strain YLK189. (C) Localization of Kcc4-CFP and Bni4-CFP to the inner and outer edges of the Cdc10-YFP septin ring, respectively. 1–3, strain YLK214; 4–6, strain YLK215. (D) Separation of Kcc4-YFP (arrowheads) and Bni4-CFP (arrows) during bud emergence. A cell of strain YLK189 was imaged before bud emergence (1 and 4) and 10 (2 and 5) and 20 (3 and 6) min later. (E) Model showing the location of Bni4 and Kcc4 during the transition of the septin ring into the septin collar upon bud emergence. Bni4 (blue) forms a larger ring that encircles the Kcc4 ring (yellow). At bud emergence, Kcc4 moves toward the growing bud, whereas Bni4 stays on the mother side of the neck. Bars, 1 μm.
Because Bni4 and Kcc4 require the septin scaffold for their association with the neck, we reasoned that these proteins should occupy the outer and inner portions of the septin ring, respectively. To test this possibility, we visualized either Bni4-CFP or Kcc4-CFP together with the septin Cdc10-YFP. Both Bni4-CFP and Kcc4-CFP occurred at the future budding site approximately simultaneous with the septin. For Bni4-CFP, all unbudded cells with polarized Cdc10-YFP also displayed colocalized Bni4-CFP (26 cells of haploid strain YLK215; 26 cells of diploid strain YLK218). For Kcc4-CFP, the same was true for nine of nine diploid cells (strain YLK217) and 38 of 48 haploid cells (strain YLK214) with polarized Cdc10-YFP. However, in the remaining 10 haploid cells, which had weak Cdc10-YFP signals, there was no detectable Kcc4-CFP. In both haploid and diploid cells, the Cdc10-YFP rings were larger than those of Kcc4-CFP but smaller than those of Bni4-CFP (Figure 1C; our unpublished data).
To follow the locations of Bni4 and Kcc4 during the transition of the septin ring in unbudded cells to the septin collar in budded cells, we performed time-lapse imaging. As shown in Figure 1D, the patch or small ring of Kcc4-YFP seemed to move into the bud at bud emergence, ultimately occupying the daughter side of the neck, whereas the previously encircling ring of Bni4-CFP remained on the mother side.
In summary, Bni4 and Kcc4 accumulate simultaneously with the septins at the cell cortex in unbudded cells, but they are asymmetrically distributed across the septin ring (Figure 1E). This asymmetry persists as the septin collar forms at bud emergence. These data show that the spatial separation between Bni4 and Kcc4 develops before, and not concomitant with, bud emergence. In the remainder of this article, we refer to the spatial separation of Bni4 and Kcc4 in both budded and unbudded cells as asymmetric localization.
Septin Ring Formation Correlates with the Establishment of Bni4/Kcc4 Asymmetry
The spatial separation of Bni4 and Kcc4 in unbudded cells may seem to exclude the hypothesis that a diffusion barrier generated by the septins imposes the asymmetry, especially because this diffusion barrier was described only in budded cells (Barral et al., 2000; Takizawa et al., 2000; Dobbelaere and Barral, 2004). However, even in unbudded cells, a hypothetical membrane-associated factor could conceivably be transported directly to the inside of the septin ring and remain trapped in this small compartment. Because directed transport of many proteins occurs by an actin-based process, one way to test this possibility is to prevent actin polymerization in unbudded cells by using Lat-A and then monitor the localization of Bni4 and Kcc4. Septins and Bni4 were shown to localize to polarized sites independently of actin (Ayscough et al., 1997; Harkins et al., 2001), but Kcc4 was not examined. If actin is necessary for the asymmetry, then the septins, Bni4, and Kcc4 should overlap fully in unbudded cells after treatment with Lat-A.
To address the role of actin, we isolated unbudded daughter cells, released them into fresh medium containing 150 μM Lat-A, and monitored the locations of Bni4-CFP and Kcc4-YFP. After 3.5 h at 24°C, 46% of the DMSO-treated control cells were budded, and most budded and unbudded cells showed normal localization of Bni4-CFP and Kcc4-YFP (Table 2; our unpublished data). As expected, 96% of the Lat-A–treated cells were unbudded after 3.5 h, and filamentous actin was absent, as shown by staining with rhodamine-phalloidin (our unpublished data). We made two unexpected observations in the Lat-A–treated cells. First, cells with polarized Bni4-CFP and polarized Kcc4-YFP both detectable were only 3 and 7% of the populations after 3.5 and 8 h, respectively (Table 2). Second, Kcc4-YFP and Bni4-CFP formed clearly defined rings in only 5% of the cells that had detectable signals (vs. 84% in the control cells); in the remaining cells, the proteins localized as small patches or larger caps of fluorescence at the cortex when examined in cross-section. In the remainder of this article, we refer to a cortical fluorescence signal that does not look like a ring as a cap, regardless of its size. Furthermore, the Bni4 and Kcc4 fluorescence signals largely colocalized in the caps of the Lat-A–treated cells (Figure 2A).
Table 2.
Effect of Lat-A treatment on the spatial organization of Kcc4 and Bni4
| DMSO
|
Lat-A
|
|||
|---|---|---|---|---|
| 3.5 h (n = 648) | 3.5 h (n = 1779) | 4.25 h (n = 948) | 8 h (n = 875) | |
| % Unbudded cells | 54 | 96 | 97 | 95 |
| No. of unbudded cells with a ring | 16 | 6 | 1 | 0 |
| Diameter of Kcc4-YFP ring (μm) | 0.92 ± 0.09 | 1.02 ± 0.21 | 1.08 | |
| Diameter of Bni4-CFP ring (μm) | 1.15 ± 0.09 | 1.46 ± 0.19 | 1.20 | |
| Kcc4-YFP/Bni4-CFP ring ratio | 0.80 ± 0.09 | 0.70 ± 0.07 | 0.90 | |
| No. of unbudded cells with a cap | 3 | 39 | 25 | 54 |
| Diameter of Kcc4-YFP cap (μm) | 0.75 ± 0.07 | 1.06 ± 0.25 | 1.22 ± 0.25 | 1.15 ± 0.28 |
| Diameter of Bni4-CFP cap (μm) | 0.99 ± 0.17 | 1.14 ± 0.22 | 1.20 ± 0.21 | 1.18 ± 0.28 |
| Kcc4-YFP/Bni4-CFP cap ratio | 0.77 ± 0.06 | 0.93 ± 0.14 | 1.02 ± 0.60 | 0.99 ± 0.15 |
| No. of unbudded cells with an indeterminate signala | 0 | 6 | 4 | 4 |
| Diameter of Kcc4 indeterminate signal (μm) | 1.11 ± 0.17 | 1.26 ± 0.25 | 1.28 ± 0.09 | |
| Diameter of Bni4 indeterminate signal (μm) | 1.28 ± 0.17 | 1.28 ± 0.05 | 1.29 ± 0.17 | |
| Kcc4/Bni4 diameter ratio for indeterminate signal | 0.87 ± 0.13 | 0.99 ± 0.21 | 1.00 ± 0.07 | |
| % of Unbudded cells with lost asymmetryb | 5 | 49 | 77 | 74 |
Unbudded cells of strain YLK189 were isolated and treated with 150 μM Lat-A as described in Materials and Methods. Only cells with both proteins visible were scored.
The Kcc4 and Bni4 signals were intermediate between rings and caps.
Lost asymmetry is defined as a ratio of Kcc4 to Bni4 signal diameter of ≥0.9.
Figure 2.
Role of F-actin in formation and/or maintenance of the septin ring and Bni4/Kcc4 asymmetry before bud emergence. (A) Unbudded cells of strain YLK189 were incubated in Lat-A–containing medium (see Materials and Methods and Table 2) for 4 (1–6) or 8 (7–9) h before imaging Bni4-CFP and Kcc4-YFP. (B) Histogram showing ratios between Kcc4-YFP and Bni4-CFP signal diameters calculated either for rings in untreated YLK189 cells (open bars and light Gaussian curve; n = 128) or for caps in the Lat-A–treated cells (black bars and dark Gaussian curve; see Table 2). The mean values were significantly different according to a t test (p < 0.01). The data for the Lat-A–treated cells are from a total of 128 cells imaged at times between 1.5 and 8 h during the incubation with Lat-A. The average ratios for the individual time points were not significantly different (Kcc4/Bni4 ratios for the individual time points were 10 cells at 1.5 h, average ratio of 1.02 ± 0.16; 39 cells at 3.5 h, average ratio of 0.93 ± 0.14; 25 cells at 4.25 h, average ratio of 1.02 ± 0.16; 54 cells at 8 h, average ratio of 0.99 ± 0.15). (C) CDC10-YFP BNI4-CFP strain YLK215 was treated as in A and incubated for 3.5 h at 24°C before imaging. (D) Strain YLK281 was transformed with a low-copy plasmid expressing Cdc3-GFP (pTD150-CDC3) and treated as in A, except that 100 μM Lat-A was used. Cells were imaged after a 4-h incubation at 24°C. Bars, 1 μm.
To assess more precisely the relative locations of Kcc4-YFP and Bni4-CFP in Lat-A–treated cells, we measured the diameters of the fluorescence signals and compared the ratios of these diameters to the ratios of diameters measured in the control cells (Figure 2B and Table 2). In the few cells that had rings, the average YFP/CFP ratio was 0.75 ± 0.09 (n = 13), similar to that in the control cells. In contrast, the average YFP/CFP ratio for the caps in the Lat-A–treated cells (0.98 ± 0.15, n = 128) was significantly larger (p < 0.01) than the average ratio for rings in either control cells that were not treated (0.81 ± 0.09, n = 128) or treated with DMSO (0.80 ± 0.09, n = 16). Sixteen percent of the DMSO-treated unbudded cells with polarized signal did show apparent caps of signal (Table 2). However, the average ratio of Kcc4-YFP to Bni4-CFP diameters in these cells was 0.77, suggesting that Bni4 and Kcc4 were still separated (note that the classification of the fluorescence signal as ring or cap is subject to error when cells are examined in only one focal plane).
The accumulation of Bni4-CFP/Kcc4-YFP caps after Lat-A treatment suggests that the actin cytoskeleton has a role in formation and/or maintenance of the septin ring. To test this, we examined the locations of Bni4-CFP and the septin Cdc10-YFP in Lat-A–treated cells of strain YLK215. As before, only a small fraction of the unbudded cells showed a polarized Bni4-CFP signal (8 and 5%, respectively, after 2.5 and 7 h in Lat-A), and this signal was largely present as caps rather than rings (Figure 2C). In contrast, significantly more cells showed a polarized Cdc10-YFP signal (27 and 37%, respectively). Approximately 10% of the unbudded cells showed more than one cap of Cdc10-YFP, a phenomenon also observed for Bni4-CFP and Kcc4-YFP, although less frequently (Figure 2, A and C, 7–9).
Because these results seemed inconsistent with those reported previously (Ayscough et al., 1997; Harkins et al., 2001; Kadota et al., 2004), we repeated our experiments by using the S288C genetic background as used in the previous studies. Strain YLK281 was transformed with a low-copy plasmid encoding either Bni4-GFP or Cdc3-GFP and treated as in the previous experiments. After 3.5 h of incubation in the presence of 100 μM Lat-A, >90% of cells in both strains remained unbudded. The percentages of cells with polarized signal for Bni4-GFP and Cdc3-GFP were higher in this strain background, reaching 34 and 70%, respectively (Table 3, lines 1 and 2; and Figure 2D). However, as in the previous experiments, the majority of signal (∼75%) for both fusion proteins occurred as caps. To confirm that the appearance of septin caps resulted from the lack of filamentous actin, we also used a strain (YLK282) that contains the Lat-A–resistant actin allele act1-117. This strain did not show Cdc3-GFP caps upon treatment with 100 μM Lat-A, and most cells had budded after 2 h of incubation (our unpublished data).
Table 3.
Effect of Lat-A on the localization of Bni4 and septins
| Line | Strain | Signal | Concentration of Lat-A (μM) | % of Unbudded cells with signal | % of Signals detected as ringsa | No. of cells scored |
|---|---|---|---|---|---|---|
| 1 | YLK281 | Bni4-GFP | 100 | 34 | 24 | 395 |
| 2 | YLK281 | Cdc3-GFP | 100 | 70 | 26 | 435 |
| 3 | KT1357 | anti-Bni4 | 100 | 9 | 44 | 1272 |
| 4 | YLK281 | anti-Bni4 | 100 | 12 | 33 | 1721 |
| 5 | YLK281 | anti-Cdc11 | 100 | 37 | 19 | 288 |
| 6 | YLK281 | anti-Cdc11 | 200 | 37 | 27 | 386 |
| 7 | YLK281 | anti-Cdc11 | 400 | 35 | 29 | 375 |
| 8 | YLK281 | anti-Cdc12 | 100 | 39 | 33 | 554 |
| 9 | YLK281 | anti-Cdc12 | 200 | 43 | 30 | 419 |
| 10 | YLK281 | anti-Cdc12 | 400 | 43 | 28 | 335 |
Unbudded cells of strains YLK281 (S288C background) and KT1357 (JC482 background) were isolated, treated with Lat-A, and prepared for indirect immunofluorescence as described in Materials and Methods. Bni4-GFP (from plasmid pLK10) and Cdc3-GFP (from plasmid pTD150-CDC3) were imaged in live cells.
The remaining signals in each case seemed to be caps.
Indirect immunofluorescence using anti-Bni4, anti-Cdc11, and anti-Cdc12 antibodies confirmed the effect of Lat-A on septin organization. DMSO-treated control cells budded and showed the expected rings of both Bni4 and septins (our unpublished data). In the Lat-A–treated cells of either strain background, polarized Bni4 was in the form of a cap in 56–67% of unbudded cells with a detectable polarized signal (Figure 3A; Table 3, lines 3 and 4). Cdc11 and Cdc12 also formed caps in most cells with a polarized septin signal (Figure 3B and Table 3, lines 5 and 8). As with the GFP fusion proteins, the percentages of cells with polarized septin signals were significantly higher than that for Bni4. The use of higher Lat-A concentrations did not significantly affect the results (Table 3, lines 6, 7, 9, and 10). To confirm that the signals observed in the Lat-A–treated cells were mostly caps and not rings, we examined the cells by using a confocal microscope in multiple focal planes. The reconstructed images of the septins show clear differences between a ring of Cdc11 (Figure 3C, 1 and 2, and Supplemental Fig 3c1.mov) and caps of Cdc11 (Figure 3C, 3–6, and Supplemental Fig 3c2.mov and Fig 3c3.mov).
Figure 3.
Localization of Bni4, Cdc11, and Cdc12 by indirect immunofluorescence. Unbudded cells of strain YLK281 were incubated in Lat-A-containing medium as described in Figure 2D. (A and B) Bni4 (A), Cdc11 (B), and Cdc12 (B) were visualized as rings or, more frequently, as caps. (C) Samples stained with anti-Cdc11 antibody were analyzed using a confocal microscope (see Materials and Methods), by using a Z-section scan to create a 3D projection image. One and 2 show an image of one focal plane and a corresponding 90° angle projection image, respectively, of a septin ring. Three to 6 show similar pairs of images of cells for which the Cdc11 signal was a cap. See also the supplementary videos showing 3D projections of a ring and caps (Fig 3c1.mov, Fig 3c2.mov, and Fig 3c3.mov). Bars, 1 μm.
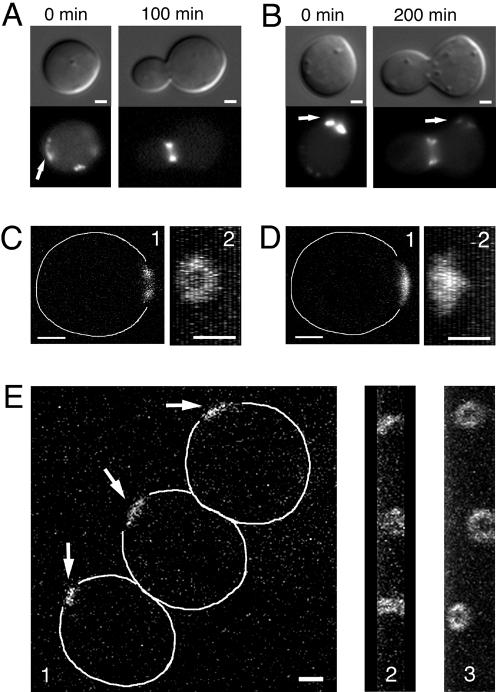
The results with Lat-A suggest that F-actin is important for the formation and/or maintenance of septin rings before bud emergence. They also suggest that the initial organization of septins into a ring in unbudded cells is crucial for the efficient spatial separation of Bni4 and Kcc4. Septin caps may be normal but transient structures that precede ring formation (Caviston et al., 2003; Longtine and Bi, 2003). If this were the case, then septin caps might rearrange into rings when F-actin is restored, and cells would presumably then bud from the site of the cap. To test this, we incubated unbudded daughter cells expressing Cdc10-GFP in Lat-A–containing medium, washed out the Lat-A, and visualized the cells by time-lapse confocal and wide-field microscopy. As before, most Lat-A–treated cells formed septin caps rather than rings (Figure 4, A, B, D, and E, and Supplemental Fig 4d.mov; a control cell is shown in Figure 4C and Supplemental Fig 4c.mov). After 2 h on Lat-A–free medium, most cells had budded again. Some cells budded from the site marked by a septin cap (Figure 4A, arrow), whereas other cells budded from a new site, accompanied by diminution of the original cap signal (Figure 4B, arrows). In cells that budded from the sites of the caps, the caps reorganized into rings before bud emergence (Figure 4E and Supplemental Fig 4e1.mov and Fig 4e2.mov). Because images were taken at 10-min intervals, these results do not provide direct evidence that septin caps normally precede the formation of rings, but they do suggest that caps can rearrange into rings under some circumstances (Caviston et al., 2003).
Figure 4.
Septin caps can rearrange into rings when F-actin is restored. Unbudded cells of CDC10-GFP strain YLK66 were incubated in medium containing 150 μM Lat-A for 4 h and washed into Lat-A-free medium. (A) A cell that budded in a location previously marked by a cap (arrow). (B) A cell that budded in a location distant from the cap, whose signal diminished during the incubation (arrows). (C–E) Images in one focal plane (1) and the corresponding 90° angle projection images (2) of a control cell treated with DMSO (C) and of cells that were treated with Lat-A for 4 h (D and E). (E, 3) A 90° angle projection image of the same cells in 1 and 2 after 2 h of incubation on Lat-A-free medium. C-E are accompanied by supplemental video materials (Fig 4c.mov, Fig 4d.mov, Fig 4e1.mov, and Fig 4e2.mov). Bars, 1 μm.
Mutants lacking the GTPase-activating proteins (GAPs) for Cdc42 (Bem3, Rga1, and Rga2) often display septin caps reminiscent of those described above (Caviston et al., 2003). To determine whether Bni4 and Kcc4 also are uniformly distributed in these caps, we imaged Bni4-CFP and Kcc4-YFP in the elongated cells (Caviston et al., 2003) of a mutant strain (JRL159). Like the septins, Kcc4-YFP and Bni4-CFP were frequently found in a cap at the site of polarized growth (Figure 5A). Although the two proteins were distributed roughly symmetrically over the cap, a close inspection revealed that they often did not completely colocalize. In particular, Kcc4-YFP was often distributed more toward the tips of the cells than was Bni4-CFP (Figure 5A, arrows). In those cells that presumably had relatively normal septin rings (Caviston et al., 2003), Kcc4-YFP and Bni4-CFP showed the normal asymmetrical distribution, with Kcc4-YFP closest to the bud tip (Figure 5A, arrowheads; cf. the congenic control cell in Figure 5B). These results support the hypothesis that the Bni4/Kcc4 asymmetry across the bud neck is associated with septin ring formation.
Figure 5.
Bni4 and Kcc4 are present as caps in a mutant lacking the Cdc42 GAPs. Unbudded cells of a bem3Δ rga1Δ rga2Δ mutant (JRL159) (A) and the congenic wild-type strain (YKL189) (B) were isolated by centrifugation of a starved population as described in Materials and Methods, incubated for 2 h at 24°C, and examined by fluorescence and DIC microscopy. Green, Bni4-CFP; red, Kcc4-YFP. Arrows and arrowheads indicate structures described in the text.
Partial Separation of Bni4 and Kcc4 in Budded Cells with Aberrant Septin Organization
The results described above suggest that the underlying structure of the septin complex is responsible for the Bni4/Kcc4 asymmetry. However, these results do not eliminate the possibility that a diffusion barrier is necessary for the initiation of asymmetry. As an alternative approach to this question, we examined the localization of Bni4-CFP and Kcc4-YFP in mutants in which septin morphology was disrupted in ways that should eliminate the diffusion barrier. Several such mutants have been described previously (Gladfelter et al., 2001; Longtine and Bi, 2003; Gladfelter et al., 2004). The affected proteins are involved in the initial assembly of the septin ring, in the conversion of the ring to the collar, or both. We examined gin4Δ, cla4Δ, and elm1Δ mutants, as well as temperature-sensitive septin mutants, for localization of Bni4 and Kcc4.
As reported previously (Longtine et al., 1998, 2000), some gin4Δ cells displayed normal morphology and septin organization, but other cells had aberrant septin organization and morphology, particularly during growth at 37°C (Figure 6A). When the gin4Δ cells were grown at 24°C, Bni4-CFP and Kcc4-YFP were asymmetrically localized in all cells in which both proteins were detected (50% of 105 cells examined; Figure 6B, 1–4). After 5 h of growth at 37°C, Bni4-CFP was detected in ∼50% of the cells (n = 341), whereas Kcc4-YFP was detected in only ∼35% of the cells. The asymmetry of the proteins was largely retained but the locations of Kcc4-YFP and Bni4-CFP were highly variable. In 36% of all small-budded and medium-budded cells, Bni4-CFP and Kcc4-YFP partially overlapped (Figure 6B, 5–8). In some cells with elongated buds, Bni4-CFP was found at the neck, whereas Kcc4-YFP was located at the bud tip (Figure 6C, 1–4). In other cells, both proteins localized in the region close to the bud tip but were still largely separated (Figure 6C, 5–8). Importantly, Bni4-CFP and Kcc4-YFP remained largely separated even in cells with bars of fluorescence that presumably correspond to the septin bars often seen in this strain (Figure 6C, 9–12).
Figure 6.
Bni4 and Kcc4 localization in gin4Δ and elm1Δ mutants. DIC, fluorescence, and merged images are shown. (A) A gin4Δ strain expressing Cdc10-GFP (YLK158) was grown at 37°C. Cdc10-GFP was often found diffusely throughout most of the bud plasma membrane (2, arrow) or as bars in the neck (2 and 4, arrowheads). In some cells with elongated buds, septins were detected both at the neck and at the tip of the bud (4, arrows). (B and C) A gin4Δ strain expressing Bni4-CFP and Kcc4-YFP (YLK202) was grown either at 24°C (B, 1–4) or for 5 h at 37°C (B, 5–8, and C). (D) An elm1Δ strain expressing Bni4-CFP and Kcc4-YFP (JRL49) was grown at 37°C. Green, Bni4-CFP; red, Kcc4-YFP. Bars, 1 μm.
Results with a cla4Δ strain (JRL55) were similar to those with gin4Δ cells, but the percentages of small- and medium-budded cells that had a partial loss of the Bni4/Kcc4 asymmetry were somewhat lower (our unpublished data). In elm1Δ cells grown at 24°C, Bni4-CFP and Kcc4-YFP seemed to localize normally, but at 37°C some cells with elongated buds contained caps (presumably corresponding to caps of septins) in which Bni4-CFP and Kcc4-YFP partially colocalized (Figure 6D). In other cells, Bni4 and Kcc4 retained their normal asymmetric distribution (our unpublished data).
Finally, we localized Bni4-CFP and Kcc4-YFP in septin mutants. Bni4 was proposed to bind primarily to the septin Cdc10 (DeMarini et al., 1997), and indeed we found that Bni4-YFP localized poorly to the neck in either cdc10Δ cells at 24°C or cdc10-1 mutant cells at 37°C (our unpublished data). This precluded colocalization studies in cdc10 mutants. We also did not evaluate colocalization in a cdc11Δ septin mutant because Kcc4 is thought to bind primarily to Cdc11 (Okuzaki and Nojima, 2001). Although Cdc3 is essential for the organization of the septin cortex at the neck (Kim et al., 1991), the morphology of cdc3-1 cells grown at 24°C was indistinguishable from that of wild type, and the asymmetry between Bni4-CFP and Kcc4-YFP was preserved (our unpublished data). However, when these cells were shifted to 28–30°C for ≥2 h, Bni4-CFP and Kcc4-YFP occupied a significant portion of the bud in many small- and medium-budded cells. Although there was substantial overlap of the two signals, Kcc4-YFP was typically located more toward the bud tip (Figure 7A, 1–4). In most large-budded cells, the signals were lost, but in some cases, both proteins were detected close to the bud tip and were still partially separated (Figure 7A, 5–8). In some cases, the localization of Kcc4-YFP was clearly aberrant but Bni4-CFP remained on the mother side of the neck (Figure 7B, 1–3). In some cells, only the Kcc4-YFP signal was detected (Figure 7B, 4–6), whereas the reverse was never observed. Temperatures higher than 30°C or longer incubation times at 30°C resulted in almost complete loss of both fluorescent signals. Similar results were observed in a cdc12-6 mutant (our unpublished data).
Figure 7.
Bni4 and Kcc4 localization in cdc3 and shs1 mutants. DIC, fluorescence, and merged images are shown. (A and B) cdc3-1 cells expressing Bni4-CFP and Kcc4-YFP (YLK271) were imaged after a 2 h incubation at 30°C (A, 1–4), a 3-h incubation at 30°C (A, 5–8), or a 6-h incubation at 28°C (B, 1–6). (C and D) shs1Δ cells expressing Bni4-YFP and Kcc4-CFP (JRL18) were imaged after a 3-h incubation at 37°C (C) or after a 2.5-h incubation at 30°C (D). Green, Bni4-CFP; red, Kcc4-YFP. Bars, 1 μm. Arrows indicate structures described in the text.
Cells lacking the nonessential septin Shs1 (Mino et al., 1998) exhibited wild-type morphology and normal asymmetric localization of Bni4-CFP and Kcc4-YFP at 24°C (our unpublished data). After 2.5 h at 30°C, most shs1Δ cells retained the spatial separation between Bni4-CFP and Kcc4-YFP despite their aberrant morphology (Figure 7D). Under these conditions, 12% of all small and medium-budded cells were scored as having a partial loss of asymmetry, as judged by the partial overlap of the two signals in the merged images (our unpublished data). Although incubation of the shs1Δ strain at 37°C caused loss of the fluorescent signals in most cells, some cells with clearly aberrant morphology still showed asymmetry between Bni4 and Kcc4 (Figure 7C).
Together, these results indicate that the Bni4/Kcc4 asymmetry can be established even when the normal organization of the septin ring is significantly perturbed in ways that would be expected to eliminate the septin-based diffusion barrier.
DISCUSSION
Several models have been proposed to explain the asymmetric location of septin-binding proteins. One possibility is that the asymmetry is initiated at the time of bud emergence when the septin ring forms a collar. Nearly 30 yr ago, Byers and Goetsch (1976) noted a periodic ∼10-nm banding pattern in electron micrographs of bud necks. These striations, together with the evidence of septin self-assembly in vitro (Field et al., 1996; Frazier et al., 1998; Kinoshita et al., 2002; Mendoza et al., 2002), support the hypothesis that septins form filaments at the neck. In the electron microscopy (EM) studies, filament-like structures were never observed in unbudded cells (Byers and Goetsch, 1976). These data, together with fluorescence recovery after photobleaching studies on GFP-tagged septins (Caviston et al., 2003; Dobbelaere et al., 2003), have led to the suggestion that septin complexes undergo a major rearrangement during the transition from the septin ring in unbudded cells to the collar in budded cells (Longtine and Bi, 2003). Although this may well be the case, our studies have revealed that Bni4 and Kcc4 bind to distinct domains of the septin array before bud emergence. Thus, the ring-to-collar transition is not responsible for the initiation of Bni4/Kcc4 asymmetry.
How then could proteins be asymmetrically distributed on the septin scaffold? As outlined in the Introduction, we can envision two mechanisms, which are not mutually exclusive. One possibility is that the septin complexes themselves have an inherent asymmetry or polarity. It has not been resolved whether septin filaments run around the neck, perpendicular to the mother/bud axis, or run parallel to the mother/bud axis (Gladfelter et al., 2001). If filaments run parallel to the long axis and do so in a polar manner, then asymmetry could be achieved by the binding of one or more proteins exclusively to one end of the polar structure. This polarity would arise upon ring formation but would not be present in the septin caps formed after Lat-A treatment or in various mutants in which the septins are largely disorganized. This structural model would not require that most of the proteins that bind asymmetrically to the ring have an inherent affinity for only one end. A single asymmetry-initiating protein that bound to one side of the ring could potentially generate or establish the overall asymmetry.
The apparent lack of filaments in unbudded cells (Byers and Goetsch, 1976) suggests that a well-defined septin higher order structure may arise only at the time of the ring-to-collar transition. This poses a difficulty for our model. However, recent EM images of yeast plasma-membrane surfaces suggest that septin rings may be arranged into a higher order structure similar to that found in septin collars (Rodal et al., 2005). In this study, immunogold labeling of HA-Cdc3 revealed binding to two classes of plasma membrane-associated ultrastructures, fibrous net-like structures that the authors call “gauzes,” which probably correspond to septin collars, and less common rings that seem to be constituted of similar cross-linked linear fibers. Purified mammalian septins form bundles of filaments that can convert into coils and rings (Kinoshita et al., 2002). Those rings have comparable diameters to the rings observed by Rodal et al. (2005). However, whereas the rings reconstituted in vitro clearly show filaments that run circumferentially, the rings imaged on yeast membranes seem to have a more complex architecture. This suggests that septin rings before bud emergence are organized in a manner that could generate different environments at the outside and inside of the ring. There is no reason to believe that septins organized circumferentially would be more dynamic than septins organized into a net-like structure, so this model does not contradict the observation that septins in unbudded cells are more dynamic than those in budded cells.
The second possible model is that Bni4/Kcc4 asymmetry is initiated by a diffusion barrier constituted by the ring in unbudded cells. This would be consistent with the loss of asymmetry observed upon Lat-A treatment, in the triple-GAP-mutant strain, and in some elm1Δ cells. However, this model seems to be contradicted by the evidence that asymmetry is preserved in gin4Δ cells, in which the septins frequently form bars that would seem unable to sustain a septin diffusion barrier. On the other hand, the dramatically altered septin structures in gin4Δ mutants are observed primarily in large-budded cells rather than in small-budded or unbudded cells. Therefore, although a diffusion barrier does not seem to be required to maintain the asymmetry, we cannot rule out the possibility that it is necessary for the establishment of the asymmetry.
The precise role of the actin cytoskeleton in cell polarization is uncertain at present. Ayscough et al. (1997) found that Cdc42 could polarize in the absence of actin polymerization, but it was subsequently reported that the polarization of an overexpressed, constitutively active form of Cdc42 was actin dependent in the absence of the spatial cues that normally define the site of polarization (Wedlich-Soldner et al., 2003). More recently, Irazoqui et al. (2005) reported that Cdc42 is initially polarized in an actin-independent manner but maintenance of the polarization requires positive feedback between Cdc42 and actin cables to counteract the dispersing effects of endocytosis. Our results do not directly address the role of actin in selecting the primary site of polarization because our strains retain the normal polarization cues. However, the failure to efficiently form and/or maintain septin rings in Lat-A–treated cells suggests that an actin-dependent process is involved in the formation and/or stabilization of the fully developed septin ring. Given the role of Cdc42 in septin ring formation (Gladfelter et al., 2002; Caviston et al., 2003), it is possible that in Lat-A–treated cells the effective delivery of active Cdc42 to the plasma membrane is impaired. Alternatively, the absence of the actin cytoskeleton could prevent a protein that acts upstream or downstream of Cdc42 from localizing to the polarization site. Distinguishing between these possibilities will require further studies.
Our observation that septin ring formation in unbudded cells is largely disrupted by Lat-A differs from published results showing that septin rings form in the absence of actin polymerization (Ayscough et al., 1997; Harkins et al., 2001; Kadota et al., 2004). The reason for the discrepancy is unknown to us, but it may reflect the complexity of the mechanisms that govern septin ring formation and particularly the involvement of actin cables in this process. The discrepancy could be due to methodological differences in the way unbudded cells were obtained and treated. Given the complex dependence of Cdc42 polarization on actin polymerization, it is also likely that the nascent septin rings in unbudded cells are unstable and/or transient in Lat-A–treated cells. We may have largely missed the time when the rings were abundant and only observed cells after the collapse of nascent septin rings.
We conclude that the septin structure that is necessary for Bni4/Kcc4 asymmetry is established early in the cell cycle, concomitant with septin ring formation. The only mutations and conditions found to date that block Bni4/Kcc4 asymmetry are those that also interfere with the formation of a normal septin ring before budding and of a septin collar at the neck after budding. In contrast, other mutations that alter septin organization but do not prevent formation of some form of septin ring and collar at the neck do not disrupt the asymmetry, as demonstrated by the septin bars in the gin4 mutant that retain Bni4/Kcc4 asymmetry (Figure 6C). This suggests that the asymmetry is due to basic structural features of septin complexes and indeed suggests that the septin complexes may be polar. Identification of mutations that eliminate the asymmetry or the reconstitution of the Bni4/Kcc4 asymmetry in vitro will allow a more thorough characterization of the phenomenon.
Supplementary Material
Acknowledgments
We thank Danny Lew for reagents and helpful discussions, Matthias Versele for the anti-Cdc12 antibody, Lucy Robinson and Amy Gladfelter for reading the manuscript, and Yves Barral for sharing information with us before publication. We are indebted to John Pringle for helpful discussions and careful editing of the manuscript. We thank the Research Core Facility at Louisiana State University Health Sciences Center for the use of the confocal microscope and the National Institutes of Health for support (GM-47789).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–09–0764) on May 18, 2005.
Abbreviations used: Lat-A, latrunculin A.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adames, N. R., and Cooper, J. A. (2000). Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman, R., and Kellogg, D. (1997). Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol. 138, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough, K. R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D. G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral, Y., Mermall, V., Mooseker, M. S., and Snyder, M. (2000). Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell 5, 841–851. [DOI] [PubMed] [Google Scholar]
- Barral, Y., Parra, M., Bidlingmaier, S., and Snyder, M. (1999). Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, E., Maddox, P., Lew, D. J., Salmon, E. D., McMillan, J. N., Yeh, E., and Pringle, J. R. (1998). Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B., and Goetsch, L. (1976). A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 69, 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon, J. F., and Tatchell, K. (1987). Characterization of Saccharomyces cerevisiae genes encoding subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 7, 2653–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston, J. P., Longtine, M., Pringle, J. R., and Bi, E. (2003). The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid, V. J., Shulewitz, M. J., McDonald, K. L., and Thorner, J. (2001). Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol. Biol. Cell 12, 1645–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini, D. J., Adams, A.E.M., Fares, H., De Virgilio, C., Valle, G., Chuang, J. S., and Pringle, J. R. (1997). A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139, 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio, C., DeMarini, D. J., and Pringle, J. R. (1996). SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology 142, 2897–2905. [DOI] [PubMed] [Google Scholar]
- Dent, J., Kato, K., Peng, X. R., Martinez, C., Cattaneo, M., Poujol, C., Nurden, P., Nurden, A., Trimble, W. S., and Ware, J. (2002). A prototypic platelet septin and its participation in secretion. Proc. Natl. Acad. Sci. USA 99, 3064–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere, J., and Barral, Y. (2004). Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305, 393–396. [DOI] [PubMed] [Google Scholar]
- Dobbelaere, J., Gentry, M. S., Hallberg, R. L., and Barral, Y. (2003). Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4, 345–357. [DOI] [PubMed] [Google Scholar]
- Fares, H., Goetsch, L., and Pringle, J. R. (1996). Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 132, 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, C. M., Al-Awar, O., Rosenblatt, J., Wong, M. L., Alberts, B., and Mitchison, T. J. (1996). A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 133, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, C. M., and Kellogg, D. (1999). Septins: cytoskeletal polymers or signalling GTPases? Trends Cell. Biol. 9, 387–394. [DOI] [PubMed] [Google Scholar]
- Frazier, J. A., Wong, M. L., Longtine, M. S., Pringle, J. R., Mann, M., Mitchison, T. J., and Field, C. (1998). Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick, D. L., and Tatchell, K. (1996). The REG2 gene of Saccharomyces cerevisiae encodes a type1 protein phosphatase-binding protein that functions with Reg1p and the Snf1p protein kinase to regulate growth. Mol. Cell. Biol. 16, 2922–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter, A. S., Bose, I., Zyla, T. R., Bardes, E. S., and Lew, D. J. (2002). Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 156, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter, A. S., Pringle, J. R., and Lew, D. J. (2001). The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4, 681–689. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A. S., Zyla, T. R., and Lew, D. J. (2004). Genetic interactions among regulators of septin organization. Eukaryot. Cell 3, 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and McCusker, J. H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- Guldener, U., Heck, S., Fielder, T., Beinhauer, J., and Hegemann, J. H. (1996). A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, P. A., and Russell, S.E.H. (2004). The pathobiology of the septin gene family. J. Pathol. 204, 489–505. [DOI] [PubMed] [Google Scholar]
- Hanrahan, J., and Snyder, M. (2003). Cytoskeletal activation of a checkpoint kinase. Mol. Cell 12, 663–673. [DOI] [PubMed] [Google Scholar]
- Harkins, H. A., Pagé, N., Schenkman, L. R., De Virgilio, C., Shaw, S., Bussey, H., and Pringle, J. R. (2001). Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol. Biol. Cell 12, 2497–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H. (1971). Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69, 265–276. [DOI] [PubMed] [Google Scholar]
- Hsu, S.-C., Hazuka, C. D., Roth, R., Foletti, D. L., Heuser, J., and Scheller, R. H. (1998). Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron 20, 1111–1122. [DOI] [PubMed] [Google Scholar]
- Irazoqui, J. E., Howell, A. S., Theesfeld, C. L., and Lew, D. J. (2005). Opposing roles for actin in Cdc42p polarization. Mol. Biol. Cell 16, 1296–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D. I., Jacobs, C. W., Pringle, J. R., Robinson, L. C., Carle, G. F., and Olson, M. V. (1987). Mapping of the Saccharomyces cerevisiae CDC3, CDC25, and CDC42 genes to chromosome XII by chromosome blotting and tetrad analysis. Yeast 3, 243–253. [DOI] [PubMed] [Google Scholar]
- Kadota, J., Yamamoto, T., Yoshiuchi, S., Bi, E., and Tanaka, K. (2004). Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 5329–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. B., Haarer, B. K., and Pringle, J. R. (1991). Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J. Cell Biol. 112, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, M., Field, C. M., Coughlin, M. L., Straight, A. F., and Mitchison, T. J. (2002). Self- and actin-templated assembly of mammalian septins. Dev. Cell 3, 791–802. [DOI] [PubMed] [Google Scholar]
- Kozubowski, L., Panek, H., Rosenthal, A., Bloecher, A., DeMarini, D. J., and Tatchell, K. (2003). A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell 14, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, J., Meyer, A., Snyder, M. P., and Barral, Y. (2002). Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 16, 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., and Li, R. (1998). Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143, 1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., and Bi, E. (2003). Regulation of septin organization and function in yeast. Trends Cell Biol. 13, 403–409. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., DeMarini, D. J., Valencik, M. L., Al-Awar, O. S., Fares, H., De Virgilio, C., and Pringle, J. R. (1996). The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8, 106–119. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., Fares, H., and Pringle, J. R. (1998). Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143, 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., Theesfeld, C. L., McMillan, J. N., Weaver, E., Pringle, J. R., and Lew, D. J. (2000). Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, J. N., Longtine, M. S., Sia, R.A.L., Theesfeld, C. L., Bardes, E.S.G., Pringle, J. R., and Lew, D. J. (1999). The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 19, 6929–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza, M., Hyman, A. A., and Glotzer, M. (2002). GTP binding induces filament assembly of a recombinant septin. Curr. Biol. 12, 1858–1863. [DOI] [PubMed] [Google Scholar]
- Mino, A., Tanaka, K., Kamei, T., Umikawa, M., Fujiwara, T., and Takai, Y. (1998). Shs1p: a novel member of septin that interacts with Spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 251, 732–736. [DOI] [PubMed] [Google Scholar]
- Nguyen, T. Q., Sawa, H., Okano, H., and White, J. G. (2000). The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J. Cell Sci. 113, 3825–3837. [DOI] [PubMed] [Google Scholar]
- Okuzaki, D., and Nojima, H. (2001). Kcc4 associates with septin proteins of Saccharomyces cerevisiae. FEBS Lett. 489, 197–201. [DOI] [PubMed] [Google Scholar]
- Richman, T. J., Sawyer, M. M., and Johnson, D. I. (1999). The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J. Biol. Chem. 274, 16861–16870. [DOI] [PubMed] [Google Scholar]
- Rodal, A. A., Kozubowski, L., Goode, B. L., Drubin, D. G., and Hartwig, J. H. (2005). Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell 16, 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G. R., and Hicks, J. B. (1986). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sheu, Y.-J., Barral, Y., and Snyder, M. (2000). Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 5235–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulewitz, M. J., Inouye, C. J., and Thorner, J. (1999). Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 7123–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, P. A., DeRisi, J. L., Wilhelm, J. E., and Vale, R. D. (2000). Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science 290, 341–344. [DOI] [PubMed] [Google Scholar]
- Theesfeld, C. L., Zyla, T. R., Bardes, E. G., and Lew, D. J. (2003). A monitor for bud emergence in the yeast morphogenesis checkpoint. Mol. Biol. Cell 14, 3280–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A. H., et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- Trimble, W. S. (1999). Septins: a highly conserved family of membrane-associated GTPases with functions in cell division and beyond. J. Membr. Biol. 169, 75–81. [DOI] [PubMed] [Google Scholar]
- Vallen, E. A., Caviston, J., and Bi, E. (2000). Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11, 593–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturi, G. M., Bloecher, A., Williams-Hart, T., and Tatchell, K. (2000). Genetic interactions between GLC7, PPZ1 and PPZ2 in Saccharomyces cerevisiae. Genetics 155, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele, M., Gullbrand, B., Shulewitz, M. J., Cid, V. J., Bahmanyar, S., Chen, R. E., Barth, P., Alber, T., and Thorner, J. (2004). Protein-protein interactions governing septin heteropentamer assembly and septin filament organization in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 4568–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele, M., and Thorner, J. (2004). Septin collar formation in budding yeast requires GTP binding and direct phosphorylation by the PAK, Cla4. J. Cell Biol. 164, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Alberti-Segui, C., Rebischung, C., and Philippsen, P. (1997). Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner, R., Altschuler, S., Wu, L., and Li, R. (2003). Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299, 1231–1235. [DOI] [PubMed] [Google Scholar]
- Wertman, K. F., Drubin, D. G., and Botstein, D. (1992). Systematic mutational analysis of the yeast ACT1 gene. Genetics 132, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall, P. J., and Momany, M. (2002). Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Yang, S., Ayscough, K. R., and Drubin, D. G. (1997). A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J. Cell Biol. 136, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.