Abstract
RasGAP bears two caspase-3 cleavage sites that are used sequentially as caspase activity increases in cells. When caspase-3 is mildly activated, RasGAP is first cleaved at position 455. This leads to the production of an N-terminal fragment, called fragment N, that activates the Ras-PI3K-Akt pathway and that promotes cell survival. At higher caspase activity, RasGAP is further cleaved at position 157 generating two small N-terminal fragments named N1 and N2. We have now determined the contribution of this second cleavage event in the regulation of apoptosis using cells in which the wild-type RasGAP gene has been replaced by a cDNA encoding a RasGAP mutant that cannot be cleaved at position 157. Our results show that cleavage of fragment N at position 157 leads to a marked reduction in Akt activity. This is accompanied by efficient processing of caspase-3 that favors cell death in response to various apoptotic stimuli. In nontumorigenic cells, fragments N1 and N2 do not modulate apoptosis. Therefore, the role of the second caspase-mediated cleavage of RasGAP is to allow the inactivation of the antiapoptotic function of fragment N so that caspases are no longer hampered in their ability to kill cells.
INTRODUCTION
Cleavage of the substrates of caspase-3 and other executioner caspases is a critical step that leads to apoptosis. Indeed, blocking these proteases with specific caspase inhibitors prevents many cell death responses (Toyoshima et al., 1997; Nicholson, 1999). Consequently, understanding the apoptotic process requires that the caspase substrates be identified, followed by the characterization of the functional roles of each cleavage event in the regulation of cell death.
The great majority of caspase substrates promotes apoptosis when cleaved by caspases (Stroh and Schulze-Osthoff, 1998; Fischer et al., 2003). There are, however, exceptions to this rule. For example, cleavage of PKCε activates the enzyme and this seems to generate an antiapoptotic signal (Basu et al., 2002). Triggering of the B-cell receptor induces the cleavage by caspases of Lyn and Fyn (Ricci et al., 2001; Luciano et al., 2003) into fragments that inhibit apoptosis. The p54 Lyn fragment may favor cell survival by modulating c-myc levels (Luciano et al., 2003). The survival pathways activated after the cleavage of Fyn or PKCε have not been characterized, however. Moreover, the ability to regulate apoptosis in cells specifically lacking the capacity of cleaving Fyn, Lyn, or PKCε has not been evaluated yet. Consequently, the importance of the cleavage of these proteins for cell survival is still incompletely understood.
RasGAP, a regulator of Ras- and Rho-dependent pathways (Campbell et al., 1998; Leblanc et al., 1998; Wen et al., 1998), bears two conserved caspase-3 cleavage sites at position 455 and 157 (Yang and Widmann, 2001; Yang et al., 2004). The first cleavage of RasGAP occurs at very low caspase-3 activity generating an N-terminal fragment, called fragment N, that is crucially required for cell survival in mild stress conditions (Yang et al., 2004). In the present study, we have determined the overall importance of the second RasGAP cleavage event in the regulation of cell death using cells in which wild-type RasGAP was replaced by a RasGAP mutant that cannot be cleaved at position 157.
MATERIALS AND METHODS
Plasmids
HA-GAP.dn3 encodes the full-length human RasGAP protein bearing an HA tag (MGYPYDVPDYAS) at the amino terminal end (Yang and Widmann, 2001). The extension .dn3 indicates that the backbone plasmid is pcDNA3 (Invitrogen, Carlsbad, CA). The plasmid HA-D157A.dn3 encodes a RasGAP mutant that cannot be cleaved at position 157 (Yang and Widmann, 2001). The plasmid encoding fragment N1 used for the production of the fragment N1-encoding lentivirus (HA-N1.lti) has been generated by subcloning the BamHI-XhoI fragment of HA-N1.dn3 (Yang and Widmann, 2001) in TRIP-PGK-ATGm-MCS-WHV opened with the same enzymes. The plasmid encoding fragment N2 used for the production of the fragment N2-encoding lentivirus (HA-N2.lti) has been generated by subcloning the KpnI-XhoI fragment of HA-N2.dn3 (Yang and Widmann, 2001) in TRIP-PGK-ATGm-MCS-WHV opened with BamHI and SalI. The plasmid used for the production of the lentivirus encoding the caspase-resistant form of fragment N (N-D157A.lti) has been described earlier (Yang et al., 2004). Plasmid HA-RasGAP (no 3′ UTR).lti, used for the production of lentiviruses encoding full-length RasGAP, was constructed as follows. A RasGAP cDNA fragment lacking the 3′ untranslated sequence was PCR amplified from plasmid HA-GAP.dn3 (Yang and Widmann, 2001) with primers CTCATGCAAGGGAAGGGCAA and CGGCGGCCGCCTACCTGACATCATTGGTTTTTGT. The amplified fragment was cut with NotI and EcoRV and subcloned in HA-GAP.dn3 opened with the same enzyme, generating plasmid HA-RasGAP (no 3′ UTR).dn3. The BamHI/XhoI insert of the latter was then subcloned in TRIP-PGK-ATGm-MCS-WHV opened with the same enzyme.
Cell Lines
The RasGAP+/+ mouse embryonic fibroblasts (MEFs; clone 12.78), the RasGAP–/– MEFs (clone 12.64), and their derivatives stably expressing wild-type RasGAP or the D455A mutant (Yang et al., 2004) were maintained in DMEM containing 10% newborn calf serum (Invitrogen, cat. no. 26010-074) at 37°C and 5% CO2. MEFs clones expressing the D157A mutant were obtained by electroporating RasGAP–/– MEFs using plasmid HA-D157A.dn3 as described (Yang et al., 2004). About 40 clones were isolated from two independent electroporations and analyzed for RasGAP expression levels. Only the clones with RasGAP expression levels similar to control RasGAP+/+ MEFs were kept for further analysis. Unless indicated otherwise, all the experiments described in this article have been repeated at least three times with similar results and have involved all the clones described in Figure 1A. U2OS cells were maintained in DMEM (Sigma Chemical Co., St. Louis, MO; cat. no. 5796) containing 15% fetal calf serum (Sigma; cat. no. F7524) at 37°C and 5% CO2. HeLa cells were maintained in RPMI 1640 containing 10% newborn calf serum (Invitrogen) at 37°C and 5% CO2.
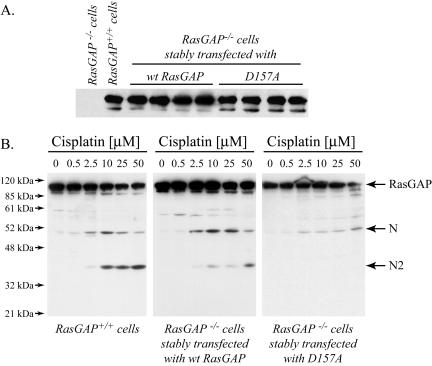
Figure 1.
Generation of cells lacking the capacity to cleave RasGAP at position 157. (A) MEFs derived from RasGAP–/– and RasGAP+/+ mice, as well as RasGAP–/– MEFs stably transfected with plasmids bearing the wild-type RasGAP cDNA or a cDNA encoding the D157A mutant of RasGAP were analyzed for their RasGAP expression levels by Western blot using an antibody directed at the SH domains of RasGAP. (B) RasGAP+/+ MEFs or representative RasGAP–/– MEF clones stably expressing a wild-type RasGAP-encoding plasmid or a plasmid encoding a RasGAP mutant that cannot be cleaved at position 157 (D157A) were subjected to the indicated concentrations of cisplatin for 24 h. The presence of RasGAP and RasGAP-derived fragments was detected by Western blotting as described above.
Chemicals and Antibodies
The anti-phospho p44/p42 ERK MAP kinase (T202/Y204) E10 monoclonal antibody (mAb) was from Cell Signaling Technology (Beverly, MA; cat. no. 9106). The antibody directed at all forms of ERK MAPKs was from Upstate Biotechnology (Lake Placid, NY; cat. no. 06-182). The anti-RasGAP antibody directed at the SH domains of RasGAP has been described before (Valius et al., 1995). The antibody recognizing the N2 fragment of RasGAP was from Alexis Biochemicals (San Diego, CA; cat. no. ALX-210-860-R100). The antibody specific for the active form of caspase-3 was from Cell Signaling Technology (cat. no. 96613). The mAb specific for the hemagglutinin (HA) tag was purchased as ascites from BabCo (Richmond, CA; cat. no. MMS-101R). This antibody was adsorbed on HeLa cell lysates to decrease nonspecific binding (Yang and Widmann, 2001). The antibodies recognizing Akt and the phosphorylated form of Akt on threonine 473 were from Santa Cruz Biotechnology (Santa Cruz, CA; cat. no. sc-8312) and from Cell Signaling Technology (cat. no. 9271), respectively. Fas ligand corresponded to a hexameric form of a fusion protein between FasL and the Fc portion of IgG1 (Holler et al., 2003) and was a generous gift from Dr. Pascal Schneider (University of Lausanne, Switzerland). Staurosporine and cisplatin were from Roche Diagnostics (Basel, Switzerland) and Sigma (cat. nos. 1055682 and P4394, respectively).
Western Blot Analysis
Cells were lysed in monoQ-c buffer (Yang and Widmann, 2001). When quantitations were not performed, Western blotting was performed as described previously (Widmann et al., 1995) using a homemade ECL reagent (Yang and Widmann, 2001). When quantitations were performed, the primary antibodies were revealed with a 1/5000 dilution of an Alexa Fluor 680–conjugated anti-rabbit antibody (Molecular Probes, Eugene, OR; cat. no. A21109) or an IRDye 800–conjugated anti-mouse antibody (Rockland, Gilbertsville, PA; cat. no. 610-132-121) and subsequently visualized with the Odyssey infrared imaging system (LICOR Biosciences, Bad Homburg, Germany). Quantitation was performed using the Odyssey infrared imaging software.
Image Acquisition of Wounds in Cell Layers
Cells were grown to confluency, wounds were generated, and cells were fixed as described (Kulkarni et al., 2000; Schlesinger et al., 2002). After fixation, the cells were mounted in Vectashield (Vector Laboratories, Burlingame, CA; cat. no. H-1000). Pictures of the wounds were taken at room temperature with a Zeiss Axioplan 2 imaging microscope equipped with a Plan-Neofluar 10×/0.30 ∞/– lens and a Zeiss AxioCam HRC camera using the Zeiss AxioVision acquisition software (Thornwood, NY).
Lentivirus
Recombinant lentivirus were produced as described (Dull et al., 1998). Briefly, 293T cells were cotransfected using the calcium phosphate DNA precipitation method (Jordan et al., 1996) with 10 μg of the lentiviral vector (TRIP-PGK-ATGm-MCS-WHV) containing the cDNA of interest (e.g., HA-N1.lti or HA-N2.lti), 2.5 μg of the envelope protein–coding plasmid (pMD.G), and 7.5 μg of the packaging construct (pCMVDR8.91). Two days after the transfection, the virus-containing medium was harvested. To determine how much of the virus preparations was needed to infect 100% of the cells, cells seeded at a 50% confluency on coverslips placed in six-well plates were cultured overnight with various volumes of the lentiviral preparations. After removal of the virus solution, the cells were maintained for 2 more days before fixation and immunocytochemical staining with antibodies directed at the HA tag born by the various constructs. The lowest volumes of the lentiviral preparations required to infect 100% of the cells were chosen for further experiments.
Infection of the cells was performed as follows. Hexadimethrine bromide (Sigma, Polybrene, cat. no. 52495) was added to cells cultured in six-well plates to a final concentration of 5 μg/ml, followed by the addition of the lentiviruses. The plates were then centrifuged 45 min at 800 × g and placed 24 h at 37°C in a 5% CO2 humidified atmosphere. The medium was then replaced with fresh medium, and the cells were further cultured for an additional 48-h period before being used in specific experiments.
Color Cell Labeling
Various MEF clones (25·104 cells in 4 ml serum-free culture medium) were stained with either 20 μM of Cell Tracker Blue CMAC (Molecular Probes, cat. no. C2110), 0.15 μM Cell Tracker Green CMFDA (Molecular Probes, cat. no. C2925) or 0.5 μM Cell Tracker Orange CMRA (Molecular Probes, cat. no. C34551) for 1 h at 37°C. The cells were then washed three times with phosphate-buffered saline, mixed in equal proportions (50 × 103 of each type of cells) in six-well plates in 2 ml of culture medium, and incubated at 37°C for 24–48 h. Living blue, green, and orange cells were then counted using an Axiovert 25 Zeiss microscope equipped with epi-fluorescence illumination, and the relative proportion of the different cell types was determined.
Apoptosis Measurements
Apoptosis was determined by scoring the number of cells displaying pycnotic nuclei. Nuclei of live cells were labeled with Hoechst 33342 (10 μg/ml final concentration) for ∼5 min, and the cells were then analyzed (at least 400 cells per condition) using an Axiovert 25 Zeiss microscope equipped with fluorescence and transmitted light optics.
Caspase Activity Measurement
Cells were lysed in monoQ-c buffer (Yang and Widmann, 2001) complemented with freshly added phenylmethylsulfonyl fluoride (1 mM final concentration). Caspase 3 activity was determined using 50 μg of the cell lysates dissolved in 3 ml of caspase-3 activity assay buffer (100 mM HEPES, 1% sucrose, 0.1% CHAPS, 2 mM dithiothreitol) in the presence of 5 μM of a fluorigenic caspase-3 substrate (Enzyme System Product, Livermore, CA; cat. no. AFC 138). After a 1-h incubation period at 37°C, the extent of cleavage of the caspase-3 substrate was measured using a Photon Technology International fluorimeter (Lawrenceville, NJ; excitation 400 nm, emission 505 nm).
RESULTS
Generation of MEF Clones Expressing the D157A Mutant of RasGAP
To specifically investigate the functional roles of the second caspase-mediated RasGAP cleavage event, we generated mouse embryonic fibroblasts (MEFs) derived from RasGAP knock-out mice in which the RasGAP mutant at position 157 (mutant D157A) was reexpressed at endogenous levels (compare the second lane with the subsequent lanes in Figure 1A). When MEFs expressing wild-type RasGAP (RasGAP+/+ cells and RasGAP–/– cells stably transfected with wild-type RasGAP) were incubated with low cisplatin concentrations, RasGAP was cleaved at position 455, as indicated by the appearance of fragment N (Figure 1B, left and middle panels). At higher cisplatin concentrations, fragment N was further processed at position 157 as indicated by the formation of fragment N2 (Figure 1B, left and middle panels). In contrast, and as expected, cells expressing the D157A mutant, while being able to cleave RasGAP at position 455, were not able to further process fragment N to the smaller N-terminal fragments, even at high cisplatin concentrations (Figure 1B, right panel).
The D157A Mutation Does Not Affect the Function of Full-length RasGAP
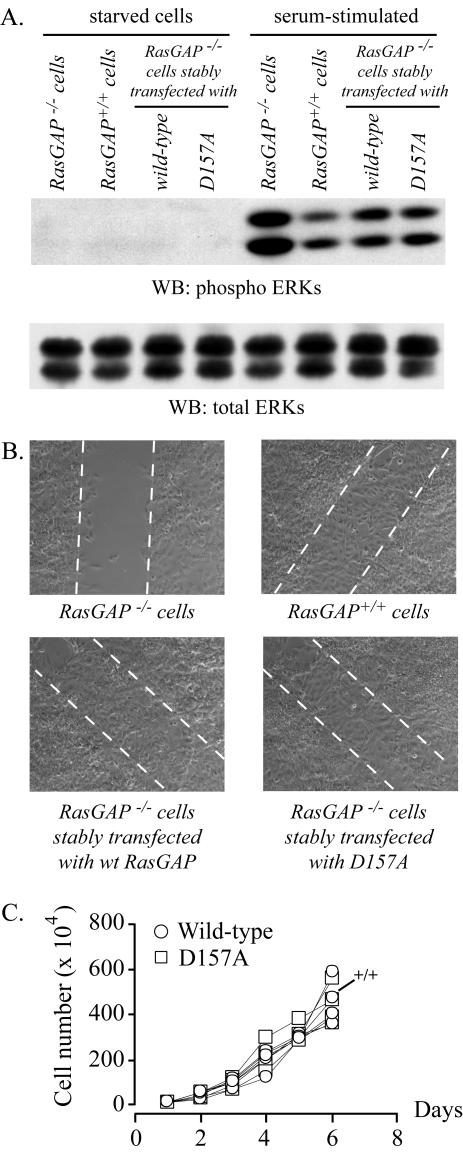
As shown above, the D157A mutation in RasGAP prevented its full processing by caspases in cells subjected to apoptotic stimuli. Conceptually, this mutation could also affect the functions of the full-length protein. To determine whether this was the case, we assessed the capacity of mutant D157A to fulfill two functions of full-length RasGAP: 1) the negative regulation of the ERK MAPK pathway mediated by the C-terminal GAP domain of the protein (Van der Geer et al., 1997) and 2) the proper cell polarization and migration mediated by the binding of the N-terminal domain of RasGAP to RhoGAP (Kulkarni et al., 2000). To do so, we measured the ability of clones expressing the wild-type and the D157A cleavage-resistant forms of RasGAP to control ERK activation in response to serum and their ability to migrate into wounds. As shown in Figure 2, A and B, there was no difference in the capacity of wild-type RasGAP and the D157A mutant to rescue the defect in ERK regulation observed in RasGAP–/– cells (i.e., increased ERK activity in response to serum) and to restore their wound-healing capacities. Additionally, the expansion rates between the various cell types were similar (Figure 2C). These results indicate that mutation D157A, while abrogating the second caspase-mediated cleavage of RasGAP, does not affect the functions of the unprocessed protein.
Figure 2.
The D157A mutation does not affect the functions of full-length RasGAP. MEFs derived from RasGAP–/– and RasGAP+/+ mice (+/+ in C), as well as RasGAP–/– MEFs stably transfected with plasmids bearing the wild-type RasGAP cDNA or a cDNA encoding the D157A mutant of RasGAP were starved for 24 h in DMEM lacking serum and stimulated for 15 min with 15% NBCS. The cells were then lysed and the extent of ERK activation and total ERK expression were evaluated by Western blotting (A). Alternatively, the cells were grown to confluency and starved for 24 h in medium lacking serum, and a wound was made with a tip as described (Kulkarni et al., 2000). Pictures of the wounds were taken after 48 h. Dotted lines indicate the edges of the wound (B). Finally, the cells were seeded in six-well plates (10,000 cells per well), and the number of cells in the wells were counted after the indicated periods of time in culture (C).
The Two Caspase Cleavage Sites of RasGAP Have Opposite Roles in the Regulation of Apoptosis
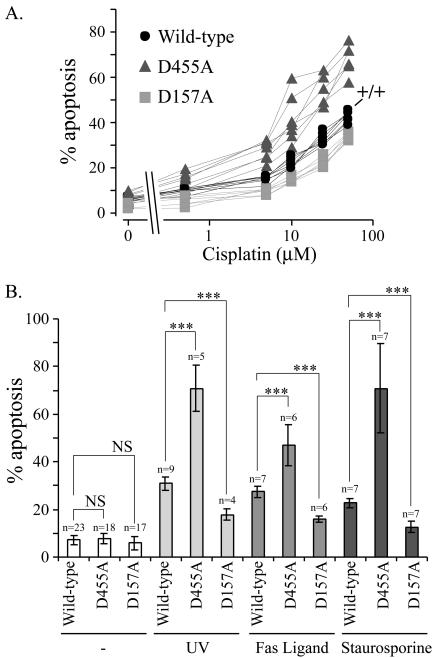
We then assessed the effect on apoptosis when RasGAP cleavage was abrogated at position 157 and compared the results to the responses obtained in cells expressing the wild-type RasGAP protein or the mutant that is totally resistant to caspases cleavage (mutant D455A). Clones expressing mutant D455A were significantly more sensitive toward cisplatin-induced apoptosis than clones expressing the wild-type protein. In contrast, clones expressing the D157A mutant were slightly more resistant to cisplatin-induced apoptosis (Figure 3A). The increased susceptibility of the D455A mutant–expressing cells to undergo apoptosis and the slightly augmented resistance of the D157A-expressing cells were also observed when other apoptosis inducers were used, including FasL that stimulates the extrinsic apoptotic pathway or UV and staurosporin that activate the intrinsic pathway (Figure 3B).
Figure 3.
The sequential cleavage of RasGAP by caspases differentially regulates apoptosis. (A) MEFs derived from RasGAP+/+ mice (indicated by +/+), and RasGAP–/– clones stably expressing plasmids encoding either wild-type RasGAP (four independent clones), the uncleavable D455A mutant or the D157A mutant (five independent clones each) were incubated with increasing concentrations of cisplatin (0, 0.5, 5, 10, 25, and 50 μM) for 24 h. The extent of apoptosis was then scored. (B) Wild-type RasGAP-, D455A-, or D157A-expressing cells were either left untreated, subjected to UV treatment (60 J/m2), or stimulated with 5 ng/ml hexameric Fas Ligand or with 1 nM staurosporine. The extent of apoptosis was scored 24 h later and expressed as the mean ± SD of the indicated number of determinations. At least four independent clones for the wild-type or mutant RasGAP-expressing cells were used. The significance of the differences was assessed by Student's t tests (NS, not significant; *** p < 0.001).
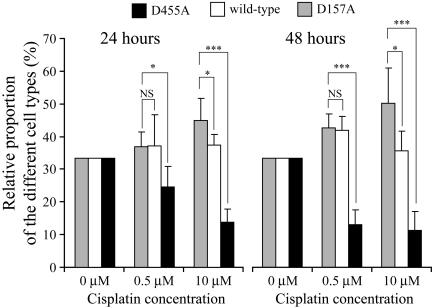
To assess whether the reduced cell death response in cells expressing the D157A mutant of RasGAP would result in a selective advantage in terms of population growth and survival, cells expressing the wild-type RasGAP protein or the two different cleavage-resistant forms were mixed in equal proportion after being labeled with different color dyes. They were then incubated with low (0.5 μM) and high (10 μM) cisplatin concentrations for 24 and 48 h, after which the number of surviving cells was scored. In the presence of either low or high cisplatin concentrations, the relative proportion of cells expressing the D455A mutant decreased in a time-dependent manner (Figure 4), as expected from their inability to produce the antiapoptotic fragment N (Yang et al., 2004). MEFs incubated with 0.5 μM cisplatin generate fragment N but do not appear to process it further into smaller fragments (Figure 1B and Yang et al., 2004). In these conditions therefore, fragment N levels should not be appreciably different between control cells and cells unable to process RasGAP at position 157, and consequently a similar survival response should be induced by low stresses in these cells. Indeed, incubation of both cell types with 0.5 μM cisplatin for 24 or 48 h did not result in a significant difference in their relative proportions (Figure 4). In contrast, in the presence of 10 μM cisplatin that induces processing of wild-type fragment N (Figure 1 and Yang and Widmann, 2001; Yang et al., 2004), the proportion of cells expressing the D157A mutant of RasGAP increased, in a time-dependent manner, relatively to the proportion of cells expressing the wild-type protein (Figure 4). These results support the notion that the persistent presence of fragment N in cells favors their survival in response to apoptotic stimuli and that this provides a selective survival advantage over cells that process RasGAP normally.
Figure 4.
Cells expressing the D157A mutant of RasGAP display selective survival capacity. Wild-type RasGAP-, D455A-, or D157A-expressing MEFs were stained with blue, green, and orange dyes, respectively, as indicated in Materials and Methods. The cells were then mixed in equal proportion and incubated with the indicated cisplatin concentrations. The proportion of the different colored cells remaining alive after 24 or 48 h was then assessed. The figure represents pooled results from six independent experiments. Data are normalized so that there are equal proportions of the three cell types in the absence of cisplatin. The significance of the differences was assessed by Student's t tests (NS, not significant; * p < 0.05; *** p < 0.001; n = 6). Similar results were obtained when another combination of color labeling of the three cell types was performed.
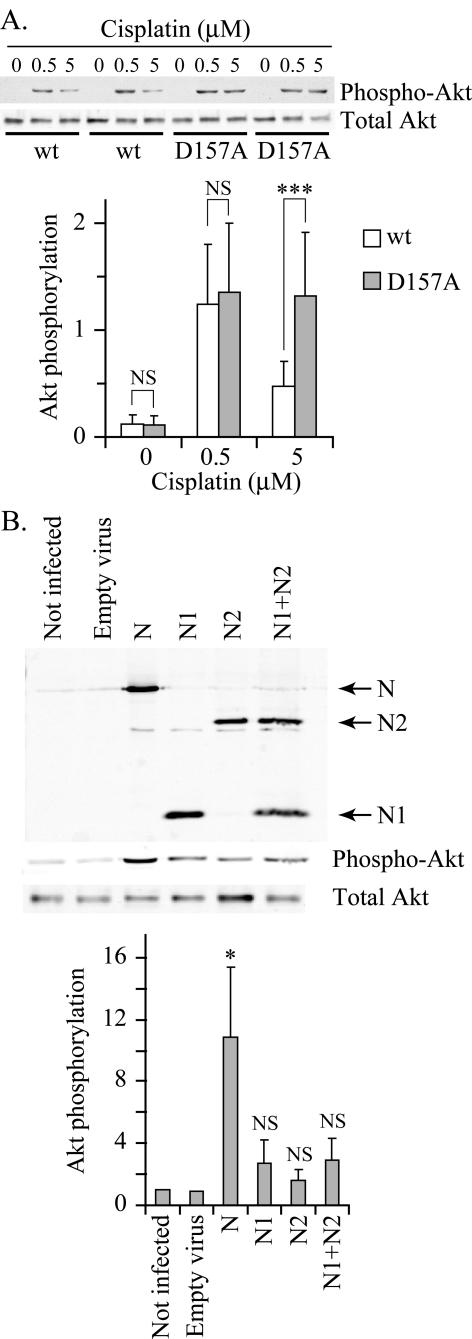
We next determined whether the second cleavage of RasGAP at position 157 leads to a decrease in Akt activity. Two experiments were performed to address this possibility. First, cells expressing wild-type RasGAP or the D157A mutant were incubated with a low cisplatin concentration (0.5 μM) that does not promote the cleavage of fragment N (Figure 1B) and with a high cisplatin concentration (5 μM) that does induce the cleavage of fragment N into fragments N1 and N2 (Figure 1B). As shown in Figure 5A, the low cisplatin dose induced similar Akt activity in both cell types. In contrast, wild-type RasGAP-expressing cells were no longer able to markedly activate Akt at the high cisplatin dose, while the cells expressing the D157A mutant still were. In the second experiment, MEFs were infected with lentiviruses encoding either fragment N or its proteolytic caspase-generated fragments N1 and N2. Figure 5B shows that fragment N induced a significant increase in Akt activity in MEFs. In contrast, the smaller N-terminal fragments, alone or in combination, only marginally activated Akt but this did not reach statistical significance (Figure 5B). Altogether these results indicate that the antiapoptotic fragment N generated by low stresses looses, when further cleaved, its ability to strongly activate Akt and consequently can no longer efficiently protect cells.
Figure 5.
Impaired down-modulation of Akt activity in cells expressing the D157A mutant of RasGAP. (A) Wild-type RasGAP- or D157A-expressing MEFs were starved in serum-free medium for 24 h. The cells were next incubated with the indicated concentrations of cisplatin for an additional 24-h period. The cells were then lysed and the extent of Akt phosphorylation and the total levels of Akt were assessed by Western blot analysis. The top part of A shows a representative blot obtained with two independent clones of each cell types. The bottom panel shows the quantitation of the phospho-Akt bands from five independent experiments performed with control MEFs, three wild-type RasGAP expressing clones and three D157A mutant expressing clones. Results were normalized to the value obtained in control MEFs stimulated with 0.5 μM cisplatin. The significance of the differences was assessed by Student's t tests (NS, not significant; *** p < 0.001). (B) MEFs were infected with lentiviruses encoding the indicated HA-tagged fragments of RasGAP as indicated in Materials and Methods. After lysis, expression levels of the different constructs was assessed by Western blot using an anti-HA antibody. Detection of total and phospho-Akt was assessed by Western blot analysis (top panel). The quantitation of the phospho-Akt signal is shown in the bottom panel. Results were normalized to the value obtained in uninfected MEFs. The significance of the differences with the value obtained with empty virus-infected cells was assessed by Student's t tests (NS, not significant; * p < 0.05; n = 5).
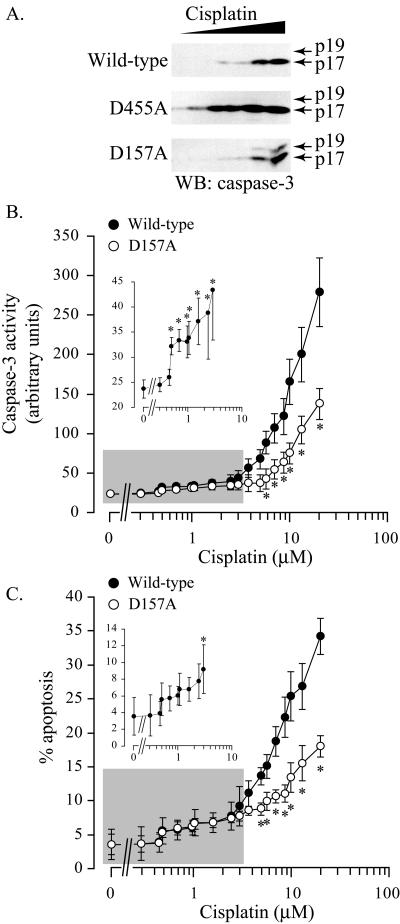
Cleavage of RasGAP at Position 157 Allows Efficient Processing and Activation of Caspase-3
Because caspase-3 is thought to be one of the main executioner caspases in cells (Slee et al., 2001), we next measured caspase-3 activation in the various clones. Activation of caspase-3 is a two-step mechanism that involves the cleavage of the zymogen p32 form, first into a partially processed p19 large subunit still bearing the prodomain, and finally into a fully processed p17 large subunit (Han et al., 1997; Zou et al., 2003). As expected from their inability to produce the antiapoptotic fragment N (Yang et al., 2004), cells expressing the uncleavable D455A RasGAP mutant processed caspase-3 into the p17 subunit at very low cisplatin concentrations (Figure 6A). Caspase-3 cleavage occurred with a similar dose response in cells expressing the D157A mutant and those expressing wild-type RasGAP. However, the partially processed p19 large subunit was detected in the former cells and not in the latter (Figure 6A). This indicates that caspase-3 is less efficiently activated in cells that cannot further process fragment N into smaller fragments. To extend this observation, the enzymatic activity of caspase-3 and the corresponding apoptotic response induced by increasing concentrations of cisplatin was measured in the two types of cells (Figure 6, B and C). Two phases were observed. Low cisplatin concentrations (0.5–2.5 μM; highlighted in gray in the figures and enlarged in the insets), induced a low, but significant, caspase activity in cells (Figure 6B, inset). This low caspase activity promotes the cleavage of RasGAP into fragment N allowing survival of stressed cells (Yang et al., 2004). Consequently, and as expected, low cisplatin concentrations did not induce a significant increase over the basal apoptotic response (Figure 6C, inset). In the presence of low cisplatin stimulation, no difference in terms of caspase activity or induction of apoptosis could be detected between wild-type RasGAP- and D157A-expressing cells (Figure 6, B and C). In contrast, at cisplatin concentrations inducing the processing of fragment N into smaller fragments (above 5 μM; Figure 1B and Yang et al., 2004), the cells expressing the D157A mutant of RasGAP induced less caspase activity (Figure 6B) and were more resistant to apoptosis (Figure 6C). The differential sensitivity toward apoptosis observed between the cells expressing the wild-type or the D157A mutant form of RasGAP correlates therefore with the extent of caspase-3 processing and activity.
Figure 6.
Caspase-3 is inefficiently processed and activated in cells expressing the D157A mutant of RasGAP. (A) Clones expressing wild-type RasGAP or the indicated cleavage-resistant mutants were stimulated with cisplatin as indicated in Figure 3A. The cells were then lysed and the presence of active caspase-3 fragments was visualized by Western blot. (B and C) Wild-type RasGAP- and D157A-expressing RasGAP–/– MEFs were seeded in six-well plates (10 × 104 cells/well) and treated the following day with the indicated concentrations of cisplatin for an additional 24-h period. The cells were then lysed, and caspase activity measured using a caspase-3 fluorigenic substrate as described in Materials and Methods (B). Alternatively, the extent of apoptosis was scored (C). Results correspond to the mean ± SEM of eight (B) or four (C) independent experiments performed with four (B) or three (C) independent clones of wild-type RasGAP-expressing cells or D157A-expressing RasGAP–/– MEFs. The insets in B and C represent enlargements of the areas highlighted in gray in the main panels. The significance of the differences between the two cell types, and, in the inset, with the nonstimulated condition, was assessed by Student's t tests (* p < 0.05).
Role of Fragments N1 and N2 in the Sensitivity to Apoptosis
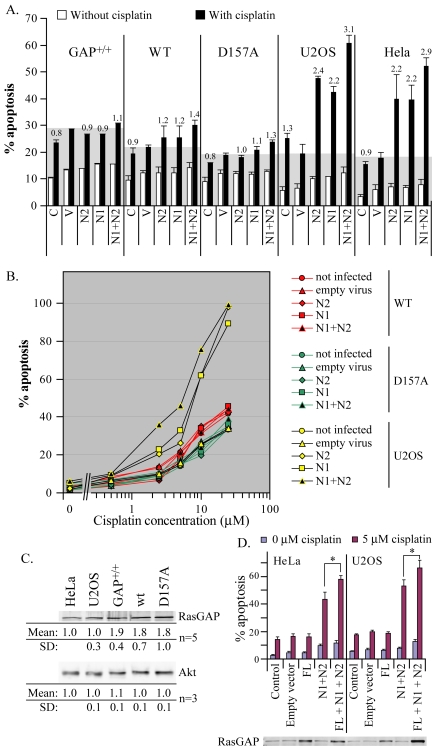
Full processing of fragment N by caspases generates the two small N-terminal fragments N1 and N2. Ectopic expression of these fragments increases the sensitivity of tumor cells, but not noncancer cells, to various genotoxins (Yang and Widmann, 2001; Michod et al., 2004). However, these observations were obtained in cells expressing the endogenous RasGAP protein and therefore the exact contribution of individual RasGAP fragments in the sensitization process could not be fully assessed as these cells can produce fragments N1 and N2 from endogenous RasGAP. To determine if the appearance of fragments N1 or N2, per se, favors apoptosis, cells that cannot produce these fragments but that express RasGAP and that can process it to fragment N (D157A mutant-expressing cells) were infected with lentiviruses encoding fragment N1 or fragment N2 (or both). The ability of the small RasGAP N-terminal fragments to sensitize these cells in response to cisplatin was then assessed. As shown in Figure 7, A and B, fragments N1 and N2, alone or in combination, neither markedly affected basal apoptosis nor apoptosis induced by cisplatin. Similar findings were observed in control MEFs (GAP+/+) and RasGAP–/– MEFs stably transfected with a vector encoding wild-type RasGAP (WT). This lack of sensitization was not due to poor infectivity as Western blot analysis demonstrated efficient expression of fragments N1 and N2 after incubation with the lentiviruses (unpublished data). The cisplatin-induced apoptotic response in the U2OS and HeLa cancer cell lines, in contrast, was increased by fragments N1 and N2 (Figure 7, A and B), demonstrating that the lentiviral preparations used here were functional in tumor cells. The differential sensitivity between cancer cells and nontumor cells toward fragments N1 and N2 could result from altered levels of Akt, a signaling protein that regulates apoptosis and that is activated by the generation of fragment N (Yang and Widmann, 2002). However, Akt expression levels were similar in the cell lines tested (Figure 7C). This suggests that sensitization of tumor cells by fragments N1 and N2 is not mediated by variations in Akt cellular levels. Expression levels of full-length RasGAP in MEFs were on average 1.8–1.9 times greater than in HeLa or U2OS tumor cells (Figure 7C). However, because of variations between experiments, these differences were not always statistically significant (significant differences were only measured between HeLa cells and control MEFs and between U2OS cells and control MEFs or wild-type RasGAP-reconstituted RasGAP–/– MEFs). It has been recently shown that small lung carcinoma cells have significantly reduced levels of full-length RasGAP and are more sensitive to etoposide compared with non–small cell lung carcinoma cell (Bartling et al., 2004). If a reduced RasGAP content in tumor cells is the cause of their sensitization to cisplatin-induced apoptosis in the presence of fragments N1 and N2, increased expression of RasGAP in these cells should abrogate this sensitization. Figure 7D shows however that ectopic expression of full-length RasGAP in tumor cells, which led to overexpression of the protein (see the lower part of the figure), did not protect them from fragments N1 and N2-induced sensitization. Rather, there was a slight, but significant, augmented sensitization in cells overexpressing RasGAP. Because these cells have higher expression of RasGAP, they have the potential to produce more fragments N1 and N2, which could explain why they are more sensitized to cisplatin-induced apoptosis than untransfected cells. Nevertheless, these results indicate that it is not because they have reduced RasGAP expression levels compared with noncancer cells that HeLa and U2OS cells are sensitized by fragments N1 and N2.
Figure 7.
Fragments N1 and N2 do not favor apoptosis in MEFs. (A) Control MEFs, wild-type RasGAP- and D157A-expressing RasGAP–/– MEFs, U2OS, and HeLa cells were left untreated (C) or were infected with an empty lentivirus (V) or with lentiviruses encoding fragments N2 or N1, alone or in combination. Two days later, the cells were incubated or not with 5 μM cisplatin for 24 h and the extent of apoptosis was scored. Results correspond to the mean ± SD of three independent experiments. Numbers over the black bars correspond to the fold-increase over the cisplatin-induced apoptosis measured in empty virus-infected cells. (B) Wild-type RasGAP- and D157A-expressing RasGAP–/– MEFs and U2OS cells were left untreated or were infected with an empty lentivirus or with lentiviruses encoding fragments N2 or N1, alone or in combination, and incubated 2 d later with the indicated concentrations of cisplatin for 24 h. The extent of apoptosis was then scored. (C) The indicated cell lines were lysed and their RasGAP and Akt content assessed by Western blot using antibodies recognizing the N2 domain of RasGAP and total Akt, respectively. The intensities of the Western blot signals were measured as indicated in Materials and Methods and normalized to the values found in HeLa cells. Results correspond to the mean ± SD of the indicated number of experiments. (D) HeLa and U2OS cells infected with viruses encoding the indicated constructs (FL, full-length RasGAP). Two days later, the cells were incubated with 0 or 5 μM cisplatin for an additional 24-h period, and the extent of apoptosis was determined (top part of the figure). The significance of the differences between the indicated conditions was assessed by Student's t tests (* p < 0.05; n = 6). Alternatively, the cells were lysed, and their RasGAP levels were visualized by Western blot using an antibody recognizing the N2 domain of RasGAP (bottom part of the figure).
In summary, our results indicate that cleavage of fragment N in normal cells fulfils only one function in the regulation of apoptosis, which is the abrogation of the strong survival signal mediated by fragment N.
DISCUSSION
Cells must continuously check their environment to determine whether they should grow, differentiate, proliferate, or undergo apoptosis. These cellular choices must be carefully balanced to maintain the organism's homeostasis. This is particularly important in the case of cell death, as exemplified by diseases characterized by pathologically increased apoptotic responses such as in Alzheimer disease or during cancer development where cells acquire resistance to apoptosis. Cells have developed a vast array of control mechanisms to regulate cell death responses. These controls can operate at the level of surface receptors (e.g., inhibition of death receptor–induced death by FLIP; Bentele et al., 2004), at the level of the mitochondria by Bcl-2 family members (Danial and Korsmeyer, 2004), and at the level of the activation of executioner caspases (Srinivasula et al., 2001; Zou et al., 2003). If these cellular control mechanisms eventually allow the activation of executioner caspases the cells generally undergo apoptosis, but other cellular responses can also occur. Indeed mild activation of several caspases, including caspase-3 and -8, is required for cell differentiation processes (De Maria et al., 1999; Fernando et al., 2002; Allombert-Blaise et al., 2003), cell activation (Algeciras-Schimnich et al., 2002; Newton and Strasser, 2003), and other cellular functions (Vercammen et al., 1998; Campbell and Holt, 2003). Cells must therefore be able to sense the activity of executioner caspases and determine, probably in a cell type-dependent manner, whether they should live or commit suicide. One mechanism allowing the cells to make these choices seems to operate at the level of RasGAP.
RasGAP bears two conserved cleavage sites at position 455 and 157 that are used sequentially as caspase activity increases (Yang and Widmann, 2001; Yang et al., 2004). The first cleavage of RasGAP occurs at very low caspase-3 activity, generating an N-terminal fragment called fragment N, which induces potent antiapoptotic signals (Yang et al., 2004) mediated by the Ras-PI3K-Akt pathway (Yang and Widmann, 2002). The antiapoptotic effectors of this pathway have not been characterized yet but proteins of the IAP family represent likely candidates because they can directly inhibit caspase activity (for further discussion see Yang et al., 2004).
We have recently demonstrated that the first RasGAP cleavage event and the resulting activation of Akt are required for cell survival in stress or adverse conditions (Yang et al., 2004). The relative importance in apoptosis of the second caspase-mediated cleavage of RasGAP has been addressed in the present study.
For this purpose, MEFs have been generated that have their wild-type RasGAP gene replaced by a cDNA encoding the RasGAP mutant that cannot be cleaved at position 157. This mutant of RasGAP can only be cleaved into fragment N, but not processed further. The cells expressing this form of RasGAP were slightly, yet significantly, more resistant toward apoptosis than control cells in response to a variety of apoptotic stimuli. Accordingly, when control cells and cells unable to process fragment N were mixed, the proportion of the latter was increased in the presence of apoptotic stimuli. The augmented resistance to apoptosis of cells expressing the D157A mutant of RasGAP was associated with a sustained activation of Akt and a reduced ability to fully process and activate caspase-3. This suggests that the function of the caspase recognition site on RasGAP at position 157 is to terminate the stimulation of Akt by fragment N in order to allow efficient activation of executioner caspases.
In noncancer cells, appearance of the small N-terminal fragments (fragments N1 and N2) do not seem to sensitize cells to apoptosis. It therefore appears that in noncancer cells the cleavage of fragment N favors the activation of caspase-3 and apoptosis solely because the survival signal induced by fragment N is shutoff. Why cancer cells, but not normal cells, are sensitized by small N-terminal RasGAP fragments (fragment N2 in particular) to be killed by genotoxins (Michod et al., 2004) is currently unknown.
Altogether, our results indicate that RasGAP cleavage at position 157 participates in the execution phase of apoptosis, like most other caspase substrates when they are cleaved. Abrogation of the cleavage at position 157 in RasGAP confers a relatively minimal protection against apoptosis, an expected finding because all the other caspase substrates involved in the destructive phase of apoptosis can still operate.
There are many caspase substrates in eukaryotic cells whose function when cleaved is to accelerate/potentiate the apoptotic response (Fischer et al., 2003). It seems therefore that it is crucial for multicellular organisms to ensure that once the decision to commit suicide has been taken, the actual dismissal of the cells occurs rapidly. An example illustrating the detrimental effect of a slow apoptotic response is the observation that delayed apoptosis in neutrophils appears to impair the resolution of inflammation and exacerbate host tissue damage (Taneja et al., 2004). Our data indicate that the cleavage of RasGAP at position 157 is one of the tools used by cells to accelerate the apoptotic process.
Acknowledgments
This work is supported by the Swiss National Science Foundation (Grant 3100-066797/1), the Oncosuisse (Grants OCS 1110-2-2001) and the Botnar Foundation (Lausanne, Switzerland).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–01–0080) on May 18, 2005.
Abbreviations used: ERK, extracellular signal-regulated kinase; HA, hemagglutinin; MEFs, mouse embryonic fibroblasts.
References
- Algeciras-Schimnich, A., Barnhart, B. C., and Peter, M. E. (2002). Apoptosis-independent functions of killer caspases. Curr. Opin. Cell Biol. 14, 721–726. [DOI] [PubMed] [Google Scholar]
- Allombert-Blaise, C. et al. (2003). Terminal differentiation of human epidermal keratinocytes involves mitochondria- and caspase-dependent cell death pathway. Cell Death Differ. 10, 850–852. [DOI] [PubMed] [Google Scholar]
- Bartling, B., Yang, J. Y., Michod, D., Widmann, C., Lewensohn, R., and Zhivotovsky, B. (2004). RasGTPase-activating protein is a target of caspases in spontaneous apoptosis of lung carcinoma cells and in response to etoposide. Carcinogenesis 25, 909–921. [DOI] [PubMed] [Google Scholar]
- Basu, A., Lu, D., Sun, B., Moor, A. N., Akkaraju, G. R., and Huang, J. (2002). Proteolytic activation of protein kinase C-epsilon by caspase-mediated processing and transduction of antiapoptotic signals. J. Biol. Chem. 277, 41850–41856. [DOI] [PubMed] [Google Scholar]
- Bentele, M., Lavrik, I., Ulrich, M., Stosser, S., Heermann, D. W., Kalthoff, H., Krammer, P. H., and Eils, R. (2004). Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J. Cell Biol. 166, 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, D. S., and Holt, C. E. (2003). Apoptotic pathway and MAPKs differentially regulate chemotropic responses of retinal growth cones. Neuron 37, 939–952. [DOI] [PubMed] [Google Scholar]
- Campbell, S. L., Khosravi-Far, R., Rossman, K. L., Clark, G. J., and Der, C. J. (1998). Increasing complexity of Ras signaling. Oncogene 17, 1395–1413. [DOI] [PubMed] [Google Scholar]
- Danial, N. N., and Korsmeyer, S. J. (2004). Cell death: critical control points. Cell 116, 205–219. [DOI] [PubMed] [Google Scholar]
- De Maria, R., Zeuner, A., Eramo, A., Domenichelli, C., Bonci, D., Grignani, F., Srinivasula, S.M., Alnemri, E.S., Testa, U., and Peschle, C. (1999). Negative regulation of erythropoiesis by caspase-mediated cleavage of GATA-1. Nature 401, 489–493. [DOI] [PubMed] [Google Scholar]
- Dull, T., Zufferey, R., Kelly, M., Mandel, R. J., Nguyen, M., Trono, D., and Naldini, L. (1998). A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando, P., Kelly, J. F., Balazsi, K., Slack, R. S., and Megeney, L. A. (2002). Caspase 3 activity is required for skeletal muscle differentiation. Proc. Natl. Acad. Sci. USA 99, 11025–11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, U., Janicke, R. U., and Schulze-Osthoff, K. (2003). Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ. 10, 76–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Z., Hendrickson, E. A., Bremner, T. A., and Wyche, J. H. (1997). A sequential two-step mechanism for the production of the mature p17, p12 form of caspase-3 in vitro. J. Biol. Chem. 272, 13432–13436. [DOI] [PubMed] [Google Scholar]
- Holler, N. et al. (2003). Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol. Cell. Biol. 23, 1428–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M., Schallhorn, A., and Wurm, F. M. (1996). Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, S. V., Gish, G., Van der Geer, P., Henkemeyer, M., and Pawson, T. (2000). Role of p120 ras-GAP in directed cell movement. J. Cell Biol. 149, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc, V., Tocque, B., and Delumeau, I. (1998). Ras-GAP controls Rho-mediated cytoskeletal reorganization through its SH3 domain. Mol. Cell. Biol. 18, 5567–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano, F., Herrant, M., Jacquel, A., Ricci, J. E., and Auberger, P. (2003). The p54 cleaved form of the tyrosine kinase Lyn generated by caspases during BCR-induced cell death in B lymphoma acts as a negative regulator of apoptosis. FASEB J. 17, 711–713. [DOI] [PubMed] [Google Scholar]
- Michod, D., Yang, J. Y., Chen, J., Bonny, C., and Widmann, C. (2004). A RasGAP-derived cell permeable peptide potently enhances genotoxin-induced cytotoxicity in tumor cells. Oncogene 23, 8971–8978. [DOI] [PubMed] [Google Scholar]
- Newton, K., and Strasser, A. (2003). Caspases signal not only apoptosis but also antigen-induced activation in cells of the immune system. Genes Dev. 17, 819–825. [DOI] [PubMed] [Google Scholar]
- Nicholson, D. W. (1999). Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 6, 1028–1042. [DOI] [PubMed] [Google Scholar]
- Ricci, J. E., Lang, V., Luciano, F., Belhacene, N., Giordanengo, V., Michel, F., Bismuth, G., and Auberger, P. (2001). An absolute requirement for Fyn in T cell receptor-induced caspase activation and apoptosis. FASEB J. 15, 1777–1779. [DOI] [PubMed] [Google Scholar]
- Schlesinger, T. K., Bonvin, C., Jarpe, M. B., Fanger, G. R., Cardinaux, J.-R., Johnson, G. L., and Widmann, C. (2002). Apoptosis stimulated by the 91-kDa caspase cleavage MEKK1 fragment requires translocation to soluble cellular compartments. J. Biol. Chem. 277, 10283–10291. [DOI] [PubMed] [Google Scholar]
- Slee, E. A., Adrain, C., and Martin, S. J. (2001). Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J. Biol. Chem. 276, 7320–7326. [DOI] [PubMed] [Google Scholar]
- Srinivasula, S. M. et al. (2001). A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410, 112–116. [DOI] [PubMed] [Google Scholar]
- Stroh, C., and Schulze-Osthoff, K. (1998). Death by a thousand cuts: an ever increasing list of caspase substrates. Cell Death Differ. 5, 997–1000. [DOI] [PubMed] [Google Scholar]
- Taneja, R., Parodo, J., Jia, S. H., Kapus, A., Rotstein, O. D., and Marshall, J. C. (2004). Delayed neutrophil apoptosis in sepsis is associated with maintenance of mitochondrial transmembrane potential and reduced caspase-9 activity. Crit. Care Med. 32, 1460–1469. [DOI] [PubMed] [Google Scholar]
- Toyoshima, F., Moriguchi, T., and Nishida, E. (1997). Fas induces cytoplasmic apoptotic responses and activation of the MKK7-JNK/SAPK and MKK6–p38 pathways independent of CPP32-like proteases. J. Cell Biol. 139, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valius, M., Secrist, J., and Kazlauskas, A. (1995). The GTPase-activating protein of ras suppresses platelet-derived growth factor β receptor signaling by silencing phospholipase c-γ1. Mol. Cell. Biol. 15, 3058–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Geer, P., Henkemeyer, M., Jacks, T., and Pawson, T. (1997). Aberrant Ras regulation and reduced p190 tyrosine phosphorylation in cells lacking p120-Gap. Mol. Cell. Biol. 17, 1840–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen, D., Beyaert, R., Denecker, G., Goossens, V., Van Loo, G., Declercq, W., Grooten, J., Fiers, W., Vandenabeele, P. (1998). Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J. Exp. Med. 187, 1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, L. P., Madani, K., Martin, G. A., and Rosen, G. D. (1998). Proteolytic cleavage of Ras GTPase-activating protein during apoptosis. Cell Death Differ. 5, 729–734. [DOI] [PubMed] [Google Scholar]
- Widmann, C., Dolci, W., and Thorens, B. (1995). Agonist-induced internalization and recycling of the glucagon-like peptide-1 receptor in transfected fibroblasts and in insulinomas. Biochem. J. 310, 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.-Y., Michod, D., Walicki, J., Murphy, B. M., Kasibhatla, S., Martin, S., and Widmann, C. (2004). Partial cleavage of RasGAP by caspases is required for cell survival in mild stress conditions. Mol. Cell. Biol. 24, 10425–10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.-Y., and Widmann, C. (2001). Antiapoptotic signaling generated by caspase-induced cleavage of RasGAP. Mol. Cell. Biol. 21, 5346–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.-Y., and Widmann, C. (2002). The RasGAP N-terminal fragment generated by caspase cleavage protects cells in a Ras/PI3K/Akt-dependent manner that does not rely on NFκB activation. J. Biol. Chem. 277, 14641–14646. [DOI] [PubMed] [Google Scholar]
- Zou, H., Yang, R., Hao, J., Wang, J., Sun, C., Fesik, S. W., Wu, J. C., Tomaselli, K. J., and Armstrong, R. C. (2003). Regulation of the Apaf-1/caspase-9 apoptosome by caspase-3 and XIAP. J. Biol. Chem. 278, 8091–8098. [DOI] [PubMed] [Google Scholar]