Abstract
The maturation of eukaryotic secretory cargo initiates cotranslationally and cotranslocationally as the polypeptide chain emerges into the endoplasmic reticulum lumen. Here, we characterized the cotranslational maturation pathway for the human type I membrane glycoprotein tyrosinase. To recapitulate the cotranslational events, including glycosylation, signal sequence cleavage, chaperone binding, and oxidation, abbreviated transcripts lacking a stop codon were in vitro translated in the presence of semipermeabilized melanocyte membranes. This created a series of ribosome/translocon-arrested chains of increasing lengths, simulating intermediates in the cotranslational folding process. Initially, nascent chains were found to associate with the heat shock protein (Hsp) 70 family member BiP. As the nascent chains elongated and additional glycans were transferred, BiP binding rapidly decreased and the lectin-based chaperone system was recruited in its place. The lectin chaperone calnexin bound to the nascent chain after the addition of two glycans, and calreticulin association followed upon the addition of a third. The glycan-specific oxidoreductase ERp57 was cross-linked to tyrosinase when calnexin and calreticulin were associated. This timing coincided with the formation of disulfide bonds within tyrosinase and the cleavage of its signal sequence. Therefore, tyrosinase maturation initiates cotranslationally with the Hsp70 system and is handed off to the lectin chaperone system that first uses calnexin before calreticulin. Interestingly, divergence in the maturation pathways of wild-type and mutant albino tyrosinase can already be observed for translocon-arrested nascent chains.
INTRODUCTION
The efficiency of the protein folding reaction in the cell is generally much higher than that observed for the renaturation of purified proteins. Several factors contribute to the increased fidelity of the cellular reaction, including the assistance of dedicated molecular chaperones and folding enzymes and the removal of malfolded substrates through degradation. An additional advantage is that cellular folding is coupled with translation, producing a vectorial process that tapers the possible ensemble of folding intermediates available during the progression to the native state. Cotranslational folding not only limits possible conformations but also provides a potential mechanism for controlling the environment a nascent chain encounters (Schnell and Hebert, 2003; Clark, 2004).
Proteins that traverse the eukaryotic secretory pathway are targeted to the endoplasmic reticulum (ER) where they are cotranslationally inserted through the membrane by way of the Sec61 translocon. Maturation and processing of the polypeptide chain occurs as the protein emerges in the ER lumen. These processes include signal sequence cleavage, transfer, and trimming of N-linked glycans; chaperone binding; disulfide bond formation; and folding (Chen et al., 1995; Hebert et al., 1997; Molinari and Helenius, 2000; Kowarik et al., 2002; Daniels et al., 2003; Schnell and Hebert, 2003). The translocon and its associated proteins assist the maturation process by providing a protective environment that sterically minimizes the opportunity for protein aggregation (Chen and Helenius, 2000). Understanding the organization of the ER translocon environment and the cotranslational maturation process is of fundamental biological concern.
Classical chaperones, including the heat shock protein (Hsp) 70 system of chaperones, bind to exposed hydrophobic domains that are hallmarks of aberrant, immature, or unassembled proteins. Hsp70s bind substrates in an adenine nucleotide-dependent manner that is regulated by J domain-containing proteins (Bukau and Horwich, 1998). The J-protein initiates the hydrolysis of ATP by the Hsp70 and also is thought to deliver the substrate to Hsp70 (Bukau and Horwich, 1998). The ER Hsp70 family member BiP serves a variety of roles in protein maturation and quality control, including protection of exposed hydrophobic domains from aggregation (Fewell et al., 2001).
In sharp contrast to the Hsp70 system, the ER also contains a lectin chaperone system that binds to exposed hydrophilic protein modifications called N-linked glycans. N-linked glycans are transferred cotranslationally to the majority of the proteins that traverse the secretory pathway. These glycans can act as maturation and quality control tags that mediate interactions with molecular chaperones or quality control sorting receptors (Helenius and Aebi, 2004). The glycan composition provides a signal that determines which factor will be recruited to assist the maturation process. Glycans trimmed of two glucoses to their monoglucosylated state bind to the lectin chaperones calnexin and calreticulin (Ou et al., 1993; Hammond et al., 1994). Alternatively, mannose-trimmed glycans, when coupled to a misfolded or native proteins, bind to EDEM or ERGIC-53, respectively (Helenius and Aebi, 2004). EDEM has been proposed to target misfolded proteins for degradation through the ER-associated protein degradation pathway, whereas ERGIC-53 selectively sorts native proteins for anterograde trafficking. Therefore, N-linked glycans play a significant role in directing the maturation and quality control processes in the early secretory pathway.
The glycoprotein tyrosinase has been extensively used as a model substrate to study the maturation process throughout the secretory pathway (Petrescu et al., 2000; Újvári et al., 2001; Francis et al., 2003). It is a melanocyte-specific protein responsible for the production of melanin in the post-Golgi organelle termed the melanosome (Lerner et al., 1949). Mutations in tyrosinase are associated with loss of pigmentation or albinism because the mutant protein is retained in the ER and subsequently degraded through ER-associated protein degradation process (Berson et al., 2000; Halaban et al., 2000; Svedine et al., 2004). In addition, due to the visual nature of the tyrosinase activity, aberrant tyrosinase trafficking has been used to identify mutations in a number of factors responsible for post-Golgi sorting (Odorizzi et al., 1998). Here, we have investigated the previously unstudied cotranslational maturation processes of tyrosinase. Initially, tyrosinase behaves as a nonglycosylated protein interacting with the ER Hsp70 family member BiP. However, once the protein becomes glycosylated, its interactions shift to the lectin-based system, including calnexin and calreticulin and their associated oxidoreductase ERp57. The temporal association with the various folding factors is characterized, and differences between the cotranslational maturation pathway of wild-type and mutant tyrosinase are uncovered. Together, this study provides us with a better understanding of the maturation process for glycoproteins and the organization of translocon-associated maturation machinery.
MATERIALS AND METHODS
Reagents
T7 transcription kit was obtained from Ambion (Austin, TX). Flexi rabbit reticulocyte lysate, RNasin, and dithiothreitol (DTT) were from Promega (Madison, WI). Restriction endonucleases and peptide N-glycosidase F (PNGase F) were from New England Biolabs (Beverly, MA). Protein A-Sepharose and Zysorbin were obtained from Zymed Laboratories (South San Francisco, CA). EasyTag [35S]Met/Cys and bifunctional sulfhydryl-to-sulfhydryl cross-linker bis-maleimidohexane (BMH) were purchased from PerkinElmer Life and Analytical Sciences (Boston, MA) and Pierce Chemical (Rockford, IL), respectively. Antibodies against the N-terminal 16 amino acids of mature human tyrosinase and C-terminal 19 amino acids of human calnexin (CNX) were produced by injecting rabbits with keyhole limpet hemocyanin-coupled peptides. Polyclonal antibodies against tyrosinase purified from hamster melanomas were provided by Dr. R. Halaban (Yale University School of Medicine, New Haven, CT). Sec61α, ERp57, and BiP antisera were generously provided by Dr. A. E. Johnson (Texas A&M Health Science Center, College Station, TX), Dr. T. Wileman (BBSRC Institute for Animal Health, Surrey, United Kingdom), and Dr. L. M. Hendershot (St. Jude Children's Research Hospital, Memphis, TN), respectively. The calreticulin (CRT) antiserum (PA3-900) was purchased from Affinity Bioreagents (Golden, CO). All other reagents were from Sigma-Aldrich (St. Louis, MO).
Plasmid Construction and Transcription
Truncations of human tyrosinase were created by linearizing tyrosinase cDNA in pSP72- Kb signal sequence (KbSS) (Újvári et al., 2001) with Eco0109, BsgI, SpeI, AflII, AluNI, ApaI, PvuII, StuI, SacI, BsrGI, and NdeI to produce the p92mer, p112mer, 142mer, 170mer, 206mer, 232mer, 297mer, 327mer, 373mer, 413mer, and the full-length (511mer), respectively. The SpeI, AflII, ApaI, StuI, and SacI digestion sites were introduced using QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The human blood coagulation factor V cDNA in the pMT2 vector was kindly provided by Dr. R. J. Kaufman (University of Michigan Medical Center, Ann Arbor, MI) (Jenny et al., 1987). A 5′ cDNA fragment of Factor V was PCR amplified and cloned into pSP72 vector. The vector was linearized using AflII, HindIII to create the 307mer and 456mer constructs, respectively. The abbreviated linearized cDNA trunctions were transcribed with Ambion T7 transcription kit at 37°C for 2 h, and the mRNAs were purified with Qiagen RNeasy kit (QIAGEN, Valencia, CA).
Cell Lines and Preparation of Semipermeabilized Melanocytes
Immortalized wild-type mouse melanocytes, B10BR cells were maintained in Ham's F-10 nutrient medium supplemented with 2 mM glutamine, 100 U/ml penicillin-streptomycin, 7% horse serum, and 50 ng/ml 12-O-tetradecanoyl phorbol-13-acetate at 37°C in 5% CO2 humidified incubator (Tamura et al., 1987). Semipermeabilized cells were prepared from 90% confluent mouse B10BR melanocyte cultures as described previously (Wilson et al., 1995; Francis et al., 2003).
Translation and Translocation
Tyrosinase truncations were translated in the presence of [35S]Met/Cys and translocated into semipermeabilized B10BR melanocytes at 27°C for the proper time to obtain the optimal fraction of ribosome-arrested proteins. The translation reaction was carried out as described previously (Újvári et al., 2001). The translocated chains were isolated by centrifugation at 18,000 × g for 5 min at 4°C upon completion. When reducing conditions are designated, 5 mM DTT also was applied. Translation products were alkylated with either 20 mM N-ethylmaleimide (NEM) or 14 mM 4-acetamido-4′-maleimidylstilbene-2,-2′-disulfonic acid (AMS; in 80 mM Tris-HCl, pH 6.8, and 1% SDS) to block free sulfhydryls when indicated. For puromycin treatment, 1 mM puromycin was added to the newly translated sample and incubated at 27°C for 10 min. For experiments using butyl-deoxinojirimycin (DNJ), the translation mixture lacking only the mRNA was adjusted to 1 mM DNJ, incubated at 27°C for 10 min, and then mRNA was added and translated for the proper time.
Immunoprecipitations
Radiolabeled tyrosinase was subjected to coimmunoprecipitation with anti-CNX, -CRT, or -BiP antisera or subjected to denaturing immunoprecipitation after chemical cross-linking with anti-Sec61α, -CNX, -CRT or -ERp57. For coimmunoprecipitation, alkylated samples were solubilized in 2% (wt/vol) 3-[3-cholamidopropyl-dimethylammonio]-1-propanesulfonate (CHAPS) in HBS (50 mM HEPES and 200 mM NaCl, pH 7.5) and precleared with Zysorbin by rotating for 1 h at 4°C. After rotation, samples were centrifuged at 18,000 × g at 4°C for 5 min. Protein A-Sepharose beads and antibody were added to the supernatant and rotated overnight at 4°C. For BiP coimmunoprecipitations, 10 U/ml apyrase also was added. Samples were washed with 0.5% CHAPS in HBS, and the beads were resuspended in gel loading buffer. For denaturing immunoprecipitation, samples were resuspended in denaturing buffer (100 mM Tris-HCl, pH 7.6, and 1% SDS) and denatured at 55°C for 45 min. IP buffer [10 mM Tris-HCl, pH 7.6, 140 mM NaCl, and 2% (vol/vol) Triton X-100] was added, and samples were rotated at 4°C for 2 h to quench the SDS. The protein A-Sepharose beads and antibody were added, and samples were rotated overnight. The samples were washed sequentially with IP buffer followed by IP buffer without Triton X-100 and then resuspended in gel loading buffer.
Chemical Cross-linking
Translations were stopped with 2 mM cycloheximide and placed on ice for 10 min. The translation products were spun down at 18,000 × g for 1 min to isolate the translocated proteins. The pellet was resuspended in salt resuspension buffer [250 mM sucrose, 100 mM KOAc, 5 mM Mg(OAc)2, and 50 mM HEPES-KOH, pH 7.9] and 0.1 mM BMH was added to cross-link possible interactions between ER chaperones and tyrosinase. The interactions were identified using denaturing immunoprecipitations.
Cetyltrimethylammonium Bromide (CTAB) Precipitation and PNGase F Digestion
The ribosome-arrested chains were precipitated with CTAB as described by Gilmore et al. (1991) after isolation of the translocated protein by sedimentation at 18,000 × g (Gilmore et al., 1991). In brief, the pellets were resuspended in 1% CTAB and 250 mM NaOAc, pH 5.4, and 0.5 U of yeast tRNA were added to accelerate the precipitation. Samples were incubated at 27°C for 20 min, and the precipitates were collected by microcentrifugation at 18,000 × g for 5 min. The pellets were then washed with 1:19 (vol/vol) HCl:acetone twice, dried at room temperature, and resuspended in gel loading buffer. For PNGase F digestion, the translocated chains isolated by microcentrifugation were resuspended in 0.5% SDS and 125 mM β-mercaptoethanol and incubated at 100°C for 10 min to denature the protein. Samples were adjusted to 50 mM sodium phosphate and 1% NP-40, pH 7.5, before digestion with 250 U of PNGase F for 12 h at 37°C.
RESULTS
Translocation and Glycosylation of Ribosome-attached Tyrosinase
To analyze the cotranslational maturation pathway of tyrosinase and probe the translocon-associated environment encountered by the nascent chain, tyrosinase truncated chains of increasing length were in vitro translated in the presence of melanocyte-derived ER membranes. The translation of abbreviated transcripts lacking a stop codon results in the production of translation intermediates, which are ribosome attached (Krieg et al., 1989; Gilmore et al., 1991). When performed in the presence of ER-derived membranes, signal sequence containing-nascent chains are arrested within the translocon enabling the characterization of translocon-associated nascent chains.
Optimal maturation of tyrosinase requires a melanocyte-specific factor (Kobayashi et al., 1998; Francis et al., 2003). Therefore, to follow the proper cotranslational maturation of tyrosinase using translocon-arrested nascent chains, the in vitro translation of tyrosinase was coupled with a semipermeabilized melanocyte (SP-melanocyte) system. SP-melanocytes were made by treating the cells with low levels of the detergent digitonin (0.002%) that has been shown to create a leaky plasma membrane while leaving the ER intact (Wilson et al., 1995). We have previously shown that digitonin-treated melanocytes support the efficient ER translocation, glycosylation, folding, and oligomerization of full-length tyrosinase (Újvári et al., 2001; Francis et al., 2003).
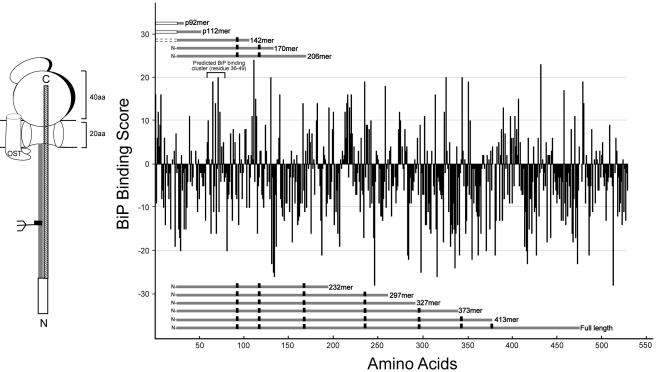
The translocation and glycosylation of tyrosinase were characterized for full-length as well as 10 truncations of increasing lengths to recapitulate the vectorial cotranslational maturation process. The truncations were termed Xmers, where X equals the number of amino acids of the mature signal sequence-cleaved protein, or pYmers, where Y equals the number of amino acids in the preprotein that includes a 24-amino acid signal sequence. In these studies, the native tyrosinase signal sequence was replaced with the murine major histocompatibility complex class I molecule KbSS, because it has been shown to increase translation and translocation by severalfold (Huppa and Ploegh, 1997). On translocation of a polypeptide into the ER lumen, N-linked glycans are predominantly transferred cotranslationally once the consensus site has emerged 11–13 amino acids deep into the lumen (Whitley et al., 1996). Therefore, the consensus site must be >70 amino acids away from the C terminus of the truncation or the ribosomal P-site to be efficiently recognized.
Although the 170mer contains three glycosylation sites, only the first two will be recognized because the third site on Asn143 is 27 amino acids away from the ribosomal P-site. This likely positions Asn143 within the ribosome, unless the nascent chain is released (Figure 9). The characteristics of the tyrosinase truncations are displayed in Table 1 for both ribosome-arrested and ribosome-released nascent chains.
Figure 9.
Summary of the results with the tyrosinase ribosome-arrested chains. Cartoons for human tyrosinase C-terminal truncations are displayed. The preprotein length is indicated as pYmer (which includes the 24-amino acid signal sequence) and the mature protein length as Xmer. Cys (circles), consensus glycosylation sequences (filled boxes) and oligosaccharides (branched structures) are designated. The glycosylation sites are numbered based on the mature protein. The distance from the ribosomal peptidyl transferase center to the OST active site is ∼70 amino acids. The table above the cartoon summarizes the results obtained from this study.
Table 1.
Characteristics of tyrosinase ribosome-arrested chains
| Molecular mass (kDa)
|
||||||
|---|---|---|---|---|---|---|
| No. of glycans
|
Translocated
|
|||||
| Truncation length (amino acids) | Ribosome-arrested | Ribosome-released | Untranslocated | Arrested | Released | Met/Cys |
| p92 | 0 | 0 | 10 | 10 | 10 | 3/6 |
| p112 | 0 | 1 | 12 | 12 | 15 | 4/10 |
| 142 | 1 | 2 | 18 | 21 | 24 | 3/10 |
| 170 | 2 | 3 | 22 | 24 | 27 | 5/10 |
| 206 | 2 | 3 | 26 | 28 | 31 | 5/10 |
| 232 | 3 | 4 | 29 | 34 | 37 | 5/12 |
| 297 | 4 | 5 | 36 | 44 | 46 | 6/14 |
| 327 | 4 | 5 | 40 | 47 | 50 | 7/15 |
| 373 | 4/5 | 5/6 | 45 | 52/55 | 55/57 | 10/15 |
| 413 | 5/6 | 6/7 | 50 | 60/62 | 62/65 | 11/15 |
| 511a | 6/7 | 6/7 | 61 | 73/76 | 73/76 | 13/16 |
Full-length protein
The proper translation time for each construct was determined to generate the maximum fraction of ribosome-arrested chains. Arrested and released chains were identified by recognition of glycosylation sites residing in the ribosome/translocon region upon release with the protein synthesis inhibitor puromycin (data not shown). The shorter nascent chains required briefer times of translation and translocation. A small fraction of released-chains persisted with the 327mer, as seen by a decrease in mobility caused by the addition of a glycan to the ribosomal sequestered glycosylation site (Figure 1, lane 16, see square; and Figure 9, 327mer).
Figure 1.
ER targeting and glycosylation of tyrosinase truncations in semipermeabilized melanocytes. Tyrosinase (TYR) truncations radiolabeled with [35S]Met/Cys were translated in the absence or presence of semipermeabilized melanocytes (SPM) and then immunoprecipitated with TYR antiserum. The translation time for each construct was optimized to gain the maximum fraction of ribosome-arrested proteins. A 17-min translation time was applied for the p92mer, p112mer, and 142mer; 30 min for the 170mer and 206mer; 40 min for the 232mer and 297mer; 50 min for the 327mer and 373mer; and 60 min for the 413mer and full-length chain, respectively. Samples were analyzed by 14% (left) and 8.5% (right) Tris-tricine SDS-PAGE, respectively, followed by autoradiography. Open and closed circles indicate untranslocated and translocated proteins, respectively. The number of glycans of the ER-translocated tyrosinase is indicated by the numbers next to the closed circles. Closed squares designate the ribosome-released protein. Note the species observed, which are smaller than the translocated and glycosylated proteins, are likely caused by ribosomal pausing or premature termination.
The 11 transcripts were translated in the absence (Figure 1, odd-numbered lanes) or presence of SP-melanocytes (Figure 1, even-numbered lanes) to determine the degree of glycosylation because each glycan adds ∼2.5 kDa. As expected for the first two constructs, p92mer and p112mer, no change in mobility was observed because the first glycan for tyrosinase is found at Asn68, which has not yet entered the lumen for either construct. The first glycan was transferred to the 142mer as seen by the decrease in mobility in the presence of SP-melanocytes (Figure 1, lanes 5 and 6). During the cotranslational synthesis of full-length tyrosinase, Asn272 is only partially recognized by the oligosaccharyl transferase (OST) due to a Pro in the Y position (Asn-X-Ser/Thr-Y) (Újvári et al., 2001). This pattern also was retained in the arrested chain system because a doublet of translocated and glycosylated bands was first observed for the 373mer, which is the first construct that can exhibit the partial recognition of Asn272. Therefore, constructs possessing the proper number of transferred glycans, ranging from zero to seven, were created to simulate the vectorial nature of the cellular cotranslational maturation process.
Initial Implementation of the Hsp70 Chaperone System
The ER Hsp70 family member BiP plays a variety of roles within the ER lumen. These functions include maintaining the permeability barrier, aiding in polypeptide translocation, acting as a molecular chaperone, activating the unfolded protein response, and targeting nonnative substrates for degradation (for review, see Fewell et al., 2001). To determine whether BiP interacts with translocon-associated nascent chains of tyrosinase, 35S-labeled tyrosinase truncations were coimmunoprecipitated with anti-BiP antibodies from ATP-depleted lysates generated with the nondenaturing detergent CHAPS (Figure 2).
Figure 2.
BiP binding to translocon-arrested tyrosinase nascent chains. Tyrosinase truncations radiolabeled with [35S]Met/Cys were translated in the presence of semipermeabilized melanocytes. Translation lysates (Lys), BiP coimmunoprecipitation products, concanavalin A precipitation products (Con-A) are indicated (A and B). Samples were analyzed by 14% (A) and 8.5% (B) nonreducing Tris-tricine SDS-PAGE, respectively, followed by autoradiography. The fraction of total tyrosinase bound by BiP from two individual experiments is indicated at the bottom of each lane. The number of glycans for each translocated construct is designated.
BiP binding commenced with the 142mer (Figure 2, lane 8). This same construct also showed the maximum BiP association, coimmunoprecipitating ∼10% of the total translocated tyrosinase. For the 142mer, ∼80 amino acids reside within the lumen of the ER and the glycan at Asn68 is present. The binding rapidly diminished with the longer constructs that contained additional glycans. BiP binding was specific because excess ATP in the BiP coimmunoprecipitation of the 142mer resulted in the loss of tyrosinase binding (data not shown). Background levels of BiP binding were observed for nascent chains longer than the 170mer.
BiP recruitment is dependent upon its ability to interact with hydrophobic amino acids in extended conformations. A BiP binding algorithm based on the primary amino acid sequence was created using the parameters established by Blond-Elguindi et al. (1993). This algorithm was used to identify regions of tyrosinase that may support BiP binding (Figure 3). Heptapeptides with a score >10 have a high probability of binding to BiP. Where the N-terminal signal sequence score exceeded 10, this hydrophobic domain is expected to remain within the translocon or lipid bilayer before its cleavage by the signal peptidase complex, hindering its accessibility to the soluble chaperone (Alder and Johnson, 2004). The first binding region with a score >10 that lies on an exposed, soluble, and lumenal domain on tyrosinase encompasses residues 36–49 in the mature protein. In accordance with the BiP coimmunoprecipitation data (Figure 2), the 142mer is the first truncation that is capable of extending this domain into the ER lumen. Although all proceeding constructs contained this domain as well as additional C-terminal regions that showed sufficient BiP binding scores (Figure 3), coimmunoprecipitation data showed that BiP binding decreased to background levels for all constructs longer than 170mer (Figure 2). The BiP binding data and the algorithm both identify an N-terminal BiP binding domain on tyrosinase that transiently interacts with the Hsp70 chaperone until further maturation occurs as the protein extends into the ER lumen.
Figure 3.
Prediction of BiP-binding regions for human tyrosinase. Overall score for each of the overlapping heptapeptides in the sequences were calculated according to the BiP binding score described by Blond-Elguindi et al. (1993) and plotted against the residue number of the first amino acid of each heptapeptide. The portion of tyrosinase truncations emerged out of the ER translocon is indicated by a gray bar for each construct with black rectangles and solid and dotted squares indicating the consensus glycosylation sites, uncleaved, and cleaved signal sequence, respectively. The predicted BiP binding cluster between residues 36–49 in the mature protein is indicated. The cartoon on the left shows the entire ribosome arrested nascent chain as a patterned bar with a signal sequence and glycan. The portion of the nascent chain within the ER lumen, ∼60 amino acids shorter than the truncation length is indicated by a gray bar.
Initiation of the Lectin-based Chaperone System
The lectin chaperones calnexin and calreticulin can bind translocating chains cotranslationally (Chen et al., 1995; Daniels et al., 2003). These interactions help to assist the maturation of nascent chains by protecting them from deleterious associations. To determine the temporal association of tyrosinase with calnexin and calreticulin, 35S-labeled tyrosinase translocation intermediates were translated and coimmunoprecipitated from SP-melanocytes with anti-calnexin and anti-calreticulin antisera under native conditions (Figure 4). In addition, concanavalin A (Con-A) precipitations were used to identify the translocated and glycosylated products.
Figure 4.
Calnexin and calreticulin binding to tyrosinase ribosome-arrested chains. Tyrosinase truncations radiolabeled with [35S]Met/Cys were translated in the presence of semipermeabilized melanocytes. Translation lysates (Lys), concanavalin A precipitation products (Con-A), coimmunoprecipitation products with CNX and CRT antisera are indicated (A and B). Samples were analyzed by 14% (A) and 8.5% (B) Tris-tricine SDS-PAGE, respectively. The glycan numbers for each construct is indicated next to the translocated protein. The binding ratio of CNX or CRT to total tyrosinase was based on three individual experiments quantified using Image Quant 1.2 software (C). White and gray bars represent CNX and CRT binding, respectively.
Binding to lectin chaperones was not observed with the three shortest constructs (Figure 4A, lanes 1–12). Weak binding to calnexin commenced with the 170mer, which possessed two glycans. The level of calnexin binding increased with polypeptide length and glycan number, reaching a maximum of ∼15% with the 327mer and the full-length chains. Calreticulin binding followed calnexin, because slight binding to calreticulin was first observed with the 206mer. This binding became more pronounced with the addition of a third glycan, eventually reaching the maximum level at 4% with the full-length protein, which contains heterogeneous tyrosinase populations with six or seven glycans.
Together, these results demonstrate that BiP initially binds the N-terminal nonglycosylated domain of tyrosinase. Once two glycans are transferred, binding to calnexin begins, which is followed by calreticulin binding after further elongation and the transfer of additional glycans. Therefore, the presence of the glycans or calnexin and calreticulin binding to these glycans inhibits BiP binding, indicating that the lectin system is the dominant chaperone system for glycoproteins.
Factor V Also Binds Calnexin before Calreticulin
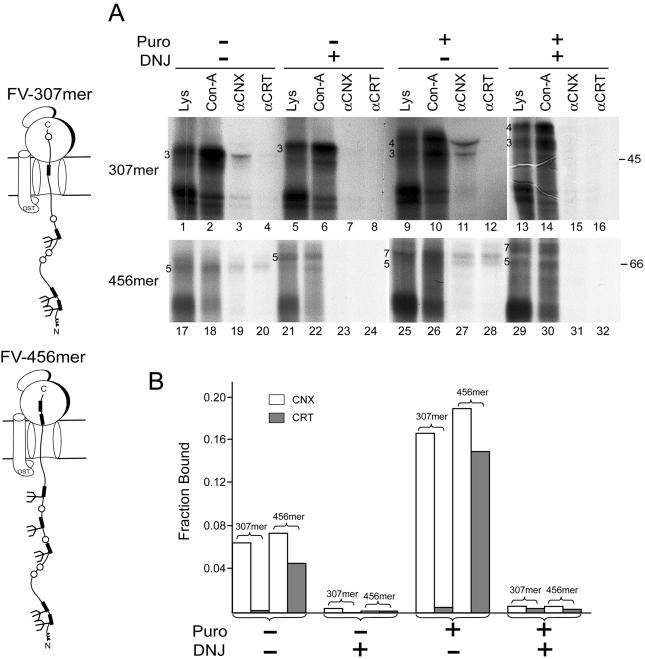
Ribosome-arrested glycoproteins have been shown to associate with the membrane-bound calnexin before the soluble calreticulin for both tyrosinase and influenza hemagglutinin (HA), with elongation and further glycan transfer being a key to the involvement of calreticulin (Daniels et al., 2003; Figure 4). To determine whether the chaperone binding order represents the organization of the translocon-associated assembly line that awaits incoming nascent chains, binding of the lectin chaperones to ribosome-arrested chains of the blood coagulation protein Factor V was studied. Factor V is a large, heavily glycosylated, and soluble protein that binds to calreticulin but not calnexin upon completion of its synthesis (Pipe et al., 1998) (Wang and Hebert, unpublished data). Since calnexin is not associated with the full-length protein, ribosome-arrested chains of Factor V should not have an inherent bias toward remaining bound to calnexin. This makes Factor V an ideal substrate to determine whether there is an order involved in lectin chaperone binding, specifically whether calnexin binds before calreticulin.
Constructs of 307 and 456 residues, which contained three and five glycans respectively, were translated in the presence of [35S]Met/Cys and rough ER-derived microsomes. Interestingly, the 307mer was only found associated with calnexin (Figure 5, lane 3). Binding to calnexin required glucose trimming because it was inhibited by the glucosidase inhibitor DNJ (Figure 5, lane 7). In addition, this pattern of binding continued after the release of the nascent chain from the ribosome with puromycin (Figure 5, lanes 9–16). For the 456mer that contains five glycans, Factor V associated with both calnexin and calreticulin in a glucose trimming-dependent manner and continued this trend, at a higher level upon release with puromycin (Figure 5, lanes 17–32). This indicated that calnexin bound Factor V before calreticulin, suggesting that calnexin is localized first in the chaperone assembly line for the lectin chaperone system.
Figure 5.
Calnexin and calreticulin binding to human Factor V ribosome-arrested chains. Factor V 307mer and 456mer radiolabeled with [35S]Met/Cys were translated in the presence of rough ER-derived microsomes with puromycin (Puro) and DNJ treatments as indicated. Translation lysates (Lys), concanavalin A precipitation products (Con-A), and coimmunoprecipitation products with CNX or CRT antisera are indicated. Samples were analyzed by 10% SDS-PAGE (A). The binding ratio of total factor V bound by CNX or CRT was quantified using Image Quant 1.2 software and is displayed in the plot (B). The cartoons on the left indicate the predicted configurations of each construct.
Chemical Cross-linking of Nascent Chains to Sec61α and ERp57
The studies described above explored interactions between nascent tyrosinase chains and the translocon-proximal factors BiP, calnexin, and calreticulin, which survived a native coimmunoprecipitation procedure. To identify weaker and possibly more transient associations, chemical cross-linking was used to trap interactions with translocation intermediates. Since folding intermediates are expected to contain free Cys residues, the cross-linker BMH was effectively used to intermolecularly link free Cys thiols before membrane lysis.
Cross-linking the 10 translocation intermediates and full-length 35S-labeled tyrosinase in SP-melanocytes with BMH trapped a variety of adducts (Figure 6). The cross-linked partners of tyrosinase were identified by determining their size and by using a variety of antibodies raised against Sec61α, calnexin, calreticulin, and the oxidoreductase ERp57 for immunoprecipitation. Here, the immunoprecipitation protocol used denaturing conditions to ensure that the components were covalently linked. Additional cross-linked species were observed with the 142mer (Figure 6, lane 14); however, the cross-linked partners could not be identified using either anti-BiP, GRP94, PDI, calnexin, or calreticulin antisera (data not shown).
Figure 6.
Interaction of Sec61α and ERp57 with tyrosinase ribosome-arrested chains detected after chemical cross-linking. Tyrosinase truncations radiolabeled with [35S]Met/Cys were translated in the presence of semipermeabilized melanocytes and subjected to cross-linking with BMH as indicated. Translation lysates (Lys), samples immunoprecipitated under denaturing conditions with Sec61α, CNX, CRT, and ERp57 antibodies are indicated. Samples from the p92mer to 327mer and from 413mer to full length were analyzed by reducing 7.5–15% gradient and 7.5% SDS-PAGE, respectively. Uncrosslinked translocated tyrosinase is indicated by the number of glycans and tyrosinase cross-linked to Sec61α and ERp57 are identified as open squares and stars, respectively.
For the three shortest translocation intermediates—p92mer, p112mer, and 142mer—a single adduct was identified. This ∼40-kDa cross-linking partner was identified as Sec61α through coimmunoprecipitation with anti-Sec61α antibodies (Figure 6, open squares). Cross-linking partners of 40 kDa that immunoprecipitated with anti-Sec61α antibodies also were identified with the 297mer. These four translocation intermediates all contain one or more Cys residues that are predicted to lie within the translocon using the molecular ruler defined above and displayed in Figure 9.
The chemical cross-linker did not trap interactions with calnexin and calreticulin. Previous studies using a similar protocol were successful in covalently linking influenza HA with calreticulin but not calnexin, because the single free Cys residue on calnexin is localized distally to the carbohydrate binding site (Schrag et al., 2001; Daniels et al., 2003). However, a 60-kDa cross-linking partner was observed with the 170mer and all longer constructs except the 232mer. This 60-kDa partner was identified by immunoprecipitation as the oxidoreductase partner of calnexin and calreticulin, ERp57 (Figure 6, lanes 24, 30, 48, 54, 60, and 66, designated by a star). Interestingly, of all the constructs 170 residues and longer, only the 232mer did not support ERp57 cross-linking. The 232mer has 10 Cys exposed to the ER lumen, which are possibly paired into five N-terminal disulfides, thereby protecting the truncation from being modified. The mutation of Cys71 to an Arg in the mature protein is associated with human albinism (Oetting and King, 1999). A similar alteration in the mouse protein results in the misfolding of tyrosinase and its subsequent degradation through the ER-associated protein degradation pathway (Halaban et al., 2000; Svedine et al., 2004). Unlike the wild-type 232mer, 232merC71R was cross-linked to ERp57 (Figure 6, compare lanes 36 and 42), indicating the availability of one or more free thiols. This strongly implies that C71 is paired with another Cys in a disulfide very early on and the cotranslational divergence of wild-type and mutant pathways may be due to an unpaired Cys or improper disulfide bond formation.
Disulfide Bond Formation
Disulfide bond formation can begin cotranslationally (Chen et al., 1995). Disulfide bonds generally create a more compact structure, which migrates faster upon nonreducing SDS-PAGE (Braakman et al., 1991). Therefore, to determine when disulfide bond formation commenced for tyrosinase, the mobility of tyrosinase arrested chains were monitored by nonreducing SDS-PAGE. Tyrosinase arrested chains were generated in the presence of SP-melanocytes under reducing (Figure 7A, even-numbered lanes) and oxidizing conditions (Figure 7A, odd-numbered lanes). The biological oxidizing agent FAD was added to create an oxidizing environment, which would support disulfide bond formation within the lumen of the ER (Francis et al., 2003). The oxidative state of tyrosinase was trapped with the alkylating agent NEM. 35S-labeled tyrosinase was then analyzed directly upon non-reducing SDS-PAGE (Figure 7A).
Figure 7.
Disulfide bond formation of tyrosinase initiates with the 142mer. Tyrosinase truncations from the p92mer to the 232mer radiolabeled with [35S]Met/Cys were translated under oxidizing (Ox) or reducing (Rd) conditions in the presence of semipermeabilized melanocytes. The translated proteins were alkylated with (A) NEM or (B) AMS and resolved on 14% nonreducing Tris-tricine SDS-PAGE, followed by autoradiography. Open and closed circles indicate the untranslocated and translocated proteins, respectively. The bands corresponding to the p92mer or p112mer was a mixture of the untranslocated and translocated proteins. The white lines were added to facilitate visualization of the mobility shifts between the oxidized and reduced proteins.
No change in mobility was observed for the two shortest constructs, p92mer and p112mer, when comparing alkylated tyrosinase translated under reducing versus oxidizing conditions (Figure 7A, lanes 1–4). However, the oxidized 142mer migrated faster than the corresponding reduced protein, and this property persisted with the longer constructs. Therefore, disulfide bond formation initiates with the 142mer. It should be noted that when alkylating agents were absent only a subtle shift was observed between the oxidized and reduced 232mer (Figure 7B, lanes 1 and 4). This indicates that oxidized tyrosinase is likely comprised of small intramolecular loops that do not significantly contribute to the compactness of the protein under nonreducing conditions.
Oxidation of the human albino mutation C71R also was explored with the 232mer using the bulky, 536-Da AMS (Figure 7B, which further enhanced oxidative differences. Interestingly, a larger mobility shift was observed for the oxidized albino Cys mutation 232merC71R in the presence of AMS than the wild-type protein (Figure 7B, lanes 1 and 2 and 5 and 6). Additional misfolding created by the absence of a Cys residue or the presence of a lone Cys residue likely caused this accentuated shift. Together, these data imply that the vectorial oxidation of tyrosinase commenced with the 142mer, and the cotranslational divergence of the oxidation pathway in the wild-type and mutant tyrosinase can be observed using ribosome-arrested chains.
Signal Sequence Cleavage
N-terminal signal sequences target secretory pathway proteins to the ER. The hydrophobic nature of the sequence produces an N-terminal membrane tether until it is cleaved, which can greatly affect the folding reaction. To determine when the 24-amino acid signal sequence was cleaved, the mobility of the four shortest constructs was monitored in the absence and presence of SP-melanocytes (Figure 8).
Figure 8.
Signal sequence cleavage of tyrosinase initiates with the 142mer. Tyrosinase p92mer, p112mer, 142mer, and 170mer radiolabeled with [35S]Met/Cys were translated in the presence of semipermeabilized melanocytes and subjected to CTAB precipitation, PNGase F digestion, and puromycin release as indicated. Samples were analyzed by 14% reducing Tris-tricine SDS-PAGE. The untranslocated (open circles), translocated (solid circles), released (solid squares), signal sequence uncleaved (UC) and cleaved (C) proteins are designated. Numbers indicate the number of glycans transferred.
No cleavage was observed for the p92mer or p112mer with the inclusion of melanocyte membranes during the translation because the translocated and untranslocated proteins migrated with equivalent mobility (Figure 8, lanes 1 and 2). The 142mer and 170mer received one and two glycans, respectively. Therefore, intermediates also were treated with the endoglycosidase PNGase F to remove the complete carbohydrate modification (Figure 8, lanes 4 and 5). The membrane-translocated 142mer and 170mer migrated further than the untranslocated signal sequence-containing proteins after deglycosylation, indicating that the signal sequence had been cleaved (Figure 8, lanes 1 and 4).
To ensure that the translation intermediate was arrested on the ribosome, the cationic detergent CTAB was used to precipitate translation products containing a C-terminal attached tRNA (Figure 8, lanes 3 and 5). In addition, the protein synthesis inhibitor puromycin was added to release intermediates from the ribosome. Release from the ribosome permitted additional glycan transfer to the p112mer, 142mer, and 170mer, indicating that a glycosylation site was protected within the translocon for the arrested chains (Figure 8, lane 6). Together, these results indicate that the 142mer is the initial tyrosinase translocon-arrested translation intermediate to have its signal sequence cleaved.
DISCUSSION
We have characterized the cotranslational maturation process for the type I membrane glycoprotein tyrosinase, and four main conclusions could be reached from our studies. First, glycans are a dominant factor in directing cotranslational chaperone interactions in the ER. Second, a substrate that possesses its first glycan at position 68 can first engage the ER Hsp70 system before being passed on to the lectin chaperone system. Next, calnexin is localized before calreticulin in the chaperone assembly line of the ER. And finally, differences between the maturation of wild-type and mutant tyrosinase can already be observed cotranslationally.
Ribosome-arrested chains have been previously used in combination with canine pancreas rough ER-derived microsomes to identify the components of the translocation machinery. In addition, we have previously used this system to map the early maturation process for HA. However, tyrosinase is a melanocyte protein that requires melanocyte-specific factors for its proper maturation. Therefore, we have developed a melanocyte-derived membrane system to monitor its maturation using semipermeabilized melanocytes. Here, in vitro-translated tyrosinase targeted to intact melanocytic ER membranes was investigated.
Tyrosinase nascent chains initially interacted with the ER translocon protein Sec61α. Interactions between Sec61α, and the p92, p112mers, and 142mers could be trapped with the thiol-reactive cross-linker BMH because the N terminus of tyrosinase is rich in Cys residues. Cross-links to Sec61α were only observed when a tyrosinase Cys residue was localized within the translocon channel, positioned 40–60 amino acids away from the ribosomal P-site (Figure 9).
Partial cleavage of the N-terminal hydrophobic signal sequence was first observed with the 142mer when the polypeptide was 166 amino acids in length (142 mature amino acids + 24 amino acids of the signal sequence) with the cleavage site positioned 142 amino acids from the ribosomal P-site. This timing is in general agreement with previously observed cleavages for HA and preprolactin where signal sequence cleavage first occurred when the cleavage site was 95 residues and 101 residues from the P-site, respectively (Nicchitta et al., 1995; Daniels et al., 2003). To optimize translation and translocation levels, the native tyrosinase signal sequence was exchanged for the signal sequence of myosin heavy chain class I Kb heavy chain (Huppa and Ploegh, 1997). This exchange does not effect the proper maturation of tyrosinase (Ujvári et al., 2001; Francis et al., 2003). However, it cannot be ruled out that the properties of the individual signal sequences influence the timing of its cleavage and later maturational events (Rutkowski et al., 2003). Cleavage of the hydrophobic signal sequence coincided with a variety of lumenal events. These events included BiP binding, glycosylation, disulfide bond formation, and cross-linking to the oxidoreductase ERp57 (Figures 1, 2, 6, and 7).
BiP in its high-affinity ADP-bound form is localized to the lumenal face of the translocon likely by the J-domain–containing translocon-associated protein Sec63p as well as through its own peptide binding domain (Meyer et al., 2000; Tyedmers et al., 2000; Alder et al., 2005). BiP acts to maintain the permeability barrier of ribosome-free translocons, and ribosome-bound translocons during the protein translocation process (Crowley et al., 1994; Hamman et al., 1998). The BiP-mediated seal is released when the nascent chain reaches the length of ∼70 amino acids, the point at which nascent chain residues first become lumenal. The positioning of BiP at the translocon exit site also optimally places BiP to interact with the emerging nascent chain.
Tyrosinase contains several potential BiP binding regions, yet only the early signal sequence cleaved translocation intermediates, the 142mer and 170mer, were found associated with BiP at significant levels (Figures 2 and 3). Maximal BiP binding was observed with the 142mer that has ∼80 residues localized within the lumen, a glycan at Asn68, eight lumenal Cys residues, and a potential BiP binding region between amino acids 36–49 (Figures 3 and 9). Interestingly, this optimal BiP binding region would be present on all the following constructs, yet BiP binding decreased for all these succeeding constructs. The lack of BiP binding to the longer constructs could be caused by the folding and sequestration of the BiP binding domain, the added hydrophilicity created by the addition of multiple glycans, or steric hindrance supported by the binding of the lectin chaperones calnexin and calreticulin.
In support of the N-terminus folding and possibly masking the BiP binding site, no free Cys are available for ERp57 cross-linking by the 232mer when the N-terminal 10 cysteine cluster is entirely present. This suggests that all 10 Cys at this point may be involved in disulfide bonds. Folding of the N-terminal region, the presence of additional glycans, and the accessibility of the lectin chaperones all likely contribute to the quick transfer from the Hsp70 system on the 142mer to the lectin system by the 206mer, with the 170mer as an intermediate stage. These results support the hypothesis proposed by Molinari and Helenius (2000) that glycoproteins with an N-linked glycan in its first 50 amino acids do not bind BiP. Here, positioning the first glycan at Asn68 on the mature protein permits initial interaction with BiP before calnexin for the ribosome-arrested chains.
The transfer of a substrate from Hsp70 system to the carbohydrate-based chaperone system is intriguing. BiP exists in a complex containing PDI; GRP94; CaBP1; ERdj3, a recently identified ER Hsp40 cochaperone; cyclophilin B; ERp72; GRP170; GT; and SDF2-L1 but not the lectin system components calnexin, calreticulin, or ERp57 (Meunier et al., 2002). In this study, BiP was observed to bind before calnexin, calreticulin, and ERp57. This sequential interaction of BiP followed by calnexin also was previously observed for full-length vesicular stomatitis virus G protein and nascent chains of Semliki Forest virus glycoproteins (Hammond and Helenius, 1994; Molinari and Helenius, 2000). The absence of the lectin components in the BiP complex, but the ability of the two systems to interchange implies that both systems are mobile within the ER.
Once the bulky hydrophilic modifications are added, the lectin-based chaperone system takes over, even though multiple BiP binding regions exist, indicative of carbohydrates being the dominant signal for chaperone binding in the ER. The membrane protein calnexin seems to be localized earlier in the chaperone assembly line than the soluble calreticulin because for both tyrosinase and HA, binding to calnexin is initiated after the transfer of two glycans followed by calreticulin with the addition of a third (Daniels et al., 2003) (Figure 4). The blood clotting protein Factor V, which post-translationally only binds calreticulin (Pipe et al., 1998), also binds calnexin first and requires longer, more glycosylated ribosome-arrested chains to associate with calreticulin (Figure 5). Together, these results are indicative of an organization of translocon-associated factors that place chaperones in the order of BiP, calnexin, and then calreticulin.
Intramolecular disulfide bond formation also commenced with the 142mer. At this point, eight Cys residues should be exposed to the oxidizing environment of the ER lumen and associations were identified with Sec61α and BiP. No association with PDI could be identified (our unpublished data). However, the longer constructs were associated with oxidoreductase ERp57. The specificity of ERp57 is determined by its association with calnexin or calreticulin (Zapun et al., 1998; Oliver et al., 1999). The ternary complex between the nascent chain, ERp57, and either calnexin or calreticulin allows electrons to be delivered to ERp57 to support substrate disulfide bond formation.
ERp57 binding persisted for all constructs longer than the 142mer, except the 232mer. The 232mer is comprised of 10 lumenal Cys residues that seem to be completely paired up in intramolecular disulfides for the wild-type protein. Interestingly, ERp57 cross-linking persisted with the 232mer for the albino mutation of tyrosinase. This mutation associated with the balb c mouse involves the alteration of Cys71 to a Ser. The mutant protein is retained within the ER and subsequently targeted to the ER-associated protein degradation process for degradation (Halaban et al., 2000; Svedine et al., 2004). Exposed thiols can mediate the retention of nonnative proteins in the ER through their availability to form intermolecular disulfides with ER-resident oxidoreductases, including PDI and ERp72 (Sitia et al., 1990; Reddy et al., 1996). Therefore, markers of aberrant protein also may include ERp57, which would combine two signals of nonnative structure: exposed hydrophobic domains for GT recognition and subsequent reglucosylation, and exposed free thiols. Alternatively, differential cross-linking to the mutant 232mer compared with the wild-type truncation may be due to the absence of the N-terminal Cys, creating an single orphan Cys residue that remains accessible for cross-linking.
This study used ribosome-arrested translocation intermediates, which enables the trapping of more transient interactions, providing a method for analyzing the translocon environment and the organization of its associated factors. The stagnant nature of these constructs could potentially result in false associations or in the identification of interactions that would not accumulate during the normal rate of protein synthesis. However, the number of substrates tested using this method and the differences observed between HA, tyrosinase, and Factor V provides support that the associations are unique to the substrate. Additional substrates will need to be studied to test the generality of the scenario described above. A more thorough understanding of the rules that govern chaperone selection will allow us to determine the order and possibly the necessity of these interactions for proper maturation by inspecting the primary sequence of a protein. This type of analysis would be beneficial in the understanding of folding diseases that can result from mutations within the protein that may result in aberrant recruitment of the proper maturation machinery. The modulation of critical chaperone levels maybe an effective strategy for the treatment of disease states associated with misfolded proteins or the efficient expression of therapeutic recombinant proteins.
Acknowledgments
We thank Drs. Linda Hendershot, Ruth Halaban, Arthur Johnson, and Thomas Wileman for providing BiP, tyrosinase, Sec61α, and ERp57 antisera, respectively. We also are grateful for the kind gift of human Factor V cDNA from Dr. Randal Kaufman. D.N.H is supported by Public Health Service Grant CA79864.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-05-0381) on June 15, 2005.
References
- Alder, N. N., and Johnson, A. E. (2004). Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J. Biol. Chem. 279, 22787-22790. [DOI] [PubMed] [Google Scholar]
- Alder, N. N., Shen, Y., Brodsky, J. L., Hendershot, L. M., and Johnson, A. E. (2005). The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 168, 389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson, J. F., Frank, D. W., Calvo, P. A., Bieler, B. M., and Marks, M. S. (2000). A common temperature-sensitive allelic form of human tyrosinase is retained in the endoplasmic reticulum at the nonpermissive temperature. J. Biol. Chem. 275, 12281-12289. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi, S., Cwirla, S. E., Dower, W. J., Lipshutz, R. J., Sprang, S. R., Sambrook, J. F., and Gething, M.-J.H. (1993). Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717-728. [DOI] [PubMed] [Google Scholar]
- Braakman, I., Hoover-Litty, H., Wagner, K. R., and Helenius, A. (1991). Folding of influenza hemagglutinin in the endoplasmic reticulum. J. Cell Biol. 114, 401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau, B., and Horwich, A. L. (1998). The hsp70 and hsp60 chaperone machines. Cell 92, 351-366. [DOI] [PubMed] [Google Scholar]
- Chen, W., and Helenius, A. (2000). Role of ribosome and translocon complex during folding of influenza hemagglutinin in the endoplasmic reticulum of living cells. Mol. Biol. Cell 11, 765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Helenius, J., Braakman, I., and Helenius, A. (1995). Cotranslational folding and calnexin binding of influenza hemagglutinin in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92, 6229-6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, P. L. (2004). Protein folding in the cell: reshaping the folding funnel. Trends Biochem. Sci. 29, 527-534. [DOI] [PubMed] [Google Scholar]
- Crowley, K. S., Liao, S., Worrell, V. E., Reinhart, G. D., and Johnson, A. E. (1994). Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell 78, 461-471. [DOI] [PubMed] [Google Scholar]
- Daniels, R., Kurowski, B., Johnson, A. E., and Hebert, D. N. (2003). N-Linked glycan direct the cotranslational maturation of influenza hemagglutinin. Mol. Cell 11, 79-90. [DOI] [PubMed] [Google Scholar]
- Fewell, S. W., Travers, K. J., Weissman, J. S., and Brodsky, J. L. (2001). The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 35, 149-191. [DOI] [PubMed] [Google Scholar]
- Francis, E., Wang, N., Parag, H., Halaban, R., and Hebert, D. N. (2003). Tyrosinase maturation and oligomerization in the endoplasmic reticulum requires a melanocyte specific factor. J. Biol. Chem. 278, 25607-25617. [DOI] [PubMed] [Google Scholar]
- Gilmore, R., Collins, P., Johnson, J., Kellaris, K., and Rapiejko, P. (1991). Transcription of full-length and truncated mRNA transcripts to study protein translocation across the endoplasmic reticulum. Methods Cell Biol. 34, 223-239. [DOI] [PubMed] [Google Scholar]
- Halaban, R., Svedine, S., Cheng, E., Smicun, Y., Aron, R., and Hebert, D. N. (2000). Endoplasmic reticulum retention is a common defect associated with tyrosinase-negative albinism. Proc. Natl. Acad. Sci. USA 97, 5889-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman, B. D., Hendershot, L. M., and Johnson, A. E. (1998). BiP maintains the permeability barrier of the ER membrane by dealing the lumenal end of the translocon before and early in translocation. Cell 92, 747-758. [DOI] [PubMed] [Google Scholar]
- Hammond, C., Braakman, I., and Helenius, A. (1994). Role of N-linked oligosaccharides, glucose trimming and calnexin during glycoprotein folding in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 91, 913-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, C., and Helenius, A. (1994). Folding of VSV G protein: sequential interaction with BiP and calnexin. Science 266, 456-458. [DOI] [PubMed] [Google Scholar]
- Hebert, D. N., Zhang, J.-X., Chen, W., Foellmer, B., and Helenius, A. (1997). The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J. Cell Biol. 139, 613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius, A., and Aebi, M. (2004). Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019-1049. [DOI] [PubMed] [Google Scholar]
- Huppa, J. B., and Ploegh, H. L. (1997). The in vitro translation and assembly of a complete T cell receptor-CD3 complex. J. Exp. Med. 186, 393-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny, R. J., Pittman, D. D., Toole, J. J., Kriz, R. W., Aldape, R. A., Hewick, R. M., Kaufman, R. J., and Mann, K. G. (1987). Complete cDNA and derived amino acid sequence of human factor V. Proc. Natl. Acad. Sci. USA 84, 4846-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., Imokawa, G., Bennett, D. C., and Hearing, V. J. (1998). Tyrosinase stabilization by Tyrp1 (the brown locus protein). J. Biol. Chem. 273, 31801-31805. [DOI] [PubMed] [Google Scholar]
- Kowarik, M., Kung, S., Martoglio, B., and Helenius, A. (2002). Protein folding during cotranslational translocation in the endoplasmic reticulum. Mol. Cell 10, 769-778. [DOI] [PubMed] [Google Scholar]
- Krieg, U. C., Johnson, A. E., and Walter, P. (1989). Protein translocation across the endoplasmic reticulum membrane: identification by photocross-linking of a 39-kD integral membrane glycoprotein as part of a putative translocation tunnel. J. Cell Biol. 109, 2033-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner, A. B., Fitzpatrick, T. B., Calkins, E., and Summerson, W. H. (1949). Mammalian tyrosinase: preparation and properties. J. Biol. Chem. 178, 185-195. [PubMed] [Google Scholar]
- Meunier, L., Usherwood, Y.-K., Chung, K. T., and Hendershot, L. M. (2002). A subset of chaperones and folding enzymes from multiprotein complexes in the endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13, 4456-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, H. A., Grau, H., Kraft, R., Kostka, S., Prehn, S., Kalies, K. U., and Hartmann, E. (2000). Mammalian Sec61 is associated with Sec62 and Sec63. J. Biol. Chem. 275, 14550-14557. [DOI] [PubMed] [Google Scholar]
- Molinari, M., and Helenius, A. (2000). Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science 288, 331-333. [DOI] [PubMed] [Google Scholar]
- Nicchitta, C. V., Murphy, E. C., Haynes, R., and Shelness, G. S. (1995). Stage- and ribosome-specific alterations in nascent chain-Sec61p interactions accompany translocation across the ER membrane. J. Cell Biol. 129, 957-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, G., Cowles, C. R., and Emr, S. D. (1998). The AP-3 complex: a coat of many colours. Trends Cell Biol. 8, 282-288. [DOI] [PubMed] [Google Scholar]
- Oetting, W. S., and King, R. A. (1999). Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum. Mutat. 13, 99-115. [DOI] [PubMed] [Google Scholar]
- Oliver, J. D., Roderick, H. L., Llewellyn, D. H., and High, S. (1999). ERp57 functions as a subunit of specific complexes formed with the ER lectins calreticulin and calnexin. Mol. Biol. Cell 10, 2573-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, W. J., Cameron, P. H., Thomas, D. Y., and Bergeron, J.J.M. (1993). Association of folding intermediates of glycoproteins with calnexin during protein maturation. Nature 364, 771-776. [DOI] [PubMed] [Google Scholar]
- Petrescu, S. M., Branza-Nichita, N., Negroiu, G., Petrescu, A. J., and Dwek, R. A. (2000). Tyrosinase and glycoprotein folding: roles of chaperones that recognize glycans. Biochemistry 39, 5229-5237. [DOI] [PubMed] [Google Scholar]
- Pipe, S. W., Morris, J. A., Shah, J., and Kaufman, R. J. (1998). Differential interaction of coagulation factor VIII and factor V with protein chaperones calnexin and calreticulin. J. Biol. Chem. 273, 8537-8544. [DOI] [PubMed] [Google Scholar]
- Reddy, P., Sparvoli, A., Fagioli, C., Fassina, G., and Sitia, R. (1996). Formation of reversible disulfide bonds with the protein matrix of the endoplasmic reticulum correlates with the retention of unassembled immunoglobulin light chains. EMBO J. 15, 2077-2085. [PMC free article] [PubMed] [Google Scholar]
- Rutkowski, D. T., Ott, C. M., Polansky, J. R., and Lingappa, V. R. (2003). Signal sequences initiate the pathway of maturation in the endoplasmic reticulum lumen. J. Biol. Chem. 278, 30365-30372. [DOI] [PubMed] [Google Scholar]
- Schnell, D. J., and Hebert, D. N. (2003). Protein modulators: multi-functional mediators of protein translocation across membranes. Cell 112, 491-505. [DOI] [PubMed] [Google Scholar]
- Schrag, J. D., Bergeron, J.J.M., Li, Y., Borisova, S., Hahn, M., Thomas, D. Y., and Cygler, M. (2001). The structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell 8, 633-644. [DOI] [PubMed] [Google Scholar]
- Sitia, R., Neuberger, M., Alberini, C., Bet, P., Fra, A., Valetti, C., Williams, G., and Milstein, C. (1990). Developmental regulation of IgM secretion: the role of the carboxy-terminal cysteine. Cell 60, 781-790. [DOI] [PubMed] [Google Scholar]
- Svedine, S., Wang, T., Halaban, R., and Hebert, D. N. (2004). Glycosidase trimming determines the sorting of tyrosinase in the early secretory pathway. J. Cell Sci. 117, 2937-2949. [DOI] [PubMed] [Google Scholar]
- Tamura, A., Halaban, R., Moellman, G., Cowan, J. M., Lerner, M. R., and Lerner, A. B. (1987). Normal murine melanocytes in culture. In Vitro Cell Dev. Biol. 23, 519-522. [DOI] [PubMed] [Google Scholar]
- Tyedmers, J., Lerner, M., Bies, C., Dudek, J., Skowronek, M. H., Haas, I. G., Heim, N., Nastainczyk, W., Volkmer, J., and Zimmermann, R. (2000). Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc. Natl. Acad. Sci. USA 97, 7214-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Újvári, A., Aron, R., Eisenhaure, T., Cheng, E., Parag, H. A., Smicun, Y., Halaban, R., and Hebert, D. N. (2001). Translation rate of human tyrosinase determines its N-linked glycosylation level. J. Biol. Chem. 276, 5924-5931. [DOI] [PubMed] [Google Scholar]
- Whitley, P., Nilsson, I., and von Heijne, G. (1996). A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 271, 6241-6244. [DOI] [PubMed] [Google Scholar]
- Wilson, R., Allen, A. J., Oliver, J., Brookman, J. L., High, S., and Bulleid, N. J. (1995). The translocation, folding, assembly and redox-dependent degradation of secretory and membrane proteins in semi-permeabilized mammalian cells. Biochem. J. 307, 679-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun, A., Darby, N. J., Tessier, D. C., Michalak, M., Bergeron, J. J., and Thomas, D. Y. (1998). Enhanced catalysis of ribonuclease B folding by the interaction of calnexin or calreticulin with ERp57. J. Biol. Chem. 273, 6009-6012. [DOI] [PubMed] [Google Scholar]