Abstract
In Aspergillus nidulans, cytoplasmic dynein and NUDF/LIS1 are found at the spindle poles during mitosis, but they seem to be targeted to this location via different mechanisms. The spindle pole localization of cytoplasmic dynein requires the function of the anaphase-promoting complex (APC), whereas that of NUDF does not. Moreover, although NUDF's localization to the spindle poles does not require a fully functional dynein motor, the function of NUDF is important for cytoplasmic dynein's targeting to the spindle poles. Interestingly, a γ-tubulin mutation, mipAR63, nearly eliminates the localization of cytoplasmic dynein to the spindle poles, but it has no apparent effect on NUDF's spindle pole localization. Live cell analysis of the mipAR63 mutant revealed a defect in chromosome separation accompanied by unscheduled spindle elongation before the completion of anaphase A, suggesting that γ-tubulin may recruit regulatory proteins to the spindle poles for mitotic progression. In A. nidulans, dynein is not apparently required for mitotic progression. In the presence of a low amount of benomyl, a microtubule-depolymerizing agent, however, a dynein mutant diploid strain exhibits a more pronounced chromosome loss phenotype than the control, indicating that cytoplasmic dynein plays a role in chromosome segregation.
INTRODUCTION
The minus-end-directed microtubule motor cytoplasmic dynein has been implicated in a variety of cellular functions, including mitosis, transport of vesicles, and distribution of nuclei and other organelles (Karki and Holzbaur, 1999). Cytoplasmic dynein is a multisubunit complex with heavy chains, intermediate chains, light intermediate chains, and light chains (King, 2000; Tynan et al., 2000). The heavy chains contain the motor activity, whereas the other components have been implicated in targeting the motor to various cargoes. The functions of cytoplasmic dynein require dynactin, another multisubunit complex that may link the motor with membranous cargos, and, in addition, enhance the processivity of the dynein motor on microtubules (Holleran et al., 1998; King and Schroer, 2000; Schroer, 2004). Besides the components of the cytoplasmic dynein and dynactin complexes, additional regulators of cytoplasmic dynein have been discovered. One of them is LIS1, the product of the causal gene for type I lissencephaly, a human disease characterized by brain malformation due to a defect in neuronal migration (Reiner, 2000; Gupta et al., 2002). Studies in fungi and in higher eukaryotic systems have indicated that LIS1 functions in the cytoplasmic dynein pathway (Xiang et al., 1995a; Geiser et al., 1997; Morris, 2000; Vallee et al., 2001; Gupta et al., 2002; Cockell et al., 2004; Rehberg et al., 2005). LIS1 physically interacts with the heavy and intermediate chains of cytoplasmic dynein and the dynamitin subunit of the dynactin complex (Faulkner et al., 2000; Niethammer et al., 2000; Sasaki et al., 2000). Two regions of the dynein heavy chain: the first AAA repeat and an N-terminal site implicated in cargo binding, directly interact with LIS1 (Tai et al., 2002). The mechanism by which LIS1 regulates dynein remains unclear.
Consistent with the multiple roles of cytoplasmic dynein in the cell, cytoplasmic dynein, dynactin and LIS1 are found at various cellular sites. Cytoplasmic dynein and dynactin have been found on organelle membranes (Roghi and Allan, 1999; Habermann et al., 2001), consistent with their roles in organizing and transporting organelles. Cytoplasmic dynein, dynactin, and LIS1 also have been localized to microtubule-plus ends in both lower and higher eukaryotic cells (Vaughan et al., 1999, 2002; Han et al., 2001; Coquelle et al., 2002; Zhang et al., 2002, 2003; Carvalho et al., 2003; Lee et al., 2003; Sheeman et al., 2003). The microtubule-plus ends may represent dynein's cargo-loading sites (Vaughan et al., 2002). In addition, plus-end dynein may be involved in regulating microtubule dynamics and/or microtubule–cortex interaction (Carminati and Stearns, 1997; Shaw et al., 1997; Han et al., 2001); Plus-end dynein also may be delivered to the cortex where dynein may play a role as a minus-end-directed motor to walk along astral microtubules for spindle positioning during mitosis (Adames and Cooper, 2000; Heil-Chapdelaine et al., 2000; Lee et al., 2003, 2005; Sheeman et al., 2003). Dynein at the cell cortex or adhesion junctions may be required for pulling or tethering microtubules for centrosome positioning during interphase and/or mediating microtubule–actin interactions (Koonce et al., 1999; Ligon et al., 2001; Dujardin and Vallee, 2002; Dujardin et al., 2003). Cytoplasmic dynein also is located at the kinetochore where it may interact with and transport checkpoint proteins away from the kinetochore, thereby allowing spindle checkpoint inactivation and entry into anaphase (Pfarr et al., 1990; Steuer et al., 1990; Echeverri et al., 1996; Howell et al., 2001; Wojcik et al., 2001). Cytoplasmic dynein and dynactin also are found at centrosomes and at mitotic spindle poles in higher eukaryotic cells, where they may focus microtubule minus ends at spindle poles during mitosis and organize cytoplasmic microtubules during interphase (Merdes et al., 1996; Heald et al., 1997; Quintyne et al., 1999; Compton, 2000; Heald, 2000; Quintyne and Schroer, 2002). In addition, cytoplasmic dynein anchored at the nuclear membrane may walk toward the centrosome, and this has been implicated in nuclear membrane breakdown during mitosis (Salina et al., 2002), separation of the daughter centrosomes (Gonczy et al., 1999), and coupling of the centrosome to the nucleus (Robinson et al., 1999; Malone et al., 2003). The perinuclear and centrosomal localizations of cytoplasmic dynein and LIS1 also have been implicated in centrosome–nucleus coupling during neuronal cell migration (Aumais et al., 2001; Shu et al., 2004; Tanaka et al., 2004). Although the multiple functions of dynein require it to be targeted to different subcellular locations, exactly how dynein is targeted to these sites remains to be investigated.
In fungi, the centrosome-equivalent organelle is the spindle pole body (SPB), which is embedded in the nuclear membrane. With its bound γ-tubulin, the SPB serves as a microtubule-organizing center for both the spindle microtubules and cytoplasmic microtubules (Oakley et al., 1990; Job et al., 2003). In the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe, cytoplasmic dynein has been found at the SPB during both interphase and mitosis (Yeh et al., 1995; Yamamoto et al., 1999; Miki et al., 2002; Sheeman et al., 2003). In the filamentous fungus Aspergillus nidulans, cytoplasmic dynein has not been detected at the SPB in interphase cells. In this study, we have investigated the localization of cytoplasmic dynein and NUDF (A. nidulans LIS1) during mitosis and have found that they both localize to the mitotic spindle poles. They localize to the spindle poles by different mechanisms, however. Whereas NUDF is located at the poles of spindles of various lengths, dynein only occurs at the poles of longer spindles, and this localization of dynein is dependent upon the function of the anaphase-promoting complex (APC). In addition, although NUDF's spindle pole localization did not require the full function of dynein, the spindle pole localization of dynein is positively regulated by NUDF. Transient benomyl treatment of mitotic cells that destroyed both astral and spindle microtubules did not eliminate the localization of dynein and NUDF at the spindle poles. Thus, dynein and NUDF are likely to bind to the SPB rather than just accumulate at the minus ends of microtubules. Importantly, a previously reported γ-tubulin mutation, mipAR63, nearly eliminated dynein's localization to the spindle poles. By contrast, the same mutation did not have an obvious effect on NUDF's spindle pole localization. The mipAR63 mutant exhibits abnormally elongated spindles accompanied by an obvious inhibition in anaphase A. We suggest that a fully functional γ-tubulin may be required for the association of regulatory proteins to the spindle poles to facilitate anaphase A progression.
MATERIALS AND METHODS
Construction of CFP-tubA and YFP-nudI/YFP-nudF Strains
Previously, we made various green fluorescent protein (GFP) constructs to observe microtubules, dynein, and NUDF in A. nidulans (Xiang et al., 2000; Han et al., 2001; Zhang et al., 2002, 2003). Because of the difference in codon preference, the commercially available GFP variants did not express well in A. nidulans. For this reason, we mutated the GFP-tubA plasmid to change GFP to cyan fluorescent protein (CFP), and the GFP-nudI/GFP-nudF plasmids to change GFP to yellow fluorescence protein (YFP), using the QuikChange multisite mutagenesis kit (Stratagene, La Jolla, CA). The change of GFP to CFP was made as described previously (Su et al., 2004). The change of GFP to YFP required five amino acid substitutions: S65G, V68L, Q69K, S72A, and T203Y. Two oligonucleotides were used for mutagenesis: M4, GTCACTACTTTCGGTTATGGTCTCAAGTGCTTTGCCAGATACCCAGATC; and M5, CCATTACCTGTCCTACCAATCTGCCCTTTC. The CFP-tubA plasmid was transformed into SO121 (pyrG89; nicB8). The YFP-nudI and YFP-nudF plasmid were transformed into GR5 (pyrG89, pyroA4 and wA3). Phenotypic analysis was used to screen for strains in which YFP-nudI or YFP-nudF was integrated at its correct locus. Because the fusions were driven by the alcA promoter that can be shut off by glucose, a strain with a site-specific integration of the plasmid would produce a nud phenotype on a YUU plate (Han et al., 2001; Zhang et al., 2002). Southern blot analyses were used to confirm that the integration indeed occurred at the right site. Strains carrying CFP-tubA and YFP-nudF both contain single integrations at the correct sites and were crossed to each other to produce a strain that carries both CFP-tubA and YFP-nudF (our unpublished data). The desired progeny were identified by their nud-like growth phenotype 2–3 d after being streaked on a YUU plate, and by their CFP-labeled microtubules under a fluorescence microscope. The original YFP-nudI strain carried the correct integration of the plasmid but also an extra integration, and this strain was crossed to the CFP-tubA strain. Two strains carrying both CFP-tubA and YFP-nudI were obtained (SL25 and SL26). Although SL25 contains the correct integration plus the extra integration, SL26 contains only the correct integration. However, these two strains exhibited nearly identical behaviors in NUDI localization at the spindle pole, indicating that the extra integration does not interfere with this study. Because SL25 contains a nutritional marker that can be used for crosses, we crossed SL25 to various mutant strains for further analyses. We also crossed the previously published GFP-nudA strain (Xiang et al., 2000) to the CFP-tubA strain and analyzed GFP-NUDA's spindle pole localization.
Introducing CFP-TUBA/YFP-NUDI, CFP-TUBA/YFP-NUDF, and CFP-H2A/GFP-TUBA Fusions to Various Mutant Backgrounds
To obtain CFP-TUBA/YFP-NUDI fusions in the nudF7 background, and the CFP-TUBA/YFP-NUDF fusions in the nudA2 background, standard genetic crosses were carried out. The desired progeny were identified based on the nud-like growth phenotype of the YFP-nudI- or YFP-nudF-carrying strain on glucose plates and the temperature-sensitive (ts) nud phenotype of the nudF7 or the nudA2 mutant on glycerol plates, and the CFP-labeled microtubules under a fluorescence microscope. To obtain CFP-TUBA/YFP-NUDI and CFP-TUBA/YFP-NUDF fusions in the bimE7 background, genetic crosses were set up. The desired progeny were identified based on the nud-like growth phenotype of the YFP-nudI- or YFP-nudF-carrying strain on glucose plates, the temperature-sensitive bimE7 mutant phenotype at 42°C, and the CFP-labeled microtubules under a fluorescence microscope. To obtain CFP-TUBA/YFP-NUDI and CFP-TUBA/YFP-NUDF fusions in the mipAR63 background, genetic crosses were set up. The desired progeny were identified based on the nud-like growth phenotype of the YFP-nudI- or YFP-nudF–carrying strain on glucose plates, the cold-sensitive mipAR63 mutant phenotype at room temperature, and the CFP-labeled microtubules under a fluorescence microscope. To obtain CFP-H2A/GFP-TUBA fusions in the bimE7 or the mipAR63 background, the desired progeny were identified from genetic crosses based on the cold-sensitive mipAR63 mutant phenotype at room temperature or the temperature-sensitive bimE7 mutant phenotype at 42°C, the CFP-labeled nuclei and the GFP-labeled microtubules under a fluorescence microscope.
Construction of Strains Containing GFP-γ-Tubulin or YFP-γ-Tubulin
Two oligonucleotides, N-γ (5′-GGGCGGCCGCTGCCTAGGTATACCCTCC-3′) and C-γ (5′-GGGCGGCCGCAGAACAATGTATGGACAG-3′) (the two underlined regions represent the NotI sites), were used to amplify a 2.1-kb region of the γ-tubulin gene, and the PCR product was digested with NotI followed by ligation to the NotI site of the LB01 plasmid that carries the alcA promoter and GFP (Liu and Morris, 2000). The resulting plasmid contains alcA-GFP fused with the γ-tubulin gene from the start codon (but ATG was changed to CTG to allow in-frame fusion with GFP) to the 3′-untranslated region. To make the YFP-γ-tubulin plasmid, we digested the YFP-nudF plasmid with NotI, and the plasmid backbone was ligated to the 2.1-kb, NotI-digested γ-tubulin fragment. These plasmids were transformed to the A. nidulans strain GR5 separately to obtain strains containing GFP-γ-tubulin or YFP-γ-tubulin. The YFP-γ-tubulin fusion was introduced to the strain expressing CFP-TUBA by genetic crosses. The CFP-TUBA and YFP-γ-tubulin fusions were introduced to the nudA2 or the nudF7 background by genetic crosses. Progeny selection was based on the nud phenotype at 42°C and microscopic observation of the CFP and YFP fusions.
Image Acquisition and Analyses
Cells were grown in ΔTC3 culture dishes (Bioptechs, Butler, PA) containing 1.5 ml of medium. Images were captured using an Olympus IX70 inverted fluorescence microscope (with a 100× objective) linked to a PCO/Cooke corporation Sensicam QE cooled charge-coupled device camera. A Bioptechs heating stage and heated objective system was used for capturing images at 32 or 42°C. Ludl Electronic Products (Hawthorne, NY) dual individual excitation and emission motorized filter wheels were used for observing YFP and CFP signals in the same living cell. Chroma 8600 filters for CFP (430-nm peak excitation with a bandwidth of 25 nm, 470-nm peak emission with a bandwidth of 30 nm) and for YFP (500-nm peak excitation with a bandwidth of 20 nm, 535-nm peak emission with a bandwidth of 30 nm) were used. Custom macros in the IPLab software were written by Jim Paladino (BioVision Technologies, Exton, PA).
RESULTS
Cytoplasmic Dynein Localizes to the Mitotic Spindle Poles in A. nidulans, and This Localization Depends on the Function of the APC
In this study, we have generally used the YFP-tagged NUDI (cytoplasmic dynein IC) to report the location of the dynein complex. In a previous study, we have shown that the functional GFP-NUDI and GFP-NUDA (cytoplasmic dynein heavy chain) fusions both localize to the plus ends of cytoplasmic microtubules and that they depend on each other for this localization (Zhang et al., 2002). In addition, in the nudA2 ts mutant grown at the restrictive temperature of 42°C for 2 d, the GFP-NUDI protein level was drastically reduced and the GFP-NUDI signal was barely detectable on Western blots. Thus, the stability of GFP-NUDI depends on the presence of the normal NUDA heavy chain (Zhang et al., 2002). Based on these results and the fact that both YFP-NUDI and GFP-NUDI are functional, we conclude that the majority of the YFP-NUDI molecules should be associated with the dynein complex and that YFP-NUDI's localization reflects the localization of the dynein complex.
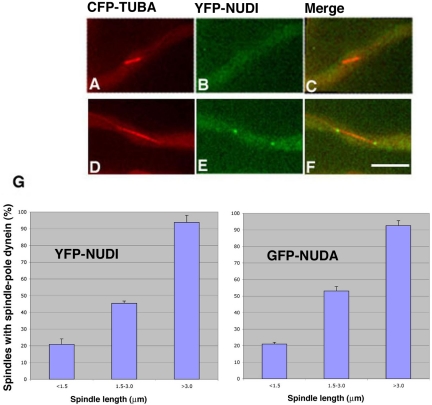
By observing living cells containing CFP-labeled microtubules and YFP-NUDI, we detected cytoplasmic dynein signals at the two poles of mitotic spindles. However, this pole localization was more obvious on longer spindles. To correlate cytoplasmic dynein's spindle pole localization with spindle lengths, randomly chosen spindles of various lengths were analyzed in an asynchronous cell population (Figure 1). The spindle pole signals of YFP-NUDI were either absent or barely detectable with spindles shorter than 1.5 μm. However, YFP-NUDI was easily observed at poles of spindles that were longer than 3 μm, suggesting that more dynein accumulates at spindle poles during spindle elongation. A similar pattern of localization also was detected using a strain containing GFP-NUDA (cytoplasmic dynein heavy chain) and CFP-TUBA (Figure 1), confirming that this mode of localization faithfully reflects the localization of the dynein complex during mitosis.
Figure 1.
YFP-NUDI (cytoplasmic dynein IC) accumulates at spindle poles during spindle elongation. YFP-NUDI's spindle pole signal is not present on a short spindle (A–C), but it is present on a longer spindle (D–F). CFP-labeled microtubules are pseudocolored red (A and D), YFP-labeled NUDI is pseudocolored green (B and E) and merged to show both colors (C and F). The percentage of spindles with the spindle pole YFP-NUDI (left) signal increases with spindle elongation (G, left). A similar profile was observed in a cell population with GFP-labeled NUDA (cytoplasmic dynein heavy chain) and CFP-labeled microtubules (G, right). SEs were calculated using three data collections, and in each collection we analyzed 60 or more spindles chosen randomly from cells grown at 32°C. Bar, 5 μm.
We next tried to correlate spindle length with cell cycle stage by measuring spindles in a ts mutant of bimE (APC1), bimE7 (Osmani et al., 1988; Engle et al., 1990; Peters, 2002; Osmani and Mirabito, 2004). Studies in several other systems indicate that the APC, a multisubunit E3 ubiquitin ligase, is required for anaphase entry by activating a protease called separase, which is required for the proteolytic cleavage of the cohesin complex that holds sister chromatids together after they replicate. The activation of separase involves two mechanisms (Peters, 2002 and references therein): 1) the degradation of securin, which binds and inhibits separase; and 2) the partial degradation of cyclin B to reduce the activity of cdk1 that negatively regulates separase via phosphorylation. For chromosomal segregation during anaphase, the APC-mediated degradation of the kinesin-like protein Xkid is also important. At the end of mitosis, the APC is required for a complete degradation of cyclin B and other mitotic regulators to effect mitotic exit (Peters, 2002; Castro et al., 2005 and references therein).
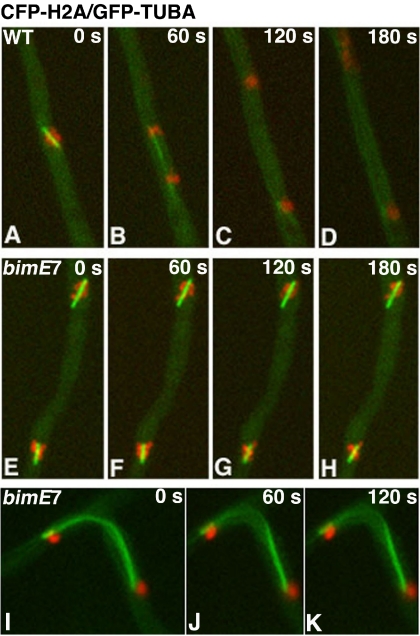
Cells carrying the bimE7 mutation and the CFP-tubA fusion were cultured overnight at the permissive temperature of 32°C and then shifted to the restrictive temperature of 42°C for 2 or 6 h before microscopic observation at 42°C. Under these conditions, a significant fraction of cells were blocked in mitosis as judged by the presence of CFP-labeled spindles. However, the spindles in the cell population were not uniform in length. To observe the bimE7-block directly, we crossed the bimE7 mutation into a strain carrying both CFP-histone-H2A and GFP-tubA (Su et al., 2004) and observed nuclei and spindles simultaneously in live cells. Typically, we found that most cells were blocked with spindles shorter than 4 μm at the restrictive temperature. Within this population of cells, shorter spindles were often surrounded by a chromatin mass, whereas relatively longer spindles had chromosomes dotted along them (Figure 2, E–H). Because a metaphase plate is not seen in A. nidulans, it is not clear whether these configurations represent a problematic anaphase or a blockage of the metaphase-to-anaphase transition. Based on the known function of the APC in anaphase entry, however, we think that the latter possibility is more likely. The cell-to-cell variability in spindle length at this block point may contribute to how easily we can observe chromosomes spread along the spindle. Occasionally, we saw cells with segregated chromosomes but containing very long spindles that failed to disassemble (Figure 2, I–K). These cells were most likely blocked at a stage before mitotic exit due to incomplete degradation of cyclin B and other mitotic regulators.
Figure 2.
Mitotic phenotype of the bimE7 mutant. Cells were grown at the permissive temperature of 32°C overnight and shifted to the restrictive temperature of 42°C for 6 h. Images were taken at 42°C. Four consecutive frames of a time-lapse image set with a 60-s pause time between frames were shown for each cell (except for the last series in I–K). CFP-labeled chromosomes are pseudocolored red, and GFP-labeled microtubules are pseudocolored green. (A–D) A wild-type cell underwent mitotic progression. (E–H) A bimE7 mutant blocked at the metaphase-to-anaphase transition. (I–K) A bimE7 mutant cell blocked before mitotic exit. One daughter nucleus was located in a branch, and the spindle was bent.
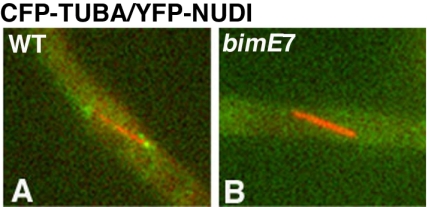
Although we could not precisely correlate the spindle lengths with the different stages of mitosis, the dependency of dynein's spindle pole localization on spindle length suggests a mitotic regulation of this localization. To test whether dynein's localization to the spindle poles requires the function of the APC, we compared YFP-NUDI's spindle pole localization in wild-type cells with that in the bimE7 mutant (Figure 3). The cells were incubated at the permissive temperature overnight and then shifted to the restrictive temperature for 6 h. The profile of dynein's spindle pole localization in wild-type cells was similar to that measured at the permissive temperature (our unpublished data). We examined ∼60 spindles in the bimE7 mutant and found that almost none of the spindles in the bimE7 mutant showed spindle pole signals (Figure 3). Some mutant spindles showed dynein signals along the spindle, but the signals were not focused at the spindle pole as that in wild-type cells. These results indicate that dynein localization to the spindle poles is abolished in the bimE7 mutant at the restrictive temperature. Importantly, not only the shorter spindles but also the spindles that were longer than 3 μm (n = 29) failed to show focused spindle pole dynein signal, whereas in wild-type cells, >90% of the spindles longer than 3 μm showed dynein's spindle pole localization. Thus, the function of the APC is required for targeting dynein to the spindle poles and/or for maintaining dynein at the poles during mitotic progression.
Figure 3.
Spindle pole accumulation of YFP-NUDI (pseudocolored green) is abolished in the bimE7 mutant at the restrictive temperature (42°C) for 6 h. Wild-type (A) and bimE7 mutant cells (B) grown under the same conditions are shown.
NUDF/LIS1 Positively Regulates Cytoplasmic Dynein's Spindle Pole Localization
LIS1, a protein required for neuronal migration during brain development, has been shown to be involved in dynein function in a variety of organisms (Morris, 2000; Vallee et al., 2001; Gupta et al., 2002). Exactly how LIS1 affects dynein's function is an important question being pursued in different experimental systems. In the budding yeast S. cerevisiae, loss of the Pac1/LIS1 protein abolishes the microtubule plus-end localization of dynein (Lee et al., 2003; Sheeman et al., 2003). However, in A. nidulans, the comet-like structures that represent the microtubule plus-end dynein are more prominent in the absence of NUDF/LIS1 (Zhang et al., 2003). Similarly, prominent dynein comets are observed in cells without the NUDF/LIS1 binding protein NUDE or its Neurospora homolog RO11 (Minke et al., 1999; Efimov, 2003). Therefore, in filamentous fungi, LIS1 may affect other aspects of dynein's activity rather than its localization to the microtubule plus ends.
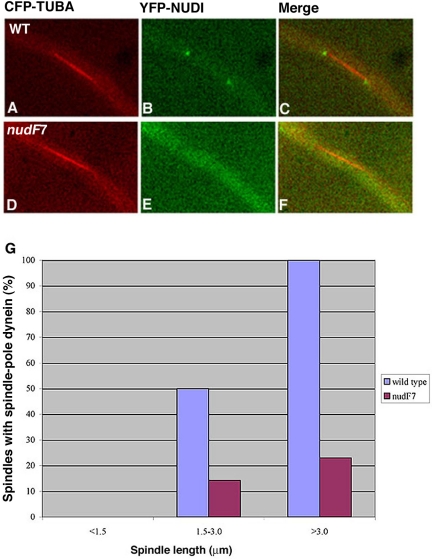
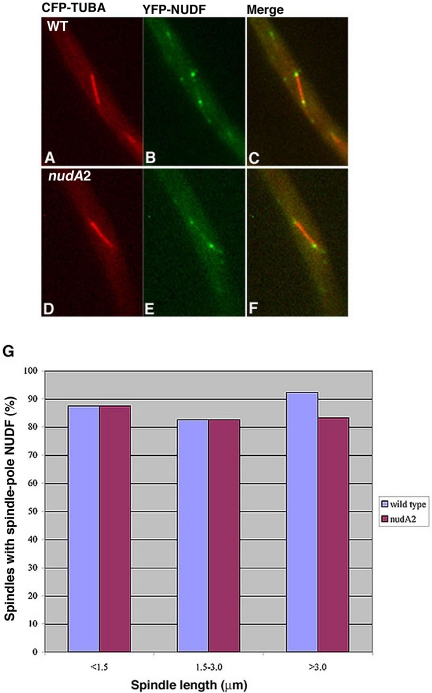
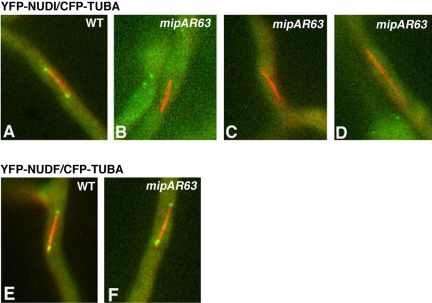
Because the spindle pole is the site associated with microtubule minus ends, we were interested in determining how loss-of-function of NUDF/LIS1 could affect cytoplasmic dynein's spindle pole association. However, these analyses are hard to perform in the nudF deletion mutant because of the clustering of spindles at the spore end, which precludes a clear view of the individual spindle poles. Moreover, the deletion mutant is severely defective in producing asexual spores at any temperature, thus, it is impossible for us to inoculate enough starting materials for the microscopic analysis that requires a large number of cells (mitotic cells only represent ∼4% of total cells under normal conditions). To circumvent this problem, we introduced the CFP-TUBA and YFP-NUDI fusions into the nudF7 mutant, which contains a temperature sensitive, loss-of-function mutation of nudF (Xiang et al., 1995a). The spores were harvested at the permissive temperature (32°C), and the cells were incubated at 32°C overnight to allow nuclei to move into the germ tube. We then shifted the cells to a restrictive temperature (42°C) for 8 h and observed many randomly chosen spindles. Compared with a wild-type control, the spindle pole dynein signal intensity was much lower in the nudF mutant, and far fewer cells with elongated spindles showed detectable spindle pole dynein signals (Figure 4). We also looked at GFP-NUDA/CFP-TUBA in the nudF7 background and obtained the same conclusion (our unpublished data). These results suggest that functional NUDF/LIS1 is important for the localization of dynein to spindle poles.
Figure 4.
Spindle pole accumulation of YFP-NUDI is significantly decreased in the nudF7 mutant. Spindles of the nudF7 mutant at the restrictive temperature (42°C) were compared with spindles of wild-type cells grown under the same condition. YFP-NUDI's spindle pole signal is present on a long spindle of the wild-type cell (A–C) but undetectable on a long spindle of the nudF7 mutant (D–F). CFP-labeled microtubules are pseudocolored red (A and D), YFP-labeled NUDI is pseudocolored green (B and E), and merged to show both colors (C and F). For image analysis, 71 wild-type spindles (left side of the two adjacent columns) and 63 mutant spindles (right side of the two adjacent columns) were chosen randomly from cells grown at 42°C with various spindle lengths (G).
NUDF/LIS1's Localization to the Spindle Poles Does Not Require the Full Function of the APC and Cytoplasmic Dynein
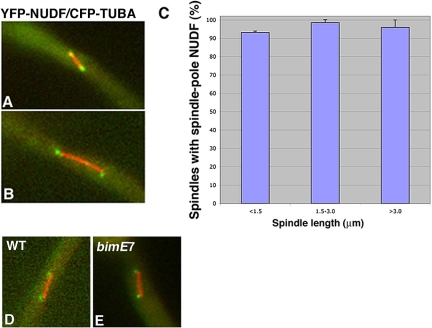
To determine whether NUDF/LIS1 in A. nidulans also is localized to the mitotic spindle poles, we observed the YFP-NUDF fusion in a strain that contains CFP-labeled microtubules. The YFP-NUDF fusion is functional like the previously described GFP-NUDF fusion (Han et al., 2001). Similar to cytoplasmic dynein, NUDF was found at the mitotic spindle poles (Figure 5). However, NUDF's spindle pole signal was detected on spindles of various lengths (Figure 5). Furthermore, NUDF's spindle pole localization was readily observed in the bimE7 mutant cells at the restrictive temperature (Figure 5), indicating that unlike dynein, NUDF localization to the spindle poles is not dependent on APC.
Figure 5.
YFP-NUDF is localized to the poles of mitotic spindles of various lengths, and this localization is APC independent. YFP-NUDF's spindle pole signal (pseudo-colored with green) is present on both short and long spindles (pseudocolored with red) (A and B). The percentage of spindles with the spindle pole YFP-NUDF signal does not change significantly with spindle elongation. SEs were calculated from two data collections, and in each collection we analyzed ∼60 spindles chosen randomly from cells grown at 32°C (C). The spindle pole accumulation of YFP-NUDF is not affected by the bimE7 mutation (D and E). A wild-type (D) and a bimE7 mutant cell (E) grown at the restrictive temperature (42°C) are shown.
Although NUDF's spindle pole signal occurs earlier than that of dynein, it is still possible that NUDF may use the dynein motor for its initial targeting to the spindle pole and then becomes associated, whereas dynein does not. To determine whether NUDF's targeting to the spindle pole is dynein dependent, we introduced the YFP-NUDF and CFP-TUBA fusions into the nudA2 mutant, which is a temperature-sensitive dynein heavy chain loss-of-function mutant (Xiang et al., 1995b). We allowed the cells to grow at the permissive temperature (32°C) overnight to let the nuclei move into the germ tube. We then shifted the culture to the restrictive temperature (42°C) for 8 h and observed the cells at 42°C. In a previous study, we have found that shifting the nudA2 mutant cells to 42°C for only 6 h is sufficient to cause an obvious defect in microtubule dynamics (Han et al., 2001). In this study, we found that after shifting the nudA2 mutant cells to 42°C for 7 or 8 h, the YFP-NUDI IC can no longer be observed at the spindle poles (our unpublished data), further indicating that dynein is not fully functional at these time points. After 8 h at the restrictive temperature, YFP-NUDF was easily observed at the spindle poles in the nudA2 mutant (Figure 6). This result suggests that NUDF localization to the spindle does not require a fully functional dynein motor.
Figure 6.
Spindle pole localization of NUDF is not affected in the nudA2 dynein heavy chain mutant. Spindles of the nudA2 mutant at the restrictive temperature (42°C) were compared with spindles of wild-type cells grown under the same conditions. YFP-NUDF's spindle pole signal is present on a spindle of the wild-type cell (A–C) and a spindle of the nudA2 mutant (D–F). CFP-labeled microtubules are pseudocolored with red (A and D), YFP-labeled NUDI is pseudocolored with green (B and E), and merged to show both colors (C and F). For image analysis, 83 wild-type (left side of the two adjacent columns) and 68 mutant (right side of the two adjacent columns) spindles of various lengths were chosen randomly from cell grown at 42°C.
The Association of Dynein and NUDF with the Spindle Poles Is Resistant to Transient Microtubule Depolymerization
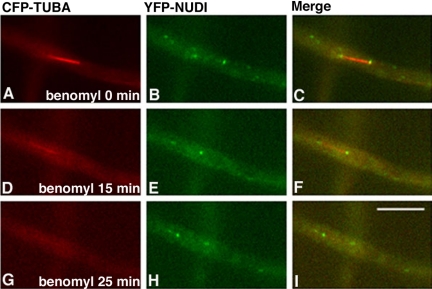
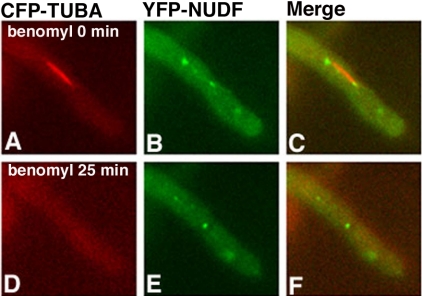
Because cytoplasmic dynein is a minus-end-directed motor, and the spindle poles are where the minus ends of microtubules locate in the cell, it is possible, in principle, that the spindle pole localization of dynein may simply be due to dynein moving along microtubules and accumulating at the minus ends. In addition, the appearance of dynein at the spindle poles may reflect dynein's localization to the plus ends of very short astral microtubules. To rule out these possibilities, we treated the cells with benomyl (2.4 μg/ml), a microtubule-depolymerizing drug, to quickly depolymerize microtubules. We found that treatment with benomyl completely depolymerized microtubules as judged by the disappearance of all CFP-labeled microtubules. There were no CFP spots left near the spindle poles, indicating that during mitosis in A. nidulans, microtubules near the SPB are not well protected by the nuclear membrane from being depolymerized by the drug. Interestingly, after the disappearance of both cytoplasmic and spindle microtubules, dynein remained at the same positions (Figure 7), suggesting that dynein is associated with the SPB. The same treatment did not abolish NUDF's spindle pole localization either, suggesting that NUDF also is associated directly with the SPB during mitosis (Figure 8).
Figure 7.
Live cell imaging demonstrating that the localization of cytoplasmic dynein to the mitotic spindle poles is resistant to transient microtubule depolymerization. CFP-labeled microtubules were pseudocolored red (A, D, and G), YFP-labeled cytoplasmic dynein IC (NUDI) was pseudocolored green (B, E, and H), and merged to show both colors (C, F, and I). Cells were grown at 32°C overnight, and images were acquired at 32°C. Benomyl was added to the culture dish to a final concentration of 2.4 μg/ml, and images were acquired after 15 min (D–F) and 25 min (G–I). Bar, 5 μm.
Figure 8.
Live cell imaging demonstrating that the localization of NUDF to the mitotic spindle poles is resistant to transient microtubule depolymerization. CFP-labeled microtubules are pseudocolored red (A and D), YFP-labeled NUDF is pseudocolored green (B and E), and merged to show both colors (C and F). Benomyl was added to the culture dish to a final concentration of 2.4 μg/ml, and images were acquired after 25 min (D–F).
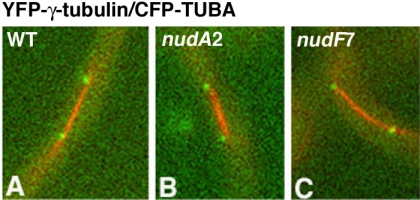
The Spindle Pole Association of Dynein but Not NUDF Is Abolished in the mipAR63 γ-Tubulin Mutant
Whether dynein and NUDF/LIS1 interact directly with a SPB component is not clear. In mammalian cells, a LIS1-binding protein, mNUDE, is located at the centrosome and interacts with several centrosomal components, including γ-tubulin (Feng et al., 2000). In A. nidulans, γ-tubulin is located at the SPB during all cell cycle stages and plays multiple roles in mitotic progression (Oakley et al., 1990; Jung et al., 2001; Prigozhina et al., 2001, 2004). A previous study in tissue culture cells and Xenopus extracts suggested that γ-tubulin may be transported to the centrosome by dynein (Young et al., 2000). In this study, we constructed a strain in which γ-tubulin is tagged at the N terminus with YFP, and we found that the YFP-γ-tubulin fusion localized to the spindle poles as expected. This localization was not affected by the nudF7 and nudA2 mutations at the restrictive temperature (Figure 9), suggesting that dynein's function is not essential for γ-tubulin's targeting to the SPB in A. nidulans. We next asked whether γ-tubulin recruits dynein or NUDF/LIS1 to the mitotic spindle poles. Toward this goal, we crossed a γ-tubulin mutant, mipAR63, with the strains expressing YFP-NUDI/CFP-TUBA and YFP-NUDF/CFP-TUBA, and obtained the progeny that carried the γ-tubulin mutation and the YFP-NUDI (or NUDF)/CFP-TUBA fusions. The rationale for using this specific γ-tubulin mutant is that this allele exhibits a partial defect in nuclear positioning in the hyphae (Jung et al., 2001), which suggests a possible link with dynein because all dynein loss-of-function mutants exhibit a severe nuclear distribution defect in A. nidulans (Xiang et al., 1995b). Because the colony size of this γ-tubulin mutant is significantly reduced at room temperature (∼25°C), we incubated the cells at 32°C overnight and then shifted them to room temperature for 3 h before observation. A previous study using fixed cells has suggested that this γ-tubulin mutant exhibited long mitotic spindles (Jung et al., 2001). Here, we measured spindle length in live cells containing YFP-NUDI/CFP-TUBA and the mipAR63 mutation and found that >60% of the total mitotic spindles were longer than 3 μm (n = 57), whereas in the wild-type background only 13.2% of the spindles were longer than 3 μm (n = 53). In wild-type cells, spindle pole signals of the YFP-NUDI dynein IC were easily observed on spindles longer than 3 μm. However, in the mipAR63 mutant, YFP-NUDI's spindle pole association was greatly diminished (Figure 10), suggesting that γ-tubulin is important for dynein's spindle pole localization. Interestingly, YFP-NUDF was easily observed at the spindle poles in the same mutant background (Figure 10), indicating that NUDF's spindle pole localization does not require the full function of γ-tubulin. In a few cells, GFP-NUDF fluorescent dots were seen to move along the spindle toward or away from one of the poles (our unpublished data), but the significance of this observation is not clear.
Figure 9.
Localization of γ-tubulin to the SPB is not affected by the nudA2 or nudF7 mutation. YFP-labeled γ-tubulin is pseudocolored green, and CFP-labeled spindles are pseudocolored red. Wild-type (A), nudA2 (B), and nudF7 (C) cells grown at the restrictive temperature (42°C) are shown.
Figure 10.
Spindle pole accumulation of YFP-NUDI but not YFP-NUDF is abolished in the mipAR63 mutant. Cells were grown at the permissive temperature (32°C) overnight and shifted to the restrictive temperature (room temperature) for 3 h. Cells containing YFP-NUDI (pseudocolored with green) in wild-type (A) and the mipAR63 mutant background (B–D), and cells containing YFP-NUDF in wild-type (E) and the mipAR63 mutant background (F), are shown.
The mipAR63 Mutant Exhibits a Severe Inhibition of Anaphase A
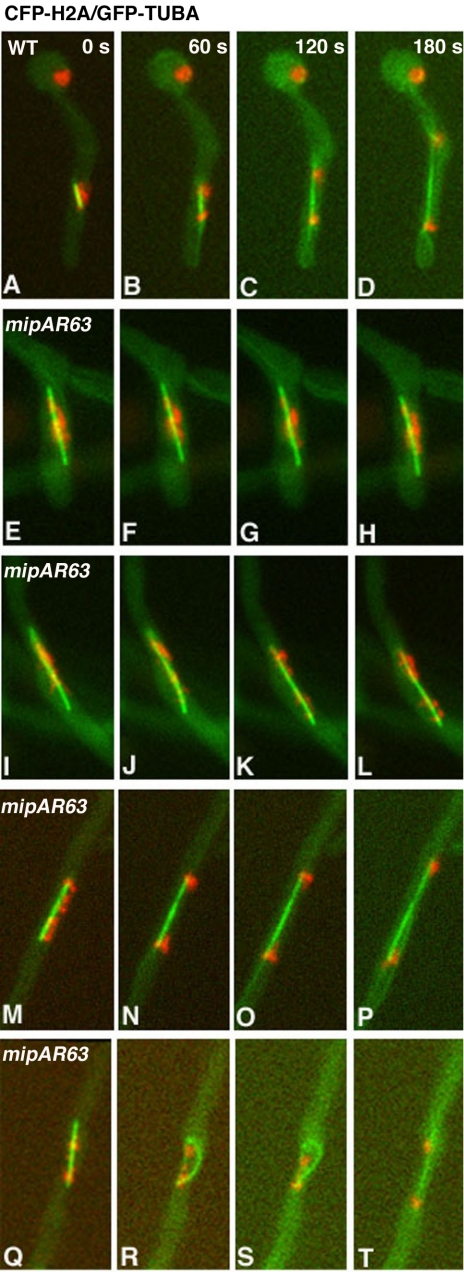
Mitotic and microtubule defects associated with the mipAR63 mutant have been noticed in a previous study (Jung et al., 2001). Because the mipAR63 mutation affected dynein's spindle pole localization during mitosis, we examined in more detail the mitotic defect associated with this mutation in live cells. By observing CFP-labeled spindles in a strain that contains YFP-nudI and mipAR63, we found that many mipAR63 mutant cells exhibited long spindles (longer than 4 μm). These long spindles did not undergo further elongation or disassembly within 5 min, whereas spindles in wild-type cells underwent dramatic elongation and disassembly during the same time period (Table 1). Because in wild-type cells spindles longer than 4 μm were usually seen after initiation of anaphase B (Su et al., 2004), we initially speculated that the mipAR63 mutants might exhibit a mitotic exit delay. To further examine the mitotic defect in a strain in which both chromosomes and microtubules are labeled, we crossed the mipAR63 γ-tubulin mutation into a strain containing GFP-tubA/CFP-Histone-H2A and observed the behaviors of the nuclei and the microtubules simultaneously during mitosis (Figure 11). Interestingly, ∼30% of the mipAR63 mutant cells (n = 50) exhibited a severe defect in either the initiation or the progression of anaphase A such that they failed to complete anaphase A within the 11-min time period of image acquisition (Figure 11, E–H, I–L). These cells were blocked with a mitotic configuration in which chromosomes were located near the middle portion or spread along the spindles. This configuration also was observed in the bimE7 APC1 mutant except that spindles in the mipAR63 γ-tubulin mutant were much longer (Figure 11, E–H, I–L). The rest of the cells did finish anaphase A during the 11 min of observation, but many of them took longer compared with wild-type cells. Some cells also took longer to disassemble spindles after anaphase A was completed (Figure 11, M–P), and a few cells exhibited abnormally curved spindles (Figure 11, Q–T). Together, our results suggest that the mipAR63 γ-tubulin mutation allows spindle elongation before chromosomal disjunction and significantly impedes anaphase A chromosomal separation.
Table 1.
Spindle length and stability in the mipAR63 mutant
| Spindles longer than 4 μm before anaphase A is completed | Spindles not disassembled 5 min after their lengths reach 4 μm | Spindles not disassembled 5 min after anaphase A is completed | |
|---|---|---|---|
| Wild type | 17.1 (n = 35) | 2.8 (n = 36) | 5.6 (n = 36) |
| mipAR63 | 71.4 (n = 49) | 52.0 (n = 50) | 17.6 (n = 34) |
Values are percentages. For these analyses, cells were grown under the same conditions as described in Figure 11 legend.
Figure 11.
mipAR63 mutation causes defects in mitotic progression. Cells were grown at the permissive temperature 32°C overnight and shifted to room temperature for 3 h. Four consecutive frames of a time lapse with a 60-s pause time between frames are shown for each cell. (A–D) Wild-type cell underwent mitotic progression. (E–H) mipAR63 mutant containing a long spindle with chromosomes associated with the middle portion. (I–L) mipAR63 mutant containing a long spindle with chromosomes along it. (M–P) mipAR63 mutant cell with a completed anaphase A but a relatively more stable spindle compared with that in wild-type cells. (Q–T) mipAR63 mutant cell that finished anaphase A with a bent spindle.
A. nidulans Dynein Plays a Nonessential Role in Chromosome Segregation
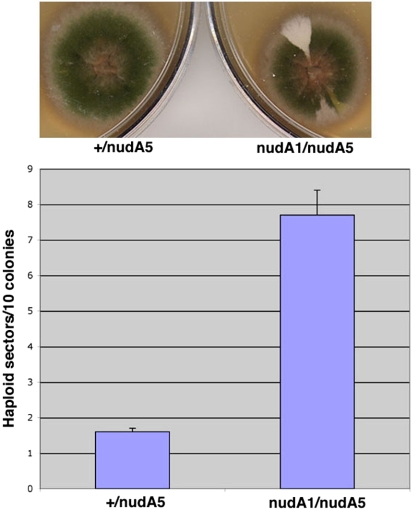
Our previous studies have suggested that A. nidulans dynein is not essential for nuclear division (Xiang et al., 1995b). To determine whether dynein plays a subtle role in chromosome segregation, we have carried out a chromosome loss assay. In A. nidulans, diploid nuclei are formed at a low frequency during vegetative growth, and a pure diploid strain can be maintained. The growth of a diploid colony is more sensitive to benomyl than that of a haploid colony (our unpublished data), suggesting that chromosome segregation in diploids is more sensitive to microtubule-depolymerizing drugs than in haploid cells. This is consistent with a higher demand for effective kinetochore–microtubule interactions with increased ploidy (Lin et al., 2001). If a diploid loses one chromosome, the resulting aneuploid often rapidly loses additional chromosomes to become a haploid (Kafer, 1977; Clutterbuck, 1992; Rischitor et al., 2004). Benomyl at a concentration of 0.7 μg/ml can induce chromosome loss, leading to the formation of haploid sectors in a diploid colony. In A. nidulans, this so-called “parasexual cycle” of haploid formation from a diploid has been routinely used for genetic mapping of genes to specifically marked chromosomes (Clutterbuck, 1992). To detect the formation of haploid sectors directly, we routinely use haploid strains with different color markers to derive a green diploid and then look for the appearance of haploid sectors with different colors such as yellow, chartreuse, white, or yellow chartreuse as a function of chromosome loss. To determine whether the dynein mutant diploid has an increased frequency of chromosome loss, we first constructed a nudA1/nudA5 diploid (nudA1 and nudA5 are both loss-of-function ts alleles of the dynein heavy chain gene as described previously in Xiang et al., 1995b). This mutant diploid exhibits a nud phenotype at a restrictive temperature of 42°C. We also constructed a nudA+/nudA5 (or +/nudA5) heterozygous diploid and a wild-type diploid as controls (Table 2). The heterozygous diploid behaves similarly to the wild-type diploid strain in growth and in chromosome loss assays (our unpublished data). Thus, we focused on the difference between the nudA1/nudA5 and the +/nudA5 diploid (control diploid) strains that contain exactly the same nutritional and color markers. We first incubated the strains at the permissive temperature (32°C) for 1 d and then shifted the plates to the restrictive temperature (42°C) for 1 d to inhibit dynein function. Because a nud mutant is defective in conidia (asexual spore) formation, we shifted the plates back to the permissive temperature (32°C) for 2 d to see the colors of the haploid sectors. In the absence of benomyl, neither the mutant diploid nor the control diploid produced enough sectors to allow a quantitative analysis. Thus, dynein loss-of-function does not seem to cause significant chromosome loss during mitosis under normal conditions. However, in the presence of a low amount of benomyl (0.4 μg/ml), the mutant diploid produced significantly more haploid sectors than the control (Figure 12; p < 0.006). This result suggests that although A. nidulans dynein is not essential for mitosis, it does play a role in chromosome segregation in diploids.
Table 2.
A. nidulans strains used in this work (all the strains have the veA1 marker)
| Strain | Genotype | Source |
|---|---|---|
| GG1 | nudA1; pabaA1; yA1 | Goldman and Morris (1995) |
| GR5 | pyrG89; wA3; pyroA4 | G. S. May |
| LO1181 | YFP-nudI-pyr4; CFP-tubA-pyr4; mipAR63; pyrG89; fwA1; pabaA1 | This work |
| R21 | PabaA1; yA1 | N. R. Morris' laboratory |
| SL12 | YFP-nudF-pyr4; CFP-tubA-pyr4; pyrG89; pyroA4 | This work |
| SL19 | YFP-nudI-pyr4; pyrG89; pyroA4; wA3 | This work |
| SL21 | GFP-nudA-pyr4; CFP-tubA-pyr4; pyrG89; pyroA4 | This work |
| SL25 | YFP-nudI-pyr4; CFP-tubA-pyr4; pyrG89; pyroA4 | This work |
| SL26 | YFP-nudI-pyr4; CFP-tubA-pyr4; pyrG89; wA3 | This work |
| SL34 | YFP-nudF-pyr4; CFP-tubA-pyr4; nudA2; pyrG89 | This work |
| SL37 | YFP-nudI-pyr4; CFP-tubA-pyr4; nudF7; pyrG89; yA1 | This work |
| SL51 | YFP-nudF-pyr4; CFP-tubA-pyr4; bimE7; pyrG89; fwA1; pyroA4 | This work |
| SL66 | YFP-nudI-pyr4; CFP-tubA-pyr4; bimE7; pyrG89; fwA1; pyroA4 | This work |
| SL71 | GFP-mipA-pyr4; pyrG89; wA3; pyroA4 | This work |
| SL73 | YFP-mipA-pyr4; pyrG89; wA3; pyroA4 | This work |
| SL89 | YFP-mipA-pyr4; CFP-tubA-pyr4; pyrG89; pyroA4; wA3 | This work |
| SL113 | YFP-mipA-pyr4; CFP-tubA-pyr4; pyrG89; nudA2 | This work |
| SL116 | YFP-mipA-pyr4; CFP-tubA-pyr4; pyrG89; nudF7; yA1 | This work |
| SL118 | GFP-tubA-pyr4; CFP-H2A-pyr4; pyrG89; nicB8 | Su et al. (2004) |
| SL141 | GFP-tubA-pyr4; CFP-H2A-pyr4; pyrG89; bimE7; fwA1; pyroA4 | This work |
| SL165 | YFP-nudF-pyr4; CFP-tubA-pyr4; mipAR63; pyrG89; fwA1; pyroA4 and pabaA1 | This work |
| SL166 | GFP-tubA-pyr4; CFP-H2A-pyr4; pyrG89; mipAR63; pyroA4 | This work |
| SL196 | GFP-nudA-pyr4; CFP-tubA-pyr4; nudF7; pyrG89; pyroA4 | This work |
| SO121 | pyrG89; nicB8 | S. A. Osmani |
| SWJ001 | bimE7; argB2; pyrG89; pabaA1; fwA1 | S. James |
| XX5 | nudA2, pyrG89, wA3 | Xiang et al. (1995b) |
| XX10 | nudA5; pyrG89; wA1; chaA1 | Xiang et al. (1995b) |
| XX21 | nudF7; pyrG89; yA1 | Xiang et al. (1995a) |
| Diploid dynein mutant (DX3) | nudA1/nudA5, other markers from the parental strains GG1 and XX10 | This work |
| Heterozygous diploid (DS2) | +/nudA5, other markers from the parental strains R21 and XX10 | This work |
| Wild-type diploid (DS1) | Markers from the parental strains R21 and GR5 | This work |
Figure 12.
Dynein mutant diploid (nudA1/nudA5) forms more haploid sectors than the control diploid (+/nudA5) in the presence of 0.4 μg/ml benomyl. Top, picture showing three haploid sectors formed from the mutant diploid colony and a control diploid colony with no sectors. KCl with a final concentration of 0.6 M was present in the complete medium YUU to enhance spore formation of the nud mutants. Bottom, quantitative analyses of haploid sector formation. SEs were calculated using two data collections. In total, 32 colonies were analyzed.
DISCUSSION
NUDF Is Important for Cytoplasmic Dynein's Spindle Pole Targeting
In this study, we have found that in the filamentous fungus A. nidulans, cytoplasmic dynein and its regulator NUDF/LIS1 both localize to the poles of mitotic spindles. This localization is in addition to their microtubule plus-end localization described in previous studies (Han et al., 2001; Zhang et al., 2002, 2003). Surprisingly, although NUDF's spindle pole localization is obvious on spindles of various lengths and is APC independent, dynein's spindle pole localization is only obvious on longer spindles and is APC dependent. Furthermore, although a γ-tubulin defect severely impairs dynein's spindle pole localization, it does not apparently affect NUDF's localization to the same site. These results suggest that NUDF's localization to the spindle poles is mechanistically different from dynein's spindle pole localization.
LIS1 is thought to be involved in spindle assembly and mitotic progression in higher eukaryotic cells (Faulkner et al., 2000; Feng et al., 2000; Cockell et al., 2004; Feng and Walsh, 2004). We have shown here that NUDF is important for dynein's spindle pole localization during mitosis in A. nidulans. One explanation for this result is that NUDF may provide a binding site for dynein at the spindle poles. Alternatively, NUDF at the plus end of a microtubule or along the microtubule may act as a dynein activator to facilitate its movement toward the spindle poles. Experiments in mammalian cells also have suggested a role of LIS1 as a positive regulator of dynein motor activity (Sasaki et al., 2000; Smith et al., 2000; Tai et al., 2002). In addition, it has been found that expression of a Nudel (a LIS1 binding protein) mutant protein impaired in LIS1 binding causes a defect in dynein's movement to the spindle poles (Yan et al., 2003). Interestingly, our current study suggests that NUDF is targeted to the spindle poles before APC activation, whereas dynein's localization at the spindle poles occurs upon or after APC activation. How NUDF is targeted to the spindle poles and how it affects dynein's localization to the same sites will require further study.
The Involvement of the APC and γ-Tubulin in Dynein's Spindle Pole Localization
The bimE7 mutation in APC1, and the mipAR63 mutation in γ-tubulin, both affect dynein's spindle pole localization, but the mechanisms of the effect are not clear. In A. nidulans, γ-tubulin and a protein in the APC complex, BIMA (a homolog of CDC27/APC3) are both localized to the SPB (Oakley et al., 1990; Mirabito and Morris, 1993). The APC is a multisubunit ubiquitin ligase E3 that is used for the degradation of securin and later of cyclin B and other regulators, thereby initiating the metaphase-to-anaphase transition and later mitotic exit (Harper et al., 2002; Peters, 2002). The temporal–spatial regulation of APC during mitosis is still not clear, although SPB-specific degradation of cyclin B during the end of metaphase has been reported previously (Clute and Pines, 1999; Huang and Raff, 1999). Compared with the APC, the function of γ-tubulin during mitosis is much less well defined. The γ-tubulin complex is involved in nucleating microtubule assembly and in minus-end capping (Gunawardane et al., 2000; Wiese and Zheng, 2000; Patel and Stearns, 2002; Job et al., 2003). Previous studies have shown that in addition to nucleating microtubule assembly, γ-tubulin is important for spindle pole separation during spindle assembly and for coordinating mitotic events in A. nidulans and the fission yeast S. pombe (Paluh et al., 2000; Prigozhina et al., 2001, 2004; Vardy et al., 2002). In the A. nidulans mipAR63 mutant, long spindles are found before anaphase A is completed, suggesting either a premature spindle elongation or a delayed chromosomal disjunction. Besides the abnormally elongated spindles, ∼30% of the cells carrying the mipAR63 γ-tubulin mutation fail to achieve anaphase A chromosome separation. This may be due to a defect of this mutant in recruiting regulatory molecules to the SPB. Alternatively, the mipAR63 γ-tubulin mutation may stabilize the plus ends of kinetochore microtubules, thereby affecting anaphase A chromosome segregation. In S. pombe, defects in the γ-tubulin complex cause the plus ends of both spindle and cytoplasmic microtubules to become more stable (Zimmerman and Chang, 2005), and a defective anaphase A also has been observed (Paluh et al., 2000). The fact that the mipAR63 mutant contains long and stable spindles is consistent with an alteration of spindle microtubule dynamics at the plus ends. How γ-tubulin mutation(s) may affect plus-end microtubule dynamics is an intriguing question that deserves to be explored further.
It is possible that the bimE7 and mipAR63 mutations affect dynein's spindle pole localization because they block the cell cycle at a specific point(s). In addition, astral microtubules are not easily visible in the mipAR63 mutant cells (our unpublished data) or in the bimE7 mutant cells (our unpublished data; Osmani et al., 2003). The deficiency in astral microtubules may contribute to the lack of dynein at the spindle poles. However, our observations suggest that dynein's spindle pole localization does not depend upon astral microtubules. In cells treated with a low amount of benomyl, astral microtubules were not visible, yet dynein's localization at the poles was clearly visible on slowly elongating spindles that most likely had passed the metaphase-to-anaphase transition (our unpublished data). It is not clear whether the spindle pole dynein comes from the kinetochore via kinetochore microtubules, from the nucleoplasm, or from the cytoplasm. In A. nidulans, the nuclear pore complex is partially disassembled during “closed” mitosis (De Souza et al., 2004), which may allow a big complex such as dynein to move in and out of the nucleus.
Possible Functions of Cytoplasmic Dynein at the Spindle Poles
Cytoplasmic dynein is known to organize the spindle poles in higher eukaryotic cells (Merdes et al., 1996; Heald et al., 1997; Compton, 2000; Heald, 2000); however, its function at the spindle poles is not clear in most fungi where the SPB is embedded in the nuclear membrane. In another filamentous fungus, Nectria hematococca, cytoplasmic dynein is required for mitotic aster formation (Inoue et al., 1998). However, this does not seem to be the case in A. nidulans because astral microtubules can be observed in dynein mutants or mutants in the dynein pathway (our unpublished data; Osmani et al., 1990).
In higher eukaryotic cells, cytoplasmic dynein has been implicated in transporting proteins to the spindle poles during mitosis. For example, dynein may transport microtubule-depolymerizing molecules such as a KinI kinesin to the spindle poles, which may be required for spindle microtubule flux during metaphase (Gaetz and Kapoor, 2004). However, the spindle pole-associated microtubule-minus-ends in fungi seem to be nondynamic during anaphase and possibly also during metaphase (Maddox et al., 2000; Zimmerman and Chang, 2005; Oakley, unpublished data). In Drosophila and in cultured cells, dynein has been implicated in moving the spindle assembly checkpoint proteins from the kinetochore toward the spindle poles for inactivating the checkpoint, which is consistent with dynein's role in the metaphase-to-anaphase transition in these cells (Howell et al., 2001; Wojcik et al., 2001; Goshima and Vale, 2003; Yan et al., 2003). In Tetrahymena thermophila, gene disruption of the ubiquitous cytoplasmic dynein results in a failure of proper chromosome segregation during micronuclear mitosis (Lee et al., 1999). But it is not clear whether this defect is caused by a failure in the inactivation of the spindle assembly checkpoint or in force generation during chromosome movement. In the budding yeast S. cerevisiae, cytoplasmic dynein plays a redundant role with the Cin8 and Kip1 kinesins in anaphase chromosome segregation (Saunders et al., 1995). However, the impact of dynein is most likely on spindle elongation during anaphase B when dynein-mediated interactions between astral microtubules and the cell cortex may help to pull the spindle poles apart (Saunders et al., 1995). This function is more likely to be achieved by dynein at the plus ends of astral microtubule rather than by dynein at the SPB (Lee et al., 2003; Sheeman et al., 2003).
In A. nidulans, cytoplasmic dynein is not essential for mitotic progression (our unpublished data; Xiang et al., 1995b), but our current study has suggested a possible role of dynein in chromosome segregation. Loss of dynein function does not cause an obvious chromosome-segregation defect under normal conditions, which is consistent with an early result from S. cerevisiae (Li et al., 1993). However, in the presence of a low amount of the microtubule drug benomyl, the dynein mutant diploid undergoes chromosome-loss-induced haploid formation with a higher frequency than that of the control diploid. Chromosome loss may be caused by a defect in kinetochore-to-microtubule attachment during prometaphase, metaphase, or anaphase, if such a defect is not caught by the spindle assembly checkpoint and corrected. Thus, dynein may play a role in kinetochore–microtubule interactions, and this role becomes more critical if kinetochore microtubules are not in their normal dynamic state. In mammalian cells, cytoplasmic dynein locates to prometaphase kinetochores (Pfarr et al., 1990; Steuer et al., 1990; Echeverri et al., 1996). Defects in dynein function perturb chromosome alignment at the metaphase plate and also cause problems in spindle assembly (Echeverri et al., 1996; Faulkner et al., 2000). In A. nidulans, mutations affecting the spindle assembly checkpoint severely exacerbate the growth defects of the dynein mutants (Efimov and Morris, 1998), which is consistent with dynein's role in spindle integrity. In fungi, both chromosomes and spindles are much smaller than the ones in higher eukaryotic cells and fungal dynein has not been shown to locate at the kinetochores. Similar to yeasts, A. nidulans kinetochores also seem to cluster in the vicinity of the SPB at the end of mitosis (Wigge and Kilmartin, 2001 and references therein; Yang et al., 2004). One possibility we cannot exclude at this stage is that a low amount of dynein molecules are located at the kinetochore and remain bound until the end of mitosis. However, it is also possible that dynein may play a role at the spindle poles in maintaining spindle integrity, thereby influencing microtubule–kinetochore interactions required for anaphase A chromosome movement.
Cytoplasmic dynein may be functionally involved in the later stages of mitosis. In S. cerevisiae, absence of the dynein heavy chain significantly decreases the SPB signal intensity of a mitotic exit network (MEN) protein Tem1p (Molk et al., 2004). In higher eukaryotic cells, dynein regulators have been found at the midbody (Karki et al., 1998) and are important for cytokinesis (Aumais et al., 2003; Zhou et al., 2003). In A. nidulans, dynein is important for specifying the position of the first septum because dynein mutants show a misplaced first septum (Liu and Morris, 2000; Liu et al., 2003). Whether septum position is related to the spindle pole-associated dynein is an interesting question. But so far, we have not observed any apparent abnormality in septum position in the mipAR63 mutant where dynein's spindle pole association is nearly abolished (our unpublished data), suggesting that dynein at the spindle poles is unlikely to be critical for septation.
Acknowledgments
We thank Dr. Steve James, Dr. Stephan A. Osmani, Aysha H. Osmani, and Dr. Jun Zhang for A. nidulans strains and plasmids. We also thank Drs. David Pellman, Carl Mann, Stephan A. Osmani, Orna Cohen-Fix, Bo Liu, and Vladimir Efimov for helpful discussions; Dr. Fred Chang for communicating unpublished results; and Drs. Carl Mann and Chou-Zen Giam for helpful readings of the manuscript. This work was supported by National Institutes of Health Grant GM-069527-01, National Science Foundation Grant MCB-0093106, and Uniformed Services University of the Health Sciences Intramural Grant R071GO (to X. X.); and National Institutes of Health Grant GM-31837 (to B.R.O.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1071) on June 1, 2005.
References
- Adames, N. R., and Cooper, J. A. (2000). Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumais, J. P., Tunstead, J. R., McNeil, R. S., Schaar, B. T., McConnell, S. K., Lin, S. H., Clark, G. D., and Yu-Lee, L. Y. (2001). NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J. Neurosci. 21, RC187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumais, J. P., Williams, S. N., Luo, W., Nishino, M., Caldwell, K. A., Caldwell, G. A., Lin, S. H., and Yu-Lee, L. Y. (2003). Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J. Cell Sci. 116, 1991–2003. [DOI] [PubMed] [Google Scholar]
- Carminati, J. L., and Stearns, T. (1997). Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, P., Tirnauer, J. S., and Pellman, D. (2003). Surfing on microtubule ends. Trends Cell Biol. 13, 229–237. [DOI] [PubMed] [Google Scholar]
- Castro, A., Bernis, C., Vigneron, S., Labbe, J. C., and Lorca, T. (2005). The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene 24, 314–325. [DOI] [PubMed] [Google Scholar]
- Clute, P., and Pines, J. (1999). Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1, 82–87. [DOI] [PubMed] [Google Scholar]
- Clutterbuck, A. J. (1992). Sexual and parasexual genetics of Aspergillus species. Biotechnology 23, 3–18. [PubMed] [Google Scholar]
- Cockell, M. M., Baumer, K., and Gonczy, P. (2004). lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J. Cell Sci. 117, 4571–4582. [DOI] [PubMed] [Google Scholar]
- Compton, D. A. (2000). Spindle assembly in animal cells. Annu. Rev. Biochem. 69, 95–114. [DOI] [PubMed] [Google Scholar]
- Coquelle, F. M., et al. (2002). LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell. Biol. 22, 3089–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza, C. P., Osmani, A. H., Hashmi, S. B., and Osmani, S. A. (2004). Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14, 1973–1984. [DOI] [PubMed] [Google Scholar]
- Dujardin, D. L., and Vallee, R. B. (2002). Dynein at the cortex. Curr. Opin. Cell Biol. 14, 44–49. [DOI] [PubMed] [Google Scholar]
- Dujardin, D. L., Barnhart, L. E., Stehman, S. A., Gomes, E. R., Gundersen, G. G., and Vallee, R. B. (2003). A role for cytoplasmic dynein and LIS1 in directed cell movement. J. Cell Biol. 163, 1205–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri, C. J., Paschal, B. M., Vaughan, K. T., and Vallee, R. B. (1996). Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov, V. P. (2003). Roles of NUDE and NUDF Proteins of Aspergillus nidulans: insights from intracellular localization and overexpression effects. Mol. Biol. Cell 14, 871–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov, V. P., and Morris, N. R. (1998). A screen for dynein synthetic lethals in Aspergillus nidulans identifies spindle assembly checkpoint genes and other genes involved in mitosis. Genetics 149, 101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engle, D. B., Osmani, S. A., Osmani, A. H., Rosborough, S., Xiang, X., and Morris, N. R. (1990). A negative regulator of mitosis in Aspergillus is a putative membrane-spanning protein. J. Biol. Chem. 265, 16132–16137. [PubMed] [Google Scholar]
- Faulkner, N. E., Dujardin, D. L., Tai, C. Y., Vaughan, K. T., O'Connell, C. B., Wang, Y., and Vallee, R. B. (2000). A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2, 784–791. [DOI] [PubMed] [Google Scholar]
- Feng, Y., Olson, E. C., Stukenberg, P. T., Flanagan, L. A., Kirschner, M. W., and Walsh, C. A. (2000). LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 28, 665–679. [DOI] [PubMed] [Google Scholar]
- Feng, Y., and Walsh, C. A. (2004). Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 44, 279–293. [DOI] [PubMed] [Google Scholar]
- Gaetz, J., and Kapoor, T. M. (2004). Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 166, 465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser, J. R., Schott, E. J., Kingsbury, T. J., Cole, N. B., Totis, L. J., Bhattacharyya, G., He, L., and Hoyt, M. A. (1997). Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol. Biol. Cell 8, 1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy, P., Pichler, S., Kirkham, M., and Hyman, A. A. (1999). Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J. Cell Biol. 147, 135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and Vale, R. D. (2003). The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162, 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, R. N., Lizarraga, S. B., Wiese, C., Wilde, A., and Zheng, Y. (2000). gamma-Tubulin complexes and their role in microtubule nucleation. Curr. Top. Dev. Biol. 49, 55–73. [DOI] [PubMed] [Google Scholar]
- Gupta, A., Tsai, L. H., and Wynshaw-Boris, A. (2002). Life is a journey: a genetic look at neocortical development. Nat. Rev. Genet. 3, 342–355. [DOI] [PubMed] [Google Scholar]
- Habermann, A., Schroer, T. A., Griffiths, G., and Burkhardt, J. K. (2001). Immunolocalization of cytoplasmic dynein and dynactin subunits in cultured macrophages: enrichment on early endocytic organelles. J. Cell Sci. 114, 229–240. [DOI] [PubMed] [Google Scholar]
- Han, G., Liu, B., Zhang, J., Zuo, W., Morris, N. R., and Xiang, X. (2001). The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11, 719–724. [DOI] [PubMed] [Google Scholar]
- Harper, J. W., Burton, J. L., and Solomon, M. J. (2002). The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16, 2179–2206. [DOI] [PubMed] [Google Scholar]
- Heald, R. (2000). Motor function in the mitotic spindle. Cell 102, 399–402. [DOI] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Habermann, A., Karsenti, E., and Hyman, A. (1997). Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine, R. A., Oberle, J. R., and Cooper, J. A. (2000). The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151, 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran, E. A., Karki, S., and Holzbaur, E. L. (1998). The role of the dynactin complex in intracellular motility. Int. Rev. Cytol. 182, 69–109. [DOI] [PubMed] [Google Scholar]
- Howell, B. J., McEwen, B. F., Canman, J. C., Hoffman, D. B., Farrar, E. M., Rieder, C. L., and Salmon, E. D. (2001). Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155, 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., and Raff, J. W. (1999). The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18, 2184–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S., Yoder, O. C., Turgeon, B. G., and Aist, J. R. (1998). A cytoplasmic dynein required for mitotic aster formation in vivo. J. Cell Sci. 111, 2607–2614. [DOI] [PubMed] [Google Scholar]
- Job, D., Valiron, O., and Oakley, B. (2003). Microtubule nucleation. Curr. Opin. Cell Biol. 15, 111–117. [DOI] [PubMed] [Google Scholar]
- Jung, M. K., Prigozhina, N., Oakley, C. E., Nogales, E., and Oakley, B. R. (2001). Alanine-scanning mutagenesis of Aspergillus gamma-tubulin yields diverse and novel phenotypes. Mol. Biol. Cell 12, 2119–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki, S., and Holzbaur, E. L. (1999). Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol. 11, 45–53. [DOI] [PubMed] [Google Scholar]
- Karki, S., LaMonte, B., and Holzbaur, E. L. (1998). Characterization of the p22 subunit of dynactin reveals the localization of cytoplasmic dynein and dynactin to the midbody of dividing cells. J. Cell Biol. 142, 1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer, E. (1977). Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19, 33–131. [DOI] [PubMed] [Google Scholar]
- King, S. M. (2000). The dynein microtubule motor. Biochim. Biophys. Acta 1496, 60–75. [DOI] [PubMed] [Google Scholar]
- King, S. J., and Schroer, T. A. (2000). Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2, 20–24. [DOI] [PubMed] [Google Scholar]
- Koonce, M. P., Kohler, J., Neujahr, R., Schwartz, J. M., Tikhonenko, I., and Gerisch, G. (1999). Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J. 18, 6786–6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Wisniewski, J. C., Dentler, W. L., and Asai, D. J. (1999). Gene knockouts reveal separate functions for two cytoplasmic dyneins in Tetrahymena thermophila. Mol. Biol. Cell 10, 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W. L., Kaiser, M. A., and Cooper, J. A. (2005). The offloading model for dynein function: differential function of motor subunits. J. Cell Biol. 168, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W. L., Oberle, J. R., and Cooper, J. A. (2003). The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. Y., Yeh, E., Hays, T., and Bloom, K. (1993). Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA 90, 10096–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon, L. A., Karki, S., Tokito, M., and Holzbaur, E. L. (2001). Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat. Cell Biol. 3, 913–917. [DOI] [PubMed] [Google Scholar]
- Lin, H., de Carvalho, P., Kho, D., Tai, C. Y., Pierre, P., Fink, G. R., and Pellman, D. (2001). Polyploids require Bik1 for kinetochore-microtubule attachment. J. Cell Biol. 155, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., and Morris, N. R. (2000). A spindle pole body-associated protein, SNAD, affects septation and conidiation in Aspergillus nidulans. Mol. Gen. Genet. 263, 375–387. [DOI] [PubMed] [Google Scholar]
- Liu, B., Xiang, X., and Lee, Y. R. (2003). The requirement of the LC8 dynein light chain for nuclear migration and septum positioning is temperature dependent in Aspergillus nidulans. Mol. Microbiol. 47, 291–301. [DOI] [PubMed] [Google Scholar]
- Maddox, P. S., Bloom, K. S., and Salmon, E. D. (2000). The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone, C. J., Misner, L., Le Bot, N., Tsai, M. C., Campbell, J. M., Ahringer, J., and White, J. G. (2003). The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115, 825–836. [DOI] [PubMed] [Google Scholar]
- Merdes, A., Ramyar, K., Vechio, J. D., and Cleveland, D. W. (1996). A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447–458. [DOI] [PubMed] [Google Scholar]
- Miki, F., Okazaki, K., Shimanuki, M., Yamamoto, A., Hiraoka, Y., and Niwa, O. (2002). The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol. Biol. Cell 13, 930–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minke, P. F., Lee, I. H., Tinsley, J. H., Bruno, K. S., and Plamann, M. (1999). Neurospora crassa ro-10 and ro-11 genes encode novel proteins required for nuclear distribution. Mol. Microbiol. 32, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Mirabito, P. M., and Morris, N. R. (1993). BIMA, a TPR-containing protein required for mitosis, localizes to the spindle pole body in Aspergillus nidulans. J. Cell Biol. 120, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk, J. N., Schuyler, S. C., Liu, J. Y., Evans, J. G., Salmon, E. D., Pellman, D., and Bloom, K. (2004). The differential roles of budding yeast Tem1p, Cdc15p, and Bub2p protein dynamics in mitotic exit. Mol. Biol. Cell 15, 1519–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, N. R. (2000). Nuclear migration. From fungi to the mammalian brain. J. Cell Biol. 148, 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer, M., Smith, D. S., Ayala, R., Peng, J., Ko, J., Lee, M. S., Morabito, M., and Tsai, L. H. (2000). NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28, 697–711. [DOI] [PubMed] [Google Scholar]
- Oakley, B. R., Oakley, C. E., Yoon, Y., and Jung, M. K. (1990). gamma-Tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell 61, 1289–1301. [DOI] [PubMed] [Google Scholar]
- Osmani, A. H., Osmani, S. A., and Morris, N. R. (1990). The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J. Cell Biol. 111, 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, A. H., Davies, J., Oakley, C. E., Oakley, B. R., and Osmani, S. A. (2003). TINA interacts with the NIMA kinase in Aspergillus nidulans and negatively regulates astral microtubules during metaphase arrest. Mol. Biol. Cell 14, 3169–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, S. A., Engle, D. B., Doonan, J. H., and Morris, N. R. (1988). Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell 52, 241–251. [DOI] [PubMed] [Google Scholar]
- Osmani, S. A., and Mirabito, P. M. (2004). The early impact of genetics on our understanding of cell cycle regulation in Aspergillus nidulans. Fungal Genet. Biol. 41, 401–410. [DOI] [PubMed] [Google Scholar]
- Paluh, J. L., Nogales, E., Oakley, B. R., McDonald, K., Pidoux, A. L., and Cande, W. Z. (2000). A mutation in gamma-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein pkl1p. Mol. Biol. Cell 11, 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, U., and Stearns, T. (2002). gamma-Tubulin. Curr. Biol. 12, R408–R409. [DOI] [PubMed] [Google Scholar]
- Peters, J. M. (2002). The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9, 931–943. [DOI] [PubMed] [Google Scholar]
- Pfarr, C. M., Coue, M., Grissom, P. M., Hays, T. S., Porter, M. E., and McIntosh, J. R. (1990). Cytoplasmic dynein is localized to kinetochores during mitosis. Nature 345, 263–265. [DOI] [PubMed] [Google Scholar]
- Prigozhina, N. L., Oakley, C. E., Lewis, A. M., Nayak, T., Osmani, S. A., and Oakley, B. R. (2004). gamma-Tubulin plays an essential role in the coordination of mitotic events. Mol. Biol. Cell 15, 1374–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina, N. L., Walker, R. A., Oakley, C. E., and Oakley, B. R. (2001). gamma-Tubulin and the C-terminal motor domain kinesin-like protein, KLPA, function in the establishment of spindle bipolarity in Aspergillus nidulans. Mol. Biol. Cell 12, 3161–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne, N. J., Gill, S. R., Eckley, D. M., Crego, C. L., Compton, D. A., and Schroer, T. A. (1999). Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne, N. J., and Schroer, T. A. (2002). Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J. Cell Biol. 159, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehberg, M., Kleylein-Sohn, J., Faix, J., Ho, T. H., Schulz, I., and Graf, R. (2005). Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol. Biol. Cell 16, 2759–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner, O. (2000). LIS1. Let's interact sometimes (part 1). Neuron 28, 633–636. [DOI] [PubMed] [Google Scholar]
- Rischitor, P. E., Konzack, S., and Fischer, R. (2004). The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae. Eukaryot. Cell 3, 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. T., Wojcik, E. J., Sanders, M. A., McGrail, M., and Hays, T. S. (1999). Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146, 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghi, C., and Allan, V. J. (1999). Dynamic association of cytoplasmic dynein heavy chain 1a with the Golgi apparatus and intermediate compartment. J. Cell Sci. 112, 4673–4685. [DOI] [PubMed] [Google Scholar]
- Salina, D., Bodoor, K., Eckley, D. M., Schroer, T. A., Rattner, J. B., and Burke, B. (2002). Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108, 97–107. [DOI] [PubMed] [Google Scholar]
- Sasaki, S., Shionoya, A., Ishida, M., Gambello, M. J., Yingling, J., Wynshaw-Boris, A., and Hirotsune, S. (2000). A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28, 681–696. [DOI] [PubMed] [Google Scholar]
- Saunders, W. S., Koshland, D., Eshel, D., Gibbons, I. R., and Hoyt, M. A. (1995). Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 128, 617–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer, T. A. (2004). Dynactin. Annu. Rev. Cell Dev. Biol. 20, 759–779. [DOI] [PubMed] [Google Scholar]
- Shaw, S. L., Yeh, E., Maddox, P., Salmon, E. D., and Bloom, K. (1997). Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 139, 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman, B., Carvalho, P., Sagot, I., Geiser, J., Kho, D., Hoyt, M. A., and Pellman, D. (2003). Determinants of S. cerevisiae dynein localization and activation. Implications for the mechanism of spindle positioning. Curr. Biol. 13, 364–372. [DOI] [PubMed] [Google Scholar]
- Shu, T., Ayala, R., Nguyen, M. D., Xie, Z., Gleeson, J. G., and Tsai, L. H. (2004). Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263–277. [DOI] [PubMed] [Google Scholar]
- Smith, D. S., Niethammer, M., Ayala, R., Zhou, Y., Gambello, M. J., Wynshaw-Boris, A., and Tsai, L. H. (2000). Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2, 767–775. [DOI] [PubMed] [Google Scholar]
- Steuer, E. R., Wordeman, L., Schroer, T. A., and Sheetz, M. P. (1990). Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature 345, 266–268. [DOI] [PubMed] [Google Scholar]
- Su, W., Li, S., Oakley, B. R., and Xiang, X. (2004). Dual-Color imaging of nuclear division and mitotic spindle elongation in live cells of Aspergillus nidulans. Eukaryot. Cell 3, 553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai, C. Y., Dujardin, D. L., Faulkner, N. E., and Vallee, R. B. (2002). Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 156, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., Serneo, F. F., Higgins, C., Gambello, M. J., Wynshaw-Boris, A., and Gleeson, J. G. (2004). Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 165, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan, S. H., Purohit, A., Doxsey, S. J., and Vallee, R. B. (2000). Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 275, 32763–32768. [DOI] [PubMed] [Google Scholar]
- Vallee, R. B., Tai, C., and Faulkner, N. E. (2001). LIS 1, cellular function of a disease-causing gene. Trends Cell Biol. 11, 155–160. [DOI] [PubMed] [Google Scholar]
- Vardy, L., Fujita, A., and Toda, T. (2002). The gamma-tubulin complex protein Alp4 provides a link between the metaphase checkpoint and cytokinesis in fission yeast. Genes Cells 7, 365–373. [DOI] [PubMed] [Google Scholar]
- Vaughan, K. T., Tynan, S. H., Faulkner, N. E., Echeverri, C. J., and Vallee, R. B. (1999). Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J. Cell Sci. 112, 1437–1447. [DOI] [PubMed] [Google Scholar]
- Vaughan, P. S., Miura, P., Henderson, M., Byrne, B., and Vaughan, K. T. (2002). A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J. Cell Biol. 158, 305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, C., and Zheng, Y. (2000). A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2, 358–364. [DOI] [PubMed] [Google Scholar]
- Wigge, P. A., and Kilmartin, J. V. (2001). The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik, E., Basto, R., Serr, M., Scaerou, F., Karess, R., and Hays, T. (2001). Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat. Cell Biol. 3, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Xiang, X., Osmani, A. H., Osmani, S. A., Xin, M., and Morris, N. R. (1995a). NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol. Biol. Cell 6, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X., Roghi, C., and Morris, N. R. (1995b). Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 92, 9890–9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X., Han, G., Winkelmann, D. A., Zuo, W., and Morris, N. R. (2000). Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr. Biol. 10, 603–606. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., West, R. R., McIntosh, J. R., and Hiraoka, Y. (1999). A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145, 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, X., Li, F., Liang, Y., Shen, Y., Zhao, X., Huang, Q., and Zhu, X. (2003). Human Nudel and NudE as regulators of cytoplasmic dynein in poleward protein transport along the mitotic spindle. Mol. Cell. Biol. 23, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Ukil, L., Osmani, A., Nahm, F., Davies, J., De Souza, C. P., Dou, X., Perez-Balaguer, A., and Osmani, S. A. (2004). Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3, 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E., Skibbens, R. V., Cheng, J. W., Salmon, E. D., and Bloom, K. (1995). Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130, 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, A., Dictenberg, J. B., Purohit, A., Tuft, R., and Doxsey, S. J. (2000). Cytoplasmic dynein-mediated assembly of pericentrin and γ tubulin onto centrosomes. Mol. Biol. Cell 11, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Han, G., and Xiang, X. (2002). Cytoplasmic dynein intermediate chain and heavy chain are dependent upon each other for microtubule end localization in Aspergillus nidulans. Mol. Microbiol. 44, 381–392. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Li, S., Fischer, R., and Xiang, X. (2003). Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol. Biol. Cell 14, 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T., Aumais, J. P., Liu, X., Yu-Lee, L. Y., and Erikson, R. L. (2003). A role for Plk1 phosphorylation of NudC in cytokinesis. Dev. Cell 5, 127–138. [DOI] [PubMed] [Google Scholar]
- Zimmerman, S., and Chang, F. (2005). Effects of {gamma}-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol. Biol. Cell 16, 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]