Abstract
Dual-specificity tyrosine-phosphorylated and regulated kinase 1A (Dyrk1A) is the human homologue of the Drosophila mnb (minibrain) gene. In Drosophila, mnb is involved in postembryonic neurogenesis. In human, DYRK1A maps within the Down syndrome critical region of chromosome 21 and is overexpressed in Down syndrome embryonic brain. Despite its potential involvement in the neurobiological alterations observed in Down syndrome patients, the biological functions of the serine/threonine kinase DYRK1A have not been identified yet. Here, we report that DYRK1A overexpression potentiates nerve growth factor (NGF)-mediated PC12 neuronal differentiation by up-regulating the Ras/MAP kinase signaling pathway independently of its kinase activity. Furthermore, we show that DYRK1A prolongs the kinetics of ERK activation by interacting with Ras, B-Raf, and MEK1 to facilitate the formation of a Ras/B-Raf/MEK1 multiprotein complex. These data indicate that DYRK1A may play a critical role in Ras-dependent transducing signals that are required for promoting or maintaining neuronal differentiation and suggest that overexpression of DYRK1A may contribute to the neurological abnormalities observed in Down syndrome patients.
INTRODUCTION
Minibrain (mnb)/dual-specificity tyrosine-phosphorylated and regulated kinase 1A (Dyrk1A) gene is a member of a growing family of protein kinases called DYRK. In Drosophila, mnb seems to play an essential role during postembryonic neurogenesis. Mutant flies are characterized by a marked reduction in size of the adult optic lobes and the central brain hemispheres. This is caused by the abnormal spacing of neuroblasts and a reduction in the production of neuronal progeny (Tejedor et al., 1995). At least seven closely related homologous mammalian kinases in the DYRK family have since been isolated (Guimera et al., 1996, 1999; Shindoh et al., 1996; Song et al., 1996, 1997; Raich et al., 2003). The DYRK family possesses serine and threonine phosphorylation activity as well as autophosphorylation activity on tyrosine residues (Kentrup et al., 1996; Becker et al., 1998; Becker and Joost, 1999; Himpel et al., 2000). Their kinase activity is dependent on the YXY motif in the activation loop of the catalytic domain, which is located at the same position as the characteristic TXY motif of the mitogen-activated protein kinases (MAPKs), suggesting an activation mechanism similar to that of the MAPK (Himpel et al., 2000). Closely related enzymes in lower eukaryotes also have been isolated, including Yak1p in Saccharomyces cerevisiae (Garrett and Broach, 1989), Pom1p in Schizosaccharomyces pombe (Bahler and Pringle, 1998), and YakA in Dictyostelium discoideum (Van Es et al., 2001). Although not much is known about their cellular functions, they all seem to be involved in the regulation of the cell growth and/or development.
In the DYRK family, DYRK1A has been the best characterized to date. The human Dyrk1A gene maps to the 21q22.2 region of chromosome 21, in the Down syndrome critical region (Rahmani et al., 1989, 1990; Delabar et al., 1993; Shindoh et al., 1996; Song et al., 1996), and transgenic mice harboring an extra copy of this gene exhibit cognitive deficits and motor abnormalities characteristic of Down syndrome (Smith and Rubin, 1997; Smith et al., 1997; Altafaj et al., 2001). The human and rodent Dyrk1A gene are ubiquitously expressed in adult and fetal tissues with high expression in the brain and the heart during development (Guimera et al., 1996, 1999; Song et al., 1996; Rahmani et al., 1998; Hämmerle et al., 2003a; Mao et al., 2003; Marti et al., 2003). DYRK1A encodes a protein of 763 or 754 amino acid residues as a result of alternative splicing. The large domain flanking the C terminus of the catalytic domain contains several striking structural features, such as a PEST region, which initiates a rapid degradation of the protein, a stretch of 13 consecutive histidine residues (amino acids 607–619), and a serine/threonine-rich segment of 14 subsequent serine/threonine residues (Kentrup et al., 1996; Becker et al., 1998; Becker and Joost, 1999; Himpel et al., 2000). Although the biological function of DYRK1A is not known, DYRK1A interacts in vivo with several transcription factors. Additionally, DYRK1A phosphorylates several substrates in vitro, including the signal transducer and activator of transcription 3 (Matsuo et al., 2001), the eukaryotic initiation factor 2Bε (Woods et al., 2001a), the microtubule-associated protein Tau (Woods et al., 2001a), the transcription factor of the Forkhead family (FHKR) (Woods et al., 2001b), dynamin (Chen-Hwang et al., 2002), glycogen synthase (Skurat and Dietrich, 2004), cyclin L2 (De Graaf et al., 2004), and 14-3-3 (Kim et al., 2004), indicating that DYRK1A may participate in several biochemicals pathways (reviewed in Galceran et al., 2003; Hämmerle et al., 2003b). In vivo, DYRK1A interacts with and activates Gli-1–dependent gene transcription (Mao et al., 2002). Moreover, DYRK1A increases FKHR-dependent glucose-6-phosphatase gene expression in hepatoma cells (von Groote-Bidlingmaier et al., 2003). DYRK1A also was recently shown to interact with Arip4 (androgen receptor-interacting protein 4), a steroid hormone receptor cofactor, and it regulates steroid hormone-induced transcription (Sitz et al., 2004). In immortalized hippocampal progenitor cells stimulated by the basic fibroblast growth factor (bFGF), DYRK1A has been shown to be activated by bFGF and stimulate the phosphorylation of the cAMP response-element binding protein to subsequently induce cAMP response element-mediated gene transcription (Yang et al., 2001). These findings suggest that DYRK1A may exert various effects during neurotrophic factor-mediated neuronal differentiation.
To elucidate a possible role of DYRK1A during neuronal differentiation, we examined the effect of DYRK1A overexpression on the activation of the MAP kinase signaling pathway in the pheochromocytoma PC12 cell line. We found that DYRK1A potentiates nerve growth factor (NGF)-mediated PC12 neuronal differentiation by up-regulating the Ras/MAP kinase signaling pathway independently of its kinase activity. Moreover, we showed that DYRK1A prolongs the kinetics of ERK activation by interacting with Ras, B-Raf, and MEK1 and by facilitating the formation of a Ras/B–Raf/MEK1 multiprotein complex.
MATERIALS AND METHODS
Plasmids
DYRK1A cDNA was isolated by PCR from a human fetal kidney cDNA library (BD Biosciences Clontech, Palo Alto, CA) by using a 5′ primer flanked with a SmaI restriction site: 5′-tcccccggggATGCATACAGGAGGAGAGAC-3′ and a 3′ primer flanked with a XhoI restriction site: 5′-ccgctcgagCGGCACGAGCTAGCTACAGGAC-3′. The cDNA was then inserted into the NheI site blunted with T4 DNA polymerase and XhoI site of pcDNA3.1/Myc-His vector (Invitrogen, Carlsbad, CA) carrying a cytomegalovirus (CMV) promoter and a C-terminal myc epitope to make myc-DYRK1A construct. myc-DYRK1A-K188R was obtained by site-directed mutagenesis by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. All constructs were verified by nucleotide sequencing by using an ABI373 automatic sequencer. Ras (provided by K. L. Guan, University of Michigan Medical School, Ann Arbor, MI), RasV12, RasN17 (provided by L. Feig, Tufts University School of Medicine, Boston, MA), HA-B-Raf, pEBG-Raf1 (GST-Raf1), and GST-Ras (provided by K. L. Guan), and HA-MEK1, HA-MEK1Δ270-307 (provided by M. J. Weber, University of Virginia School of Medicine, Charlottesville, VA), have been described previously (Feig and Cooper, 1988; Catling et al., 1995; Robinson and Cobb, 1997; Sugimoto et al., 1997; Li et al., 2000).
Antibodies
DYRK1A antibodies used in this study include a monoclonal antibody generated against the carboxy terminus (amino acid residues 588–746 of Rat DYRK1A; BD Transduction Laboratories, Lexington, KY) and a rabbit polyclonal antibody generated against a glutathione S-transferase (GST) fusion protein containing a fragment of human DYRK1A encoding amino acid residues 1–129 (Rahmani et al., 1998). MEK1, B-Raf, and GST tag antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA); myc tag and hemagglutinin (HA) tag antibodies were from Roche Diagnostics (Indianapolis, IN); Tubulin-βIII and Actin antibodies were from Sigma (St. Louis, MO); Ras antibodies were from Calbiochem (San Diego, CA); and extracellular signal-regulated kinase (ERK) and phospho-ERK antibodies were from Cell Signaling Technology (Beverly, MA).
Cell Culture and Transfection
HeLa cells were grown in DMEM supplemented with 10% fetal calf serum (FCS). PC12 cells were grown in DMEM supplemented with 10% horse serum and 5% FCS on poly-d-lysine–coated plates. Cells were transfected using Lipofectamine 2000 (Invitrogen) for 6 h in Opti-MEM reduced serum medium. After transfection, cells were allowed to recover in normal medium for 24 h. Transfected cells were then serum starved in 0.1% serum-containing DMEM for 20 h before stimulation. PC12 cells were transfected with myc-DYRK1A and myc-DYRK1A-K188R constructs, selected in medium containing G418, and drug-resistant cell lines were established. All cell lines were screened fore the presence of myc-tagged DYRK1A constructs by immunoblot analysis.
PC12 Differentiation Assays
PC12 cells (1 × 105) in 60-mm-diameter plates were transfected at 40% confluence with 1.5 μg of myc-DYRK1A or myc-DYRK1A-K188R plasmids and 0.5 μg of a CMV-βGal reporter vector. After 24 h, transfected cells were serum starved for 20 h, treated with 50 ng/ml NGF for the indicated times, stained for LacZ gene expression, and examined by light microscopy. The percentage of βGal-positive differentiated cells was determined by calculating the number of βGal-positive cells with neurites longer than two cell body diameters compared with the total number of βGal-positive cells counted. More than 200 blue cells were counted per condition in three independent experiments.
Immunoprecipitation and Immunoblotting
PC12 cells were cultured in 100-mm-diameter plates to 70% confluence and transiently transfected with 36 μl of Lipofectamine 2000 and 12 μg of total DNA (6 μg of myc-DYRK1A or myc-DYRK1A-K188R plasmids and 6 μg of the indicated plasmids). Forty-eight hours posttransfection, cells were lysed in 1 ml of lysis buffer L (10 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40, 10% glycerol, 1 mM EDTA, 2 mM NaF, 2 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride) for 30 min at 4°C. Lysates were clarified by centrifugation at 15,000 × g for 15 min and precleared by incubation with protein G or protein A-Sepharose for 1 h at 4°C. Supernatants were then incubated overnight with the relevant antibody and protein A or protein G-Sepharose, washed three times with buffer L, and then resolved by SDS-PAGE followed by immunoblotting on a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA), and processed as described by the manufacturer. For immunoblotting analysis, cells were lysed in lysis buffer L and clarified by centrifugation as described above. The protein concentration of the supernatant was measured using a Bio-Rad protein assay, and then the same amount of protein was resolved by SDS-PAGE and immunoblotted on a PVDF membrane. Proteins were detected by enhanced chemiluminescence assay (ECL Plus; Amersham Biosciences, Piscataway, NJ).
GST-Fusion Protein Expression
GST-fusion constructs were introduced into DH5α bacteria, and protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactoside at 37°C for 3 h. Bacteria were lysed in phosphate-buffered saline (PBS) containing 1% Triton X-100, and 50 mM EDTA. After sonication and centrifugation, cleared lysates were incubated with glutathione-agarose beads (Amersham Biosciences) for 2 h at 4°C with agitation. Beads were washed four times with PBS and 1% Triton X-100 and eluted according to the manufacturer's instructions.
GST-Fusion Binding Assays
Bacterially expressed and purified GST-Ras protein was immobilized on glutathione-agarose beads. GST-Ras proteins were preloaded with 5′-O-(3-thio)triphosphate (GTPγS) or GDP by incubating GST-Ras beads with 1 mM GTPγS or 1 mM GDP for 20 min at 37°C in binding buffer (25 mM Tris, pH 7.5, 100 mM NaCl, 2 mM EDTA, 5 mM dithiothreitol [DTT]). The beads were incubated for 3 h at 4°C with 5 mg of extracts from untransfected PC12 cells or PC12 cells transfected with myc-DYRK1A. Cell extracts were obtained as described above except that the lysis buffer L was supplemented with 5 mM MgCl2 and 5 mM DTT. After four washes in lysis buffer L supplemented with 5 mM MgCl2 and 5 mM DTT, the proteins associated with GST-Ras were eluted in Laemmli buffer. Samples were subjected to SDS-PAGE and then to Western blot analysis by using B-Raf or myc-tag antibodies.
Transactivation Assays
For reporter gene assays, 1 μg of 5x-Gal4-TATA/luciferase and 70 ng of a plasmid encoding the Gal-Elk1 transactivation domain (Elk/gal) were used according to the manufacturer (Stratagene) in combination with other plasmids as indicated. CMV-βGal construct was transfected as an internal control for gene expression. Cells were transfected as described in 60-mm-diameter plates. Twenty-four hours posttransfection, cells were serum starved for 20 h and either left unstimulated or stimulated with 100 ng/ml NGF for the indicated times before being collected (48 h posttransfection). Luciferase activity was detected with a luminometer, and βGal expression was detected by absorbance at A420. Results were normalized to βGal activity. Values are presented as the mean ± SEM (bars) of three independent experiments carried out in triplicate.
Inhibition of DYRK1A Expression by RNA Interference (RNAi)
To design small interfering RNA (siRNA) to knockdown DYRK1A, regions of the cDNAs that are identical in rats (Dyrk1A; GenBank accession no. NM012791), mice (Dyrk1A; GenBank accession no. U58497), and humans (Dyrk1A; GenBank accession no. AF108830) were chosen. The siRNAs sequences were as follows: 5′-AACCACCAGGAACCCGUAAACdTdT-3′ (nucleotides 1376–1396) and 5′-AAUGGAGCUAUGGACGUUAAUdTdT-3′ (nucleotides 2212–2232) for targeting Dyrk1A; and 5′-CGACAUUGGCGUAAGUGAAdTdT-3′ (nucleotides 2372–2390) for targeting LacZ. The 23-nucleotide RNAs (sense and antisense strand) were chemically synthesized, deprotected, gel purified, and annealed by Proligo. For silencing the endogenous DYRK1A expression, PC12 cells cultured in 60-mm-diameter plates were transfected with an empty control vector, 200 pmol of LacZ siRNA duplex, or equal amounts of the two DYRK1A siRNAs duplex (100 pmol each) using Lipofectamine 2000. Forty-eight hours posttransfection, cells were lysed in lysis buffer L and clarified by centrifugation. The protein concentration of the supernatant was measured using a Bio-Rad protein assay. For transactivation assays, an empty control vector, LacZ-siRNA (200 pmol), or pools of equal amounts of the two DYRK1A siRNAs (100 pmol each) were used.
Statistical Analysis
Significant differences between two groups were analyzed by using Student's t test. Differences between two means with a p < 0.05 were regarded as significant. All values were expressed as means ± SEM of at least three independent experiments carried out in triplicate.
RESULTS
DYRK1A Overexpression Potentiates Neurite Outgrowth in PC12 Cells
To investigate the role played by DYRK1A in neuronal cells, we overexpressed DYRK1A in PC12 cells. PC12 cells are an ideal model system for the differentiation of neuronal cells because they respond to NGF by extending neurites and developing many properties similar to those of sympathetic neurons (Greene and Tischler, 1976). PC12 cells were transiently transfected with either a control vector, myc-DYRK1A, or a kinase-dead form of DYRK1A (myc-DYRK1A-K188R) expression plasmid together with a CMV-βGal reporter plasmid. DYRK1A-K188R, mutated in its ATP-binding site, has been shown to be enzymatically inactive (Kentrup et al., 1996). Cells were then treated with NGF for various times and stained for LacZ gene expression. After 24 h, the DYRK1A-transfected cells displayed an enhanced induction of neurite outgrowth in response to NGF (p < 0.0001) compared with the control-transfected PC12 cells (Figure 1A). This enhanced induction lasted several days after NGF treatment. Surprisingly, in PC12 cells transfected with a kinase-dead mutant form of DYRK1A (DYRK1A-K188R), the neurite outgrowth also was enhanced (p < 0.0001) compared with the control-transfected cells (Figure 1A). The enhanced neurite outgrowth observed in DYRK1A-K188R–transfected cells were not significantly different from that observed in DYRK1A-transfected cells (p > 0.05).
Figure 1.
DYRK1A promotes neurite outgrowth and neuronal differentiation in PC12 cells. (A) PC12 cells were transiently transfected with either an empty vector, myc-DYRK1A, or myc-DYRK1A-K188R plasmids in combination with a CMV-βGal reporter plasmid. After 24 h, cells were serum starved for 20 h and then treated with 50 ng/ml NGF for the indicated times, stained for LacZ gene expression, and examined by light microscopy. Percentages of βGal-positive differentiated cells were determined by the ratio between the number of βGal-positive cells with neurites longer than two cell body diameters, and the total number of βGal-positive cells was counted. More than 200 blue cells were counted per condition in three independent experiments. Error bars represent means ± SEM. For comparisons indicated by brackets, three asterisks indicate a p value <0.0001. Percentages of differentiated cells in DYRK1A and DYRK1A-K188R–transfected cells were not significantly different (p > 0.05). (B) Vector-transfected, DYRK1A, and DYRK1A-K188R stable PC12 cell lines were stimulated with 50 ng/ml NGF. Cells were photographed 2 d after treatment. The experiment was repeated with two different stable DYRK1A- and DYRK1A-K188R–expressing clones with similar results. The percentage of cells that exhibited neurites longer than two cell body diameters was determined. More than 20 random fields were examined. Data are means ± SEM of values from three separate experiments. For comparisons indicated by brackets, three asterisks indicate a p value of <0.0001. Percentages of differentiated cells in DYRK1A and DYRK1A-K188R stable cell lines were not significantly different (p > 0.05). (C) PC12 cells were transiently transfected with either an empty vector, myc-DYRK1A, or myc-DYRK1A-K188R plasmids. After 24 h, transfected cells were serum starved for 20 h and then either left unstimulated or stimulated with 50 ng/ml NGF for 3 d, and analyzed for expression of the differentiation marker Tubulin-βIII by immunoblotting (WB). Actin was used as an internal control to verify equal loading of proteins. Densitometric quantitation of relative Tubulin-βIII expression is indicated underneath each lane of the top panel. These experiments were repeated three times, and representative data are shown. Sizes of molecular weight markers are indicated on the right.
PC12 cell lines that stably express myc-tagged DYRK1A or DYRK1A-K188R were generated. Three stables cell lines were isolated for each construct and examined for cell differentiation. By 2 d of NGF treatment, the neurite projections extending from the DYRK1A and the DYRK1A-K188R cells had well-defined growth cones and were 3 to 4 times the diameter of the cell body. In contrast, the neurite projections in vector-transfected cells were approximately one cell body length (Figure 1B). These data indicate that DYRK1A enhances the neurite outgrowth in PC12 cells independently of its kinase activity.
Neuronal differentiation of PC12 cells is characterized not only by morphological changes (neurite outgrowth) but also synthesis of neuronal proteins (Greene and Tischler, 1976). To further confirm that the promotion of neurite outgrowth induced by DYRK1A was accompanied by neuronal differentiation of PC12 cells, the expression of a neuron-specific marker of neuronal differentiation was examined. β-Tubulin isoform III (Tubulin-βIII) is synthesized exclusively by neurons and increases in conjunction with the rate of neuronal differentiation (Greene and Tischler, 1976). After 3 d of NGF treatment, a marked increase in the expression of Tubulin-βIII protein was detected in PC12 cells transfected with either myc-DYRK1A or myc-DYRK1A-K188R expression plasmids. DYRK1A-K188R–transfected cells displayed slightly reduced levels of Tubulin-βIII compared with DYRK1A-expressing cells. However, levels were still higher than those observed in control vector-transfected cells (Figure 1C).
DYRK1A Up-Regulates Ras/MAP Kinase Signaling
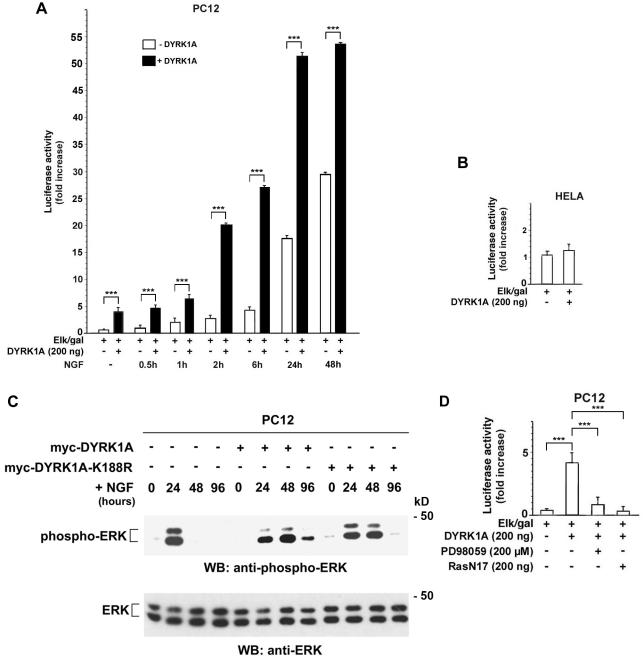
Because PC12 differentiation often correlates with sustained MAPK activity (Qui and Green, 1992), the effect of DYRK1A on the Ras/MAP kinase signaling pathway was examined. PC12 cells were transiently transfected with myc-DYRK1A or a control vector in combination with reporter plasmids that read out ERK-dependent activation of the transcription factor Elk1. Transfection with DYRK1A resulted in activation of the Ras/MAP kinase signaling cascade (Figure 2A). This activation was further enhanced when transfected PC12 cells were treated with NGF for various times, indicating that DYRK1A and NGF act synergistically to increase ERK-dependent Elk1 activation (Figure 2A). However, DYRK1A had no effect on ERK activity in HeLa and Cos-7 cells (Figure 2B; our unpublished data). Previously reported data also showed no effect of DYRK1A on ERK activity in NIH 3T3 cells (Mao et al., 2002). These findings indicate that the effect of DYRK1A on the Ras/MAP kinase pathway is cell line specific (discussed in Figure 6D).
Figure 2.
DYRK1A up-regulates Ras/MAPK signaling in neuronal cells. (A, B, and D) PC12 or HeLa cells were cotransfected with 1 μg of 5x-Gal4-TATA/luciferase, 70 ng of Gal4-Elk1 (Elk/gal), 1 μg of CMV-βGal, and the indicated expression plasmids. Transfected cells were serum starved for 20 h and either left unstimulated or stimulated with 100 ng/ml NGF for the indicated times (A) or treated with 200 μM PD98059 for 30 min (D) before being collected (48 h posttransfection). (B) Transfected HeLa cells were serum starved for 20 h before being collected (48 h posttransfection). Values obtained with DYRK1A were not significantly different from the ones obtained with the empty control vector (p > 0.05). Activation of Elk1 is reflected by luciferase activity normalized to βGal activity and is described as fold increase. Error bars represent means ± SEM. For comparisons indicated by brackets (A and D), three asterisks indicate a p value <0.0001. (C) myc-DYRK1A, myc-DYRK1A-K188R, or control vector-transfected PC12 cells were serum starved for 20 h and then stimulated with 50 ng/ml NGF for the indicated times. Cell lysates were prepared and examined by immunoblot (WB) analysis by using phospho-ERK and ERK antibodies. Immunoblotting with ERK antibody confirmed that equivalent amount of ERK were present in cell lysates of transfected cells. The results shown are representative of three independent experiments. Sizes of molecular weight markers are indicated on the right.
Figure 6.
DYRK1A associates with B-Raf but not with Raf1. (A–C) PC12 cells were transiently transfected with the described constructs and analyzed by coimmunoprecipitation (IP) followed by immunoblotting (WB) with anti-myc, anti-HA, or anti-GST tag antibodies. As a control, myc-tag antibody coimmunoprecipitated HA-B-Raf in cells expressing myc-DYRK1A and HA-B-Raf but not in cells expressing HA-B-Raf alone (A, lane 1). In the same manner, HA-tag antibody coimmunoprecipitated myc-DYRK1A in cells expressing myc-DYRK1A and HA-B-Raf but not in cells expressing myc-DYRK1A alone (B, lane 1). Whole cell lysates of transfected cells were analyzed by Western blotting (WB) with the appropriate antibody to verify equal expression of all constructs. The results shown are representative of three independent experiments. Sizes of molecular weight markers are indicated on the right. (D) HeLa cells were cotransfected with 1 μg of 5x-Gal4-TATA/luciferase, 70 ng of Gal4-Elk1 (Elk/gal), 1 μg of CMV-βGal, and the indicated expression plasmids. Cells were serum starved for 20 h before being collected (48 h posttransfection). Activation of Elk1 is reflected by luciferase activity normalized to βGal activity and is described as fold increase. Error bars represent means ± SEM. For comparisons indicated in brackets, two asterisks indicate a p value <0.001. Values obtained with DYRK1A or B-Raf were not significantly different from the ones obtained with the empty control vector (p > 0.05).
The activation state of ERK was then determined by probing the lysates with an antibody that specifically recognizes activated (phosphorylated) ERK. Compared with vector-transfected cells, myc-DYRK1A– and myc-DYRK1A-K188R–transfected PC12 cells exhibited a longer sustained ERK activation in response to NGF stimulation (Figure 2C).
To determine where in the Ras/MAP kinase cascade DYRK1A acts, inhibitor studies were carried out. In transient transfection assays, DYRK1A-induced ERK-dependent Elk1 activation was fully blocked by inhibiting the upstream kinase MEK1 with PD98059 (Figure 2D). When DYRK1A or DYRK1A-K188R PC12-transfected cells were pretreated with PD98059 for 30 min before stimulation, NGF-induced neurite outgrowth also was completely blocked (our unpublished data). These findings demonstrate that the stimulatory effect of DYRK1A requires MEK activity. A dominant negative form of Ras (RasN17) also inhibited the DYRK1A-mediated ERK-dependent Elk1 activation, indicating that Ras also was necessary for the stimulatory effect of DYRK1A (Figure 2D).
DYRK1A Enhances Ras-mediated, B-Raf-mediated, and MEK1-mediated Stimulatory Effects on ERK-dependent Elk1 Activation
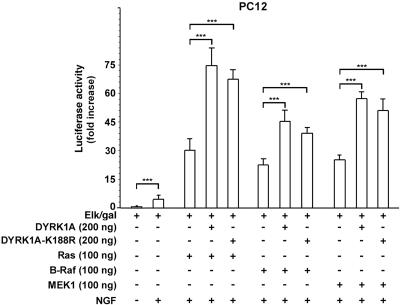
The effect of DYRK1A on Ras-, B-Raf-, and MEK1-mediated ERK-dependent Elk1 activation was addressed by reporter gene assays as described above. As shown in Figure 3, stimulation of cells with NGF alone resulted in a strong induction of ERK-dependent Elk1 activation of 5-fold (p < 0.0001). The expression of either Ras, B-Raf, or MEK1 led to a further increase in Elk1 activation of 6-, 4.5-, and 5.1-fold respectively (p < 0.0001). The expression of DYRK1A and Ras, B-Raf, or MEK1 together led to an even stronger increase of 15-, 9-, and 11.4-fold, respectively, over that obtained with NGF alone (p < 0.0001). The expression of DYRK1A-K188R together with Ras, B-Raf, or MEK1 resulted in a similar increase of 13.5-, 7.8-, and 9.6-fold, respectively (p < 0.0001), which was not significantly different from the one observed with the expression of DYRK1A (p > 0.05). Thus, these findings indicate a kinase-independent synergistic effect of DYRK1A on Ras-mediated, B-Raf-mediated, and MEK1-mediated ERK-dependent Elk1 activation.
Figure 3.
DYRK1A acts synergistically with Ras, B-Raf, and MEK1 to increase ERK-dependent Elk1 activation. PC12 cells were cotransfected with 1 μg of 5x-Gal4-TATA/luciferase, 70 ng of Gal4-Elk1 (Elk/gal), 1 μg of CMV-βGal, and the indicated expression plasmids. Cells were serum starved for 20 h and stimulated with 100 ng/ml NGF for 6 h before being collected (48 h posttransfection). Activation of Elk1 is reflected by luciferase activity normalized to βGal activity and is described as fold increase. Error bars represent means ± SEM. For comparisons indicated by brackets, three asterisks indicate a p value <0.0001. Values obtained with DYRK1A and DYRK1A-K188R were not significantly different (p > 0.05).
DYRK1A Interacts with Ras
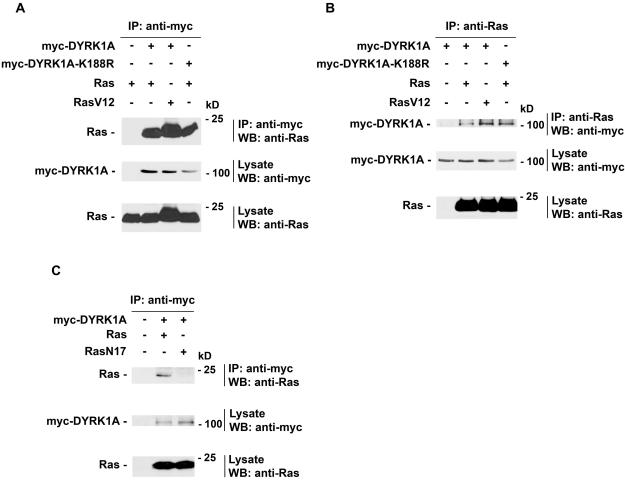
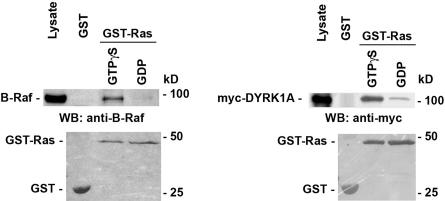
The cooperative effect observed between DYRK1A and Ras led us to test whether they could form a complex in vivo. For this, wild-type Ras was coexpressed with myc-DYRK1A or myc-DYRK1A-K188R in PC12 cells. Cell lysates of transfected cells were analyzed by coimmunoprecipitation and protein immunoblotting with specific antibodies to myc-tagged DYRK1A constructs and Ras. DYRK1A and DYRK1A-K188R coprecipitated with wild-type Ras (Figure 4A), and reciprocal immunoprecipitation confirmed the interaction between DYRK1A and Ras (Figure 4B). DYRK1A also interacted with RasV12, an activated form of Ras which is stabilized in the GTP-bound state (reviewed in Wittinghofer and Nassar, 1996) (Figure 4, A and B). However, DYRK1A did not associate with the dominant negative form of Ras, RasN17 (Figure 4C). RasN17 have been shown to have a preferential affinity for GDP versus GTP binding in vitro (Feig and Cooper, 1988) and in vivo (Stewart and Guan, 2000). We further examined this interaction in vitro. Recombinant GST-Ras protein was loaded with GTPγS, a nonhydrolysable GTP analog, or guanosine 5′-diphosphate (GDP) and incubated with protein extracts from untransfected PC12 cells or PC12 cells transfected with myc-DYRK1A. The amount of B-Raf or myc-DYRK1A bound proteins was determined by immunoblotting with anti-B-Raf or anti-myc tag antibodies. B-Raf was used as a positive control for the experiment. As shown in Figure 5, myc-DYRK1A interacted with the GTPγS-bound form of GST-Ras preferentially over the GDP-bound form of GST-Ras. myc-DYRK1A did not interact with the control GST protein. As expected, B-Raf interacted only with the GTPγS-bound form of GST-Ras. These data indicate that DYRK1A preferentially binds to Ras in its active GTP-bound conformation.
Figure 4.
DYRK1A associates with Ras. PC12 cells were transiently transfected with the described constructs and analyzed by coimmunoprecipitation (IP) followed by Western blotting (WB) with anti-myc or anti-Ras. (A and B) DYRK1A and DYRK1A-K188R interacted with Ras and RasV12. As a control, myc-tag antibody coimmunoprecipitated Ras in cells expressing myc-DYRK1A and Ras but not in cells expressing Ras alone (A, lane 1). (C) DYRK1A did not interact with RasN17. Whole cell lysates of transfected cells were analyzed by WB with the appropriate antibody to verify equal expression of all constructs. The results shown are representative of three independent experiments. Note that in the lysate of untransfected PC12 cells (Figure 4, B and C, bottom, lane 1), a signal corresponding to the endogenous Ras could be seen when the immunoblot with anti-Ras was exposed a longer time (our unpublished data). Sizes of molecular weight markers are indicated on the right.
Figure 5.
DYRK1A preferentially interacts with GTP-bound Ras. Lysate from PC12 cells untransfected or transfected with myc-DYRK1A were incubated with the glutathione-Sepharose resin carrying GTPγS-bound, GDP-bound forms of GST-Ras or GST. B-Raf and myc-DYRK1A were detected by Western immunoblotting with anti-B-Raf and anti-myc antibodies, respectively (top). GST-fusion proteins were detected by Ponceau S staining of the polyvinylidene difluoride membrane (bottom). These experiments were repeated three times, and representative data are shown. Sizes of molecular weight markers are indicated on the right.
DYRK1A Associates with B-Raf
Because the effect of DYRK1A on Ras/MAP kinase signaling pathway was observed only in neuronal cells that specifically express high levels of B-Raf, we addressed the possibility that DYRK1A might interact with B-Raf. PC12 cells were transiently cotransfected with myc-tagged DYRK1A or DYRK1A-K188R and HA-tagged B-Raf expression plasmids. Cell lysates were analyzed by coimmunoprecipitation and protein immunoblotting as described previously. As shown in Figure 6, A and B, B-Raf interacted with DYRK1A as well as DYRK1A-K188R, indicating that the kinase activity of DYRK1A is not necessary for DYRK1A/B-Raf interaction. However, DYRK1A did not interact with Raf1 (Figure 6C), which indicates that DYRK1A associates specifically with B-Raf.
The inability of DYRK1A to activate the Ras/MAP kinase pathway in HeLa (Figure 2B), Cos-7 (our unpublished data) and NIH 3T3 cells (Mao et al., 2002) could be explained by the fact that DYRK1A does not interact with Raf1, whereas it does interact with the neuron-specific Raf isoform B-Raf. Indeed, HeLa, Cos-7, and NIH 3T3 cells predominantly express Raf1 but not B-Raf, whereas PC12 cells express high levels of both Raf isoforms (Vossler et al., 1997). To address this hypothesis, HeLa cells were transiently cotransfected with HA-B-Raf and myc-DYRK1A expression plasmids in combination with reporter plasmids that read out ERK-dependent activation of the transcription factor Elk1 and processed as described above. As shown in Figure 6D, expression of B-Raf by itself did not result in a significant Elk1 activation (p > 0.05). However, coexpression of B-Raf and DYRK1A led to a significant induction of ERK-dependent Elk1 activation of threefold (p < 0.001). These data indicate that DYRK1A uses the specific neuronal isoform B-Raf to enhance ERK-dependent Elk1 activation.
DYRK1A Interacts with MEK1
Because DYRK1A associates with Ras and B-Raf, we asked whether DYRK1A interacts with MEK1. To test this hypothesis, PC12 cells were transiently transfected with myc-tagged DYRK1A, or myc-tagged DYRK1A-K188R together with HA-tagged MEK1, and analyzed by coimmunoprecipitation as described above. As shown in Figure 7, lanes 1 and 2, DYRK1A interacted with MEK1. MEK1 has a consensus DYRK1A substrate sequence (RPRT292P) (Becker et al., 1998; Himpel et al., 2000) in its C-terminal proline-rich region (aa 270–307). Therefore, we tested whether DYRK1A could interact with HA-MEK1Δ270–307, an MEK1 deletion mutant that lacks the C-terminal proline-rich sequence. DYRK1A still interacted with MEK1Δ270-307 mutant (Figure 7, lane 4). Moreover, the kinase-deficient mutant DYRK1A-K188R also associated with MEK1 and MEK1Δ270-307 mutant (Figure 7, lanes 3 and 5), indicating that the effect of DYRK1A on MEK1-mediated ERK-dependent Elk1 activation was independent of its kinase activity.
Figure 7.
DYRK1A associates with MEK1. PC12 cells were transiently transfected with the described constructs and analyzed by coimmunoprecipitation (IP) followed by Western blotting (WB) with anti-myc or anti-HA. As a control, myc-tag antibody coimmunoprecipitated HA-MEK1A in cells expressing myc-DYRK1A and HA-MEK1 but not in cells expressing HA-MEK1 alone (lane 1). Whole cell lysates of transfected cells were analyzed by WB with the appropriate antibody to verify equal expression of all constructs. The results shown are representative of three independent experiments. Sizes of molecular weight markers are indicated on the right.
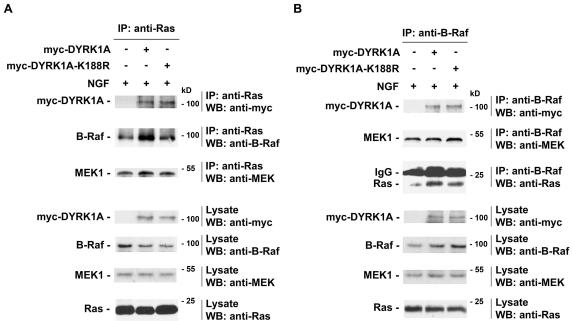
DYRK1A Associates with Ras, B-Raf, and MEK1 in PC12 Cells
Because DYRK1A interacted independently with Ras, B-Raf, and MEK1, we next examined whether DYRK1A also existed as part of a multiprotein complex. PC12 cells were transiently transfected with an empty control vector, myc-DYRK1A, or myc-DYRK1A-K188R expression vectors. Transfected cells were serum starved for 20 h and then stimulated with NGF for 5 min before being collected and lysed. Endogenous Ras or B-Raf immunoprecipitates were then examined for the presence of associated myc-DYRK1A protein and endogenous MEK1, B-Raf, and Ras proteins, respectively. Endogenous Ras was found to be associated with endogenous B-Raf in control-, DYRK1A-, or DYRK1A-K188R–transfected cells (Figure 8A). myc-DYRK1A and myc-DYRK1A-K188R were found to be associated with endogenous Ras in DYRK1A- and DYRK1A-K188R–transfected cells, respectively (Figure 8A). However, Ras/B–Raf association was significantly enhanced in DYRK1A- or DYRK1A-K188R–transfected cells (Figure 8, A and B). MEK1 was detected as part of the Ras/B-Raf complex in control-, DYRK1A-, or DYRK1A-K188R–transfected stimulated cells (Figure 8A). Reciprocal immunoprecipitation confirmed the fact that endogenous B-Raf interacted with endogenous MEK1 and Ras in control-, DYRK1A-, or DYRK1A-K188R–transfected cells (Figure 8B). Endogenous B-Raf interacted with myc-DYRK1A and myc-DYRK1A-K188R in DYRK1A- and DYRK1A-K188R–transfected cells, respectively (Figure 8B). However, the amount of MEK1 protein associated with endogenous Ras or endogenous B-Raf in DYRK1A or DYRK1A-K188R–transfected cells was not enhanced (Figure 8, A and B). Therefore, the association between Ras and B-Raf in cells treated with NGF can be significantly enhanced by DYRK1A or DYRK1A-K188R, whereas B-Raf interaction with MEK1 in cells treated with NGF cannot be enhanced by DYRK1A or DYRK1A-K188R. Together, these findings demonstrate that ectopic expression of DYRK1A enhances the formation of a DYRK1A/Ras/B–Raf/MEK1 multiprotein complex, which leads to sustained ERK activation and increased PC12 neuronal differentiation. Moreover, these effects are independent of DYRK1A kinase activity.
Figure 8.
DYRK1A enhances the formation of a multiprotein complex containing Ras, B-Raf, and MEK1 proteins. (A and B) PC12 cells were transiently transfected with either an empty control vector, myc-DYRK1A, or myc-DYRK1A-K188R expression plasmids. Transfected cells were serum starved for 20 h and then stimulated with 50 ng/ml NGF for 5 min before being collected (48 h posttransfection). Cell lysates were analyzed by coimmunoprecipitation (IP) followed by Western blotting (WB) with the indicated antibodies. Whole cell lysates of transfected cells were analyzed by WB with the appropriate antibody to verify equal expression of proteins in each condition. The results shown are representative of three independent experiments. Sizes of molecular weight markers are indicated on the right.
Down-Regulation of Endogenous DYRK1A Affects NGF-mediated Elk1 Transactivation
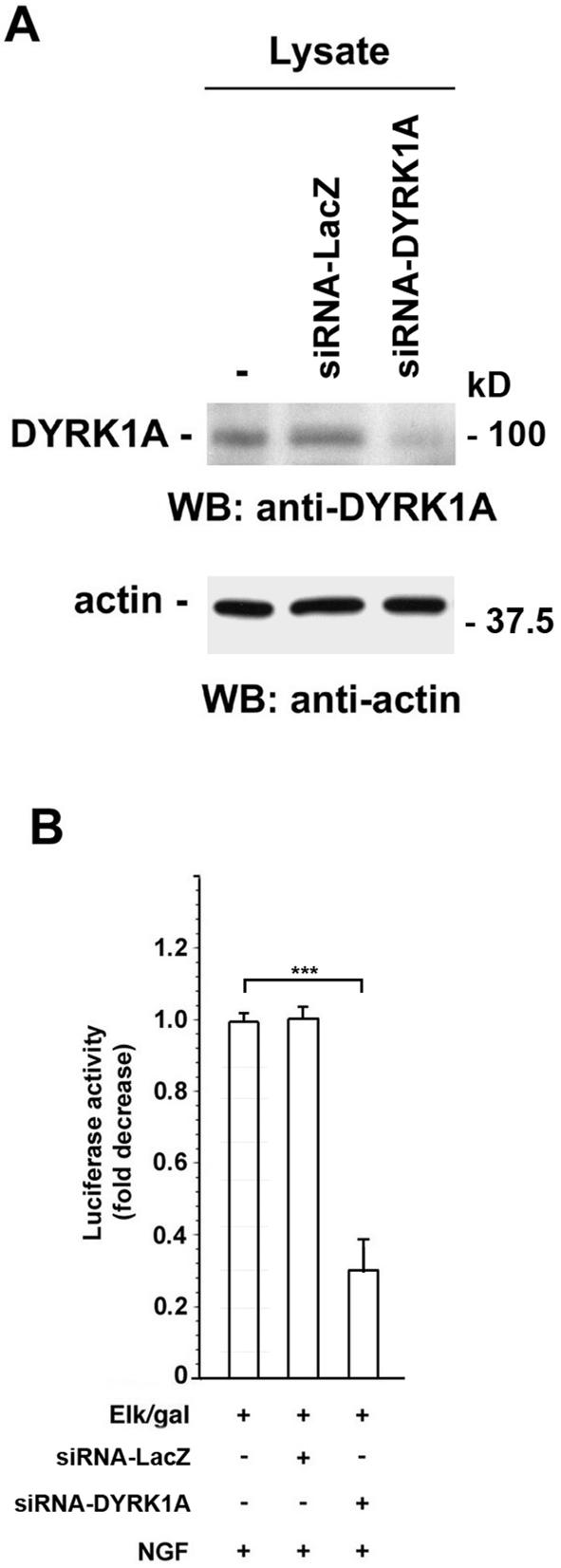
To investigate the biological function of endogenous DYRK1A on NGF-mediated Elk1 activation, endogenous expression of DYRK1A was inhibited by RNAi. PC12 cells were transiently transfected with either siRNAs directed against LacZ as a negative control (siRNA-LacZ) or DYRK1A (siRNA-DYRK1A). Transfected cells were lysed and immunoblotted with anti-DYRK1A antibody. As shown in Figure 9A, expression of endogenous DYRK1A in DYRK1A-siRNA–transfected cells was strongly decreased in comparison with control LacZ-siRNA–transfected cells or vector-transfected cells. The effect of down-regulation of endogenous DYRK1A on NGF-mediated Elk1 transactivation was assessed using luciferase transactivation assays as described above (Figure 2A). A shown in Figure 9B, down-regulation of endogenous DYRK1A expression by DYRK1A-siRNA reduced the stimulatory effect of NGF on Elk1 activation by 70% (p < 0.0001), whereas expression of LacZ siRNA (control) did not show any effect (p > 0.05). These data suggest that the potentiating effect of DYRK1A on NGF-mediated Elk1 activation also is mediated by endogenous DYRK1A.
Figure 9.
Effect of endogenous DYRK1A down-regulation on NGF-mediated Elk1 activation. (A) PC12 cells were transiently transfected with an empty control vector, siRNA for LacZ, or siRNA for DYRK1A. Forty-eight hours posttransfection, total cell lysates were analyzed by Western blot for the expression of endogenous DYRK1A by using DYRK1A antibody. The membrane was stripped and reprobed with anti-actin antibody to verify equal loading of proteins in each condition. Sizes of molecular weight markers are indicated on the right. (B) PC12 cells were cotransfected with 1 μg of 5x-Gal4-TATA/luciferase, 70 ng of Gal4-Elk1 (Elk/gal), 1 μg of CMV-βGal, and an empty control vector or the indicated siRNA (200 pmol) as indicated. Transfected cells were serum starved for 20 h and then stimulated with 100 ng/ml NGF for 6 h before being collected (48 h posttransfection). Activation of Elk1 reflected luciferase activity normalized to βGal activity and is described as fold decrease. The value obtained with the empty control vector was set to 1. Bars represent means ± SEM. For comparisons indicated in brackets, three asterisks indicate a p value <0.0001. Values obtained with siRNA-LacZ were not significantly different from the ones obtained with the empty control vector (p > 0.05).
DISCUSSION
This study shows that transient expression of DYRK1A potentiates the neuronal differentiation of PC12 cells as evidenced by increased neurite outgrowth and neuronal protein expression. Luciferase reporter assays showed that DYRK1A expression up-regulates Ras/MAP kinase signaling in a cell type-specific manner and that NGF stimulation has a synergistic effect on this up-regulation. NGF-mediated PC12 cell differentiation correlates with a sustained activation of the Ras/MAP kinase pathway (Qui and Green, 1992). Based on our observations, the ability of DYRK1A to augment neurite outgrowth could be mediated by the Ras/MAP kinase pathway. First, ERK-dependent Elk1 activation used as a read out for monitoring ERK activation is blocked by the MEK inhibitor PD98059 and by expression of a dominant negative form of Ras, RasN17. Second, the sustained ERK activation observed in response to NGF treatment lasts longer in DYRK1A-transfected cells than in control-transfected cells. Third, down-regulation of endogenous DYRK1A expression in PC12 cells by expression of siRNA directed against DYRK1A inhibits NGF-mediated Elk1 activation.
Our data show that the kinase activity of DYRK1A is not necessary for the enhancing effect of DYRK1A on NGF-mediated ERK-dependent Elk1 activation. Similarly, DYRK1A has been shown to stimulate FKHR-mediated transactivation of the glucose-6-phosphatase gene independently of its kinase activity (von Groote-Bidlingmaier et al., 2003). More recently, DYRK1A was found to stimulate steroid hormone-induced transactivation by interacting with Arip4 in a kinase-independent manner (Sitz et al., 2004). Although DYKR1A has been shown to phosphorylate several proteins in vitro (reviewed in Galceran et al., 2003; Hämmerle et al., 2003b), these findings suggest that DYRK1A also may possess kinase-independent biological functions.
The data reported herein demonstrate that DYRK1A interacts preferentially with GTP-bound Ras. Furthermore, DYRK1A associates with B-Raf. This interaction seems to be specific to B-Raf because DYRK1A does not bind to Raf1, another Raf isoform. Moreover, we showed that the inability of DYRK1A to activate the Ras/MAP kinase signaling pathway in HeLa or Cos-7 cells can be explained by the fact that DYRK1A is unable to associate with Raf1, whereas it does interact with the neuron-specific Raf isoform, B-Raf. Finally, DYRK1A associates also with MEK1 in a kinase-independent manner. It is not clear whether B-Raf directly interacts with DYRK1A. Indeed, DYRK1A has been shown to interact with 14-3-3, an adapter protein (Kim et al., 2004). Moreover, 14-3-3 interacts with B-Raf (Papin et al., 1996). Therefore, 14-3-3 may serve as a bridge between DYRK1A and B-Raf. Further studies will be required to address this issue.
The Ras/MAP kinase pathway is a key signaling cascade in eukaryotic cells. Its function has been implicated in many diverse signal transduction events ranging from cell growth, differentiation, and transformation (reviewed in Robinson and Cobb, 1997). This kinase cascade seems to be spatially organized in a signaling complex that is nucleated by Ras proteins (Moodie et al., 1993). The regulation of the Ras/Raf/MEK/ERK module is complex and may include associations with scaffolding and regulatory proteins (reviewed in Kolch, 2000). Our data suggest a model whereby DYRK1A sustains ERK activation in PC12 cells by facilitating the formation of a complex with Ras, B-Raf, and MEK1. By overexpressing DYRK1A in PC12 cells, additional DYRK1A–Ras-B–Raf–MEK1 complexes might form. This would then accelerate and extend the activation of ERK such that the critical threshold of MAPK activity is achieved more rapidly in cells stimulated with NGF. DYRK1A also could recruit another protein involved in differentiation and/or MAPK activation. B-KSR1, a brain-specific variant of KSR1, has been shown to form a complex with MEK and ERK in PC12 cells and to enhance ERK activation and NGF-mediated neuronal differentiation (Muller et al., 2000). Because DYRK1A does not interact with ERK (Rahmani, unpublished observation), it will be interesting to investigate whether B-KSR1 may form a bridge between DYRK1A and MEK1 to orchestrate the formation of a complete Ras–B-Raf–MEK–ERK module. Nevertheless, these data suggest that DYRK1A may function in promoting or maintaining a differentiated phenotype by bringing Ras, B-Raf, and MEK factors together in a protein complex to enhance signaling efficiency.
In Drosophila, DYRK1A is required for the proliferation of distinct neuronal cell types during postembryonic development (Tejedor et al., 1995). In situ hybridization studies show that DYRK1A is highly expressed in brain gray matter, spinal cord, and retina in developing murine embryos (Song et al., 1996; Rahmani et al., 1998; Hämmerle et al., 2003a; Mao et al., 2003; Marti et al., 2003) and in the cerebral cortex, cerebellum, and pyramidal cell layer in the hippocampus in adult mice (Guimera et al., 1996). DYRK1A has been considered a good candidate gene for the Down syndrome phenotypic abnormalities (reviewed in Epstein, 2000; Vicari et al., 2000; Nadel, 2003) due to its localization in the Down syndrome critical region of chromosome 21 (Rahmani et al., 1989, 1990; Delabar et al., 1993; Shindoh et al., 1996; Song et al., 1996), its overexpression in the brain of Down syndrome patients (Guimera et al., 1999), and the neurobehavioral alterations shown by transgenic mice overexpressing the gene (Smith and Rubin, 1997; Smith et al., 1997; Altafaj et al., 2001). The recent observation that the size of dendrites in some brain areas is reduced in DYRK1A haploinsufficient mice indicates that DYRK1A dose reduction could affect the length and the complexity of the dendrites (Fotaki et al., 2002). Moreover, brains of patients with Down syndrome present size reduction, increased astrocyte number, delayed myelination, and several abnormal neuronal differentiation processes (reviewed in Coyle et al., 1986; Becker et al., 1991; Raz et al., 1995). Thus, our finding that overexpression of DYRK1A potentiates the neurite outgrowth of PC12 cells stimulated with NGF supports the fact that DYRK1A may be involved in signaling mechanisms that regulate dendritic differentiation.
In conclusion, the high levels of DYRK1A expression in neurons, together with the multicomplex formation of DYRK1A with Ras, B-Raf, and MEK1 in neuronal cells, indicate that DYRK1A may play a critical role in Ras-dependent transducing signals that are required for promoting or maintaining neuronal differentiation or that may be involved in the normal functioning of the central nervous system. Interestingly, the hippocampus, which is a brain region that plays a critical role in learning and memory, has been shown to be affected in Down syndrome patients (Vicari et al., 2000). This raises the possibility that DYRK1A may participate in the regulation of critical cellular events occurring in the adult brain, such as synaptic plasticity required for learning and memory.
Acknowledgments
We thank L. Feig, K. L. Guan, and M. J. Weber for generous gifts of plasmids. We thank A. K. Sobering and F. Schweisguth for critical reading of the manuscript. This work was supported in part by the Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Université René Descartes and Faculté de Médecine Necker-Enfants Malades, Association Française contre les Myopathies, Fondation Jérôme Lejeune, Institut Necker, and Ecole Normale Supérieure.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1085) on May 25, 2005.
Abbreviations used: DYRK1A, dual-specificity tyrosine-phosphorylated and regulated kinase 1A; FKHR, Forkhead in rhabdomyosarcoma; GDP, guanosine 5′-diphosphate; GST, glutathione S-transferase; GTPγS, 5′-O-(3-thio)triphosphate; MAPK, mitogen-activated protein kinase; mnb, minibrain; NGF, nerve growth factor; siRNA, small interfering RNA.
References
- Altafaj, X., et al. (2001). Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum. Mol. Genet. 10, 1915–1923. [DOI] [PubMed] [Google Scholar]
- Bahler, J., and Pringle, J. R. (1998). Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 12, 1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, L., Mito, T., Yakashima, S., and Onodera, K. (1991). Growth and development of the brain in Down syndrome. In: The Morphogenesis of Down Syndrome, New York: Wiley-Liss, 133–152. [PubMed]
- Becker, W., and Joost, H. G. (1999). Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog. Nucleic Acids Res. Mol. Biol. 62, 1–17. [DOI] [PubMed] [Google Scholar]
- Becker, W., Weber, Y., Wetzel, K., Eirmbter, K., Tejedor, F. J., and Joost, H. G. (1998). Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J. Biol. Chem. 273, 25893–25902. [DOI] [PubMed] [Google Scholar]
- Catling, A. D., Schaeffer, H.-J., Reuter, C.W.M., Reddy, G. R., and Weber, M. J. (1995). A proline-rich sequence unique to MEK1 and MEK2 is required for Raf binding and regulates MEK function. Mol. Cell. Biol. 15, 5214–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Hwang, M. C., Chen, H. R., Elzinga, M., and Hwang, Y. W. (2002). Dynamin is a minibrain kinase/dual specificity Yak1-related kinase 1A substrate. J. Biol. Chem. 277, 17597–17604. [DOI] [PubMed] [Google Scholar]
- Coyle, J. T., Oster-Granite, M. L., and Gearhart, J. D. (1986). The neurobiologic consequences of Down syndrome. Brain Res. Bull. 16, 773–787. [DOI] [PubMed] [Google Scholar]
- De Graaf, K., Hekerman, P., Spelten, O., Herrmann, A., Packman, L. C., Bussow, K., Muller-Newen, G., and Becker, W. (2004). Characterization of cyclin L2, a novel cyclin with an arginine/serine-rich (RS) domain: phosphorylation by DYRK1A and colocalization with splicing factors. J. Biol. Chem. 279, 4612–4624. [DOI] [PubMed] [Google Scholar]
- Delabar, J. M., Theophile, D., Rahmani, Z., Chettouh, Z., Blouin, J. L., Prieur, M., Noel, B., and Sinet, P. M. (1993). Molecular mapping of twenty-four features of Down syndrome on chromosome 21. Eur. J. Hum. Genet. 1, 114–124. [DOI] [PubMed] [Google Scholar]
- Epstein, C. J. (2000). Down syndrome (trisomy 21). In: The Metabolic and Molecular Bases of Inherited Disease, ed. C. R. Scriver and W. S. Sly, New York: McGraw-Hill Book Co., 1223–1256
- Feig, L. A., and Cooper, G. M. (1988). Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol. Cell. Biol. 8, 3235–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotaki, V., Dierssen, M., Alcantara, S., Martinez, S., Marti, E., Casas, C., Visa, J., Soriano, E., Estivill, X., and Arbones, M. L. (2002). Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol. Cell. Biol. 22, 6636–6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galceran, J., De Graaf, K., Tejedor, F. J., and Becker, W. (2003). The MNB/DYRK1A protein kinase: genetic and biochemical properties. J. Neural Transm. Suppl. 67, 139–148. [DOI] [PubMed] [Google Scholar]
- Garrett, S., and Broach, J. (1989). Loss of Ras activity in saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 3, 1336–1348. [DOI] [PubMed] [Google Scholar]
- Greene, L. A., and Tischler, A. S. (1976). Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA 73, 2424–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera, J., Casas, C., Estivill, X., and Pritchard, M. (1999). Human minibrain homologue (MNBH/DYRK1): characterization, alternative splicing, differential tissue expression, and overexpression in Down syndrome. Genomics 57, 407–418. [DOI] [PubMed] [Google Scholar]
- Guimera, J., Casas, C., Pucharcos, C., Solans, A., Domenech, A., Planas, A. M., Ashley, J., Lovett, M., Estivill, X., and Pritchard, M. A. (1996). A human homologue of Drosophila minibrain (MNB) is expressed in the neuronal regions affected in Down syndrome and maps to the critical region. Hum. Mol. Genet. 5, 1305–1310. [DOI] [PubMed] [Google Scholar]
- Hämmerle, B., Carnicero, A., Elizalde, C., Ceron, J., Martinez, S., and Tejedor, F. J. (2003a). Expression patterns and subcellular localization of the Down syndrome candidate protein MNB/DYRK1A suggest a role in late neuronal differentiation. Eur. J. Neurosci. 17, 2277–2286. [DOI] [PubMed] [Google Scholar]
- Hämmerle, B., Elizalde, C., Galceran, J., Becker, W., and Tejedor, F. J. (2003b). The MNB/DYRK1A protein kinase: neurobiological functions and Down syndrome implications. J. Neural Transm. Suppl. 67, 129–137. [DOI] [PubMed] [Google Scholar]
- Himpel, S., Tegge, W., Frank, R., Leder, S., Joost, H. G., and Becker, W. (2000). Specificity determinants of substrate recognition by the protein kinase DYRK1A. J. Biol. Chem. 275, 2431–2438. [DOI] [PubMed] [Google Scholar]
- Kentrup, H., Becker, W., Heukelbach, J., Wilmes, A., Schrmann, A., Huppertz, C., Kainulainen, H., and Joost, H. G. (1996). Dyrk, a dual specificity protein kinase with unique structural features. whose activity is dependent on tyrosine residues between subdomains VII and VIII. J. Biol. Chem. 271, 3488–3495. [DOI] [PubMed] [Google Scholar]
- Kim, D., et al. (2004). Regulation of Dyrk1A kinase activity by 14–3-3. Biochem. Biophys. Res. Commun. 323, 499–504. [DOI] [PubMed] [Google Scholar]
- Kolch, W. (2000). Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351, 289–305. [PMC free article] [PubMed] [Google Scholar]
- Li, W., Han, M., and Guan; K.-L. (2000). The leucine-rich repeat protein SUR-8 enhances MAPK kinase activation and forms a complex with Ras and Raf. Genes Dev. 14, 895–900. [PMC free article] [PubMed] [Google Scholar]
- Mao, J., Maye, P., Kogerman, P., Tejedor, F. J., Toftgard, R., Xie, W., Wu, G., and Wu, D. (2002). Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J. Biol. Chem. 277, 35156–35161. [DOI] [PubMed] [Google Scholar]
- Mao, R., Zielke, C. L., Zielke, H. R., and Pevsner, J. (2003). Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics 81, 457–467. [DOI] [PubMed] [Google Scholar]
- Marti, E., Altafaj, X., Dierssen, M., de la Luna, S., Fotaki, V., Alvarez, M., Perez-Riba, M., Ferrer, I., and Estivill, X. (2003). Dyrk1A expression pattern supports specific roles of this kinase in the adult central nervous system. Brain Res. 964, 250–263. [DOI] [PubMed] [Google Scholar]
- Matsuo, R., Ochiai, W., Nakashima, K., and Taga, T. (2001). A new expression cloning strategy for isolation of substrate-specific kinases by using phosphorylation site-specific antibody. J. Immunol. Methods 247, 141–151. [DOI] [PubMed] [Google Scholar]
- Moodie, S. A., Willumsen, B. M., Weber, M. J., and Wolfman, A. (1993). Complexes of Ras/GTP with Raf-1 and mitogen-activated protein kinase kinase. Science 260, 1658–1661. [DOI] [PubMed] [Google Scholar]
- Muller, J., Cacace, A. M., Lyons, W. E., Mc Gill, C. B., and Morrison, D. K. (2000). Identification of B-KSR1, a novel brain specific isoform of KSR1 that functions in neuronal signalling. Mol. Cell. Biol. 20, 5529–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadel, L. (2003). Down's syndrome: a genetic disorder in biobehavioral perspective. Genes Brain Behav. 2, 156–166. [DOI] [PubMed] [Google Scholar]
- Papin, C., Denouel, A., Calothy, G., and Eychène, A. (1996). Identification of signalling proteins interacting with B-Raf in the yeast two-hybrid system. Oncogene 12, 2213–2221. [PubMed] [Google Scholar]
- Qui, M. S., and Green, S. H. (1992). PC12 cell neuronal differentiation is associated with prolonged p21ras activity and consequent prolonged ERK activity. Neuron 9, 705–717. [DOI] [PubMed] [Google Scholar]
- Rahmani, Z., Blouin, J. L., Creau-Goldberg, N., Watkins, P. C., Mattei, J. F., Poissonnier, M., Prieur, M., Chettouh, Z., Nicole, A., and Aurias, A. (1989). Critical role of the D21S55 region on chromosome 21 in the pathogenesis of Down syndrome. Proc. Natl. Acad. Sci. USA 86, 5958–5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani, Z., Blouin, J. L., Creau-Goldberg, N., Watkins, P. C., Mattei, J. F., Poissonnier, M., Prieur, M., Chettouh, Z., Nicole, A., and Aurias, A. (1990). Down syndrome critical region around D21S55 on proximal 21q22.3. Am. J. Med. Gene Suppl. 7, 98–103. [DOI] [PubMed] [Google Scholar]
- Rahmani, Z., Lopes, C., Rachidi, M., and Delabar, J. M. (1998). Expression of the mnb (dyrk) protein in adult and embryonic mouse tissues. Biochem. Biophys. Res. Commun. 253, 514–518. [DOI] [PubMed] [Google Scholar]
- Raich, W. B., Moorman, C., Lacefield, C. O., Lehrer, J., Bartsch, D., Plasterk, R.H.A., Kandel, E. R., and Hobert, O. (2003). Characterization of Caenorhabditis elegans homologs of the Down syndrome candidate gene DYRK1A. Genetics 163, 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz, N., Torres, I. J., Briggs, S. D., Spencer, W. D., Thornton, A. E., Loken, W. J., Gunning, F. M., McQuain, J. D., Driesen, N. R., and Acker, J. D. (1995). Selective neuroanatomic abnormalities in Down's syndrome and their cognitive correlates: evidence from MRI morphometry. Neurology 45, 356–366. [DOI] [PubMed] [Google Scholar]
- Robinson, M. J., and Cobb, M. H. (1997). Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9, 180–186. [DOI] [PubMed] [Google Scholar]
- Shindoh, N., Kudoh, J., Maeda, H., Yamaki, A., Minoshima, S., Shimizu, Y., and Shimizu, N. (1996). Cloning of a human homolog of the Drosophila minibrain/rat Dyrk gene from the Down syndrome critical region of chromosome 21. Biochem. Biophys. Res. Commun. 225, 92–99. [DOI] [PubMed] [Google Scholar]
- Sitz, J. H., Tigges, M., Baumgärtel, K., Khaspekov, L. G., and Lutz, B. (2004). Dyrk1A potentiates steroid hormone-induced transcription via the chromatin remodeling factor Arip4. Mol. Cell. Biol. 24, 5821–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurat, A. V., and Dietrich, A. D. (2004). Phosphorylation of Ser640 in muscle glycogen synthase by DYRK family protein kinases. J. Biol. Chem. 279, 2490–2498. [DOI] [PubMed] [Google Scholar]
- Smith, D. J., and Rubin, E. M. (1997). Functional screening and complex traits: human 21q22.2 sequences affecting learning in mice. Hum. Mol. Genet. 6, 1729–1733. [DOI] [PubMed] [Google Scholar]
- Smith, D. J., et al. (1997). Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat. Genet. 16, 28–36. [DOI] [PubMed] [Google Scholar]
- Song, W. J., Chung, S. H., and Kurnit, D. M. (1997). The murine Dyrk protein maps to chromosome 16, localizes to the nucleus, and can form multimers. Biochem. Biophys. Res. Commun. 231, 640–644. [DOI] [PubMed] [Google Scholar]
- Song, W. J., et al. (1996). Isolation of human and murine homologues of the Drosophila minibrain gene: human homologue maps to 21q22.2 in the Down syndrome “critical region.” Genomics 38, 331–339. [DOI] [PubMed] [Google Scholar]
- Stewart, S., and Guan, K. L. (2000). The dominant negative Ras mutant, N17Ras, can inhibit signalling independently of blocking Ras activation. J. Biol. Chem. 275, 8854–8862. [DOI] [PubMed] [Google Scholar]
- Sugimoto, T., Stewart, S., Han, M., and Guan, K.-L. (1997). The kinase suppressor of Ras (KSR) modulates growth factor and Ras signaling by uncoupling elk-1 phosphorylation from MAP kinase activation. EMBO J. 17, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejedor, F., Zhu, X. R., Kaltenbach, E., Ackermann, A., Baumann, A., Canal, I., Heisenberg, M., Fischbach, K. F., and Pongs, O. (1995). Ninibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron 14, 287–301. [DOI] [PubMed] [Google Scholar]
- Van Es, S., Weening, K. E., and Devreotes, P. N. (2001). The protein YakA regulates g-protein-linked signaling responses during growth and development of Dictyostelium. J. Biol. Chem. 276, 30761–30765. [DOI] [PubMed] [Google Scholar]
- Vicari, S., Bellucci, S., and Carlesimo, G. A. (2000). Implicit and explicit memory: a functional dissociation in persons with Down syndrome. Neuropsychologia 38, 240–251. [DOI] [PubMed] [Google Scholar]
- von Groote-Bidlingmaier, F., Schmoll, D., Orth, H. M., Joost, H. G., Becker, W., and Barthel, A. (2003). DYRK1 is a co-activator of FKHR (FOXO1a)-dependent glucose-6-phosphatase gene expression. Biochem. Biophys. Res. Commun. 300, 764–769. [DOI] [PubMed] [Google Scholar]
- Vossler, M. R., Yao, H., York, R. D., Pan, M. G., Rim, C. S., and Stork, P. J. (1997). cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell 89, 73–82. [DOI] [PubMed] [Google Scholar]
- Wittinghofer, A., and Nassar, N. (1996). How Ras-related proteins talk to their effectors. Trends Biochem. Sci. 21, 488–491. [DOI] [PubMed] [Google Scholar]
- Woods, Y. L., Cohen, P., Becker, W., Jakes, R., Goedert, M., Wang, X., and Proud, C. G. (2001a). The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bepsilon at Ser539 and the microtubule-associated protein tau at Thr 212, potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem. J. 355, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods, Y. L., Rena, G., Morrice, N., Barthel, A., Becker, W., Guo, S., Unterman, T. G., and Cohen, P. (2001b). The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem. J. 355, 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, E. J., Ahn, Y. S., and Chung, K. C. (2001). Protein kinase Dyrk1 activates cAMP response element-binding protein during neuronal differentiation in hippocampal progenitor cells. J. Biol. Chem. 276, 39819–39824. [DOI] [PubMed] [Google Scholar]