Abstract
Alphavirus infection results in the shutoff of host protein synthesis in favor of viral translation. Here, we show that during Semliki Forest virus (SFV) infection, the translation inhibition is largely due to the activation of the cellular stress response via phosphorylation of eukaryotic translation initiation factor 2α subunit (eIF2α). Infection of mouse embryo fibroblasts (MEFs) expressing a nonphosphorylatable mutant of eIF2α does not result in efficient shutoff, despite efficient viral protein production. Furthermore, we show that the SFV translation enhancer element counteracts the translation inhibition imposed by eIF2α phosphorylation. In wild-type MEFs, viral infection induces the transient formation of stress granules (SGs) containing the cellular TIA-1/R proteins. These SGs are disassembled in the vicinity of viral RNA replication, synchronously with the switch from cellular to viral gene expression. We propose that phosphorylation of eIF2α and the consequent SG assembly is important for shutoff to occur and that the localized SG disassembly and the presence of the enhancer aid the SFV mRNAs to elude general translational arrest.
INTRODUCTION
A hallmark of infection with many lytic viruses is the shutoff of host cell protein synthesis. Many viruses commandeer the cellular translational apparatus to increase the yield of viral protein and dampen cellular innate immune signaling. However, in some cases, this translational inhibition can itself be seen as a cellular defense mechanism, to which viruses have evolved certain levels of tolerance. An example of this is the activation of the cellular double-stranded RNA-activated protein kinase R (PKR) by many viral infections and the multifarious viral countermeasures (Gale et al., 2000; Kaufman, 2000). PKR, activated by dimerization and autophosphorylation, phosphorylates the eukaryotic translation initiation factor 2α subunit (eIF2α). Phospho-eIF2α restricts the activity of the guanine nuclear exchange factor eIF2B, which is responsible for the formation of the GTP–eIF2–tRNAiMet ternary complex. Because translation of most cellular and viral mRNA proceeds via ternary complex-dependent mechanisms, this leads to a global decrease in protein synthesis.
Semliki Forest virus (SFV) is an RNA virus of the Alphavirus genus, family Togaviridae. The viral particles are enveloped and contain a single-stranded positive-sense 42S RNA genome. The 5′ two-thirds of the capped and polyadenylated genome codes for the four viral nonstructural proteins (nsp1–4), whereas the structural protein-coding regions are carried on a subgenomic 26S mRNA, transcribed from the 3′ one-third of the 42S RNA. Alphavirus infection of permissive cells causes shutoff between 2 and 4 h postinfection (hpi), the only proteins efficiently produced thereafter being virus encoded, expressed from the 26S mRNA. This property has contributed to the widespread use of recombinant alphaviruses as expression vectors (Xiong et al., 1989; Liljeström and Garoff, 1991; Pushko et al., 1997). Despite much interest in applying these vectors to biotechnology, the mechanism by which this important step in the alphavirus life cycle is achieved has remained largely unknown.
Several models have been proposed for the mechanism of alphavirus-mediated shutoff (Strauss and Strauss, 1994). It was thought that the efficient replication of viral RNA (vRNA) led to high concentrations of cytoplasmic 26S mRNA molecules, which outcompete cellular mRNAs for the available translation machinery (Kääriäinen and Ranki, 1984). Furthermore, it was thought that the consequent reduction in intracellular rNTP pools could lead to shutoff (Wengler, 1980). However, the observation that 26S mRNA-defective mutants of Sindbis alphavirus (SIN) cause efficient shutoff in infected cells suggested instead that the earlier events in 42S RNA synthesis are important for this to occur (Frolov and Schlesinger, 1994a). Other functions attributed to the viral structural proteins themselves (van Steeg et al., 1984; Elgizoli et al., 1989) also were suggested to be the causative agents. The advent of several alphavirus-based replicon systems showed this not to be the case (Xiong et al., 1989; Liljeström and Garoff, 1991; Pushko et al., 1997). Each of these systems involves the use of replication-competent alphavirus RNAs that elicit efficient shutoff while lacking the structural protein-coding regions. Other well studied viral systems have translation mechanisms conferring independence to certain eIFs, which are then cleaved or otherwise inactivated (Belsham and Jackson, 2000), leading to preferential vRNA translation and host shutoff. Although, the SFV 26S mRNA has been shown to have a low requirement for both eIF4E (the cap-binding protein) and eIF4B (van Steeg et al., 1981), neither of these proteins is known to be inactivated in alphavirus-infected cells. Despite these uncertainties, it remains apparent that shutoff is linked with vRNA replication, because noncytopathic vectors that do not induce complete translation inhibition tend to have mutations in the replicase region, particularly nsp2 (Agapov et al., 1998; Frolov et al., 1999; Perri et al., 2000; Frolova et al., 2002). Recently, results by Frolov and coworkers (Gorchakov et al., 2004) showed that shutoff in SIN-infected cells is mediated by both PKR-dependent and -independent mechanisms, as revealed by infection of wild-type (wt) and PKR-/- cells with wt and nsp2 mutant recombinant SIN.
The 5′ end of SFV and SIN structural protein coding regions have been shown to contain a unique genetic element known as the translation enhancer (Frolov and Schlesinger, 1994b; Sjöberg et al., 1994). Recombinant viral vectors that lack the enhancer, while eliciting efficient host cell shutoff, produce approximately 10-fold less protein than those that contain the enhancer (Frolov and Schlesinger, 1994b; Sjöberg et al., 1994). These regions are predicted by RNA sequence analysis to contain very stable hairpin loops composed of mainly G:C base pairs. The translational enhancement by this element was shown to occur only in infected cells (Frolov and Schlesinger, 1994b; Sjöberg and Garoff, 1996), suggesting that its presence is an adaptation to an altered translational environment therein. Although the mechanism of translational enhancement by this element is not entirely clear, it does seem to be carried out at the initiation step of translation (Frolov and Schlesinger, 1996).
General translational inhibition also is seen in cells under conditions of environmental stress, such as heat shock. The mechanism of shutoff under these conditions is better understood. During heat shock, cells shut down the expression of normal housekeeping genes to allow the selective expression of stress response factors (e.g., heat-shock proteins [HSPs]). eIF2α becomes phosphorylated by one of a small range of kinases, leading to reduction in ternary complex levels. The RNA binding protein TIA-1 and the related protein TIAR (collectively referred to as TIA-1/R) are then included into noncanonical preinitiation complexes known as 48S* complexes (Kedersha et al., 2002). Aggregation of TIA-1/R then sequesters the components of these complexes into cytoplasmic structures known as stress granules (SGs) (Kedersha et al., 1999; Gilks et al., 2004). In a process referred to as mRNA triage, the mRNA is believed to be stored, pending either degradation or resumption of normal translation in the absence of stress (Anderson and Kedersha, 2002). Consistent with this model, it has been shown that during heat shock, the HSP chaperone HSP70 mRNA molecules are excluded from SGs (Kedersha and Anderson, 2002) and are efficiently translated.
TIA-1/R are multifunctional proteins and in addition to their role in SG assembly, they are involved in other diverse activities, such as translational silencing (Kedersha et al., 1999; Piecyk et al., 2000), alternative splicing regulation (Förch et al., 2000), viral replication (Iseni et al., 2002; Li et al., 2002), and Fas-mediated apoptosis (Taupin et al., 1995). Although TIA-1-/- and TIAR-/- mice have been produced previously (Beck et al., 1998; Piecyk et al., 2000), the presence of either TIA-1/R protein in the cells is essential for viability (Le Guiner et al., 2003), illustrated by the fact that double (TIA-1-/-TIAR-/-) knockout mice cannot be generated (Kedersha and Anderson, unpublished results).
We were interested to study the role of this stress-induced translation inhibition mechanism in the host shutoff observed during SFV infection and possible mechanisms of viral escape. Our results suggest that the inhibition of protein production seen during conditions of environmental stress and during SFV infection is caused by similar mechanisms. The virus seems to have adapted to this response by preventing the formation of the cytosolic SGs on the 26S mRNA molecules. Furthermore, we show that the translation enhancer only functions in the presence of high levels of phospho-eIF2α, suggesting that it is also an adaptation to this response. Many viruses have mechanisms to inhibit the cellular defense capacity of PKR by inhibiting its activation (Kaufman, 2000). This work shows that SFV also inhibits this pathway but at a point downstream of eIF2α phosphorylation.
MATERIALS AND METHODS
Cells and Viruses
Previously described mouse embryo fibroblast (MEF) cell lines (Scheuner et al., 2001; Li et al., 2002) were cultured in DMEM (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (Sigma) and 2 mM l-glutamine (complete medium). In each experiment with these cells, wt controls were of the same genetic background. Stocks of wt SFV were generated from the pSP6-SFV4 infectious clone as described previously (Liljeström et al., 1991). The pSFV-b7lacZ and pSFV-b7EGFP clones contain the lacZ and EGFP genes, respectively, fused to the translation enhancer sequence (Sjöberg et al., 1994). Stocks of SFV1-lacZ (Liljeström and Garoff, 1991), SFV-b7lacZ, SFV1-EGFP, and SFV-b7EGFP recombinant virus particles were prepared as described previously (Smerdou and Liljeström, 1999). Titers of viruses were quantified by either plaque assay (wt) or by immunofluorescence (recombinants) on wt MEF cells. SFV-b7EGFP was UV-inactivated using Amersham Biosciences (Piscataway, NJ) UV Cross-linker at 2000 kJ/cm2 for 30 s. SFV stocks were used for infection as follows: cell monolayers were washed with phosphate-buffered saline (PBS), and virus was added in 300 μl of Glasgow minimum essential medium with 0.2% bovine serum albumin with periodic shaking for 1 h at 37°C. Virus solutions were then removed, and cells were washed with PBS before prewarmed complete medium was added. Where indicated, cells were stressed by the addition of sodium arsenite to 0.5 mM in complete medium.
Plasmids
The enhanced green fluorescent protein (EGFP) gene was cloned into BamHI/KpnI-cut pcDNA3.1 to give pcDNA-EGFP. For pcDNA-b12A-EGFP, a fragment encoding the SFV 26S mRNA 5′-untranslated region, the enhancer sequence and the foot-and-mouth disease virus 2A autoprotease in frame with the EGFP gene was constructed by overlap PCR and ligated into BamHI/KpnI-cut pcDNA3.1. Transient transfection was performed with Lipofectamine (Invitrogen, Carlsbad, CA).
Western Blotting
For analysis of protein by Western blotting, cells were lysed on ice in lysis buffer (1% NP-40, 50 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 2 mM EDTA), clarified by centrifugation at 6000 × g for 5 min in a microcentrifuge at 4°C, separated by SDS-PAGE, transferred to Hybond-P (Amersham Biosciences) membranes, and blotted with rabbit polyclonal antibodies specific for either phospho-eIF2α (phosphorylated on serine 51; BioSource International, Camarillo, CA) or total eIF2α (Santa Cruz Biotechnology, Santa Cruz, CA). The secondary antibody was horseradish peroxidase-conjugated goat anti-rabbit serum (BD Biosciences, San Jose, CA). Chemiluminescence was detected using the ECL reagents (Amersham Biosciences). Membranes were stripped by washing in 100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris-HCl, pH 6.7, for 30 min at 50°C, washed, blocked, and probed again as described above.
Protein Expression Assays
To control for different cell lines growth rates in the total protein expression experiments, MEFs were seeded at equal numbers into 35-mm dishes (Falcon; BD Biosciences Discovery Labware, Bedford, MA) 4 h before the experiments. For metabolic labeling of virus-infected cultures, cells were incubated first in methionine-free minimum essential medium supplemented with 2 mM l-glutamine (starvation medium) for 20 min. Newly produced proteins were then labeled by incubation in starvation medium supplemented with 50 μCi/ml [35S]methionine (Amersham Biosciences) for 10 min (pulse medium). Cells were then washed in cold PBS, lysed on ice in lysis buffer, and clarified by centrifugation at 6000 × g for 5 min in a microcentrifuge at 4°C. Supernatants were then analyzed on 10% SDS-PAGE gels, dried, and subjected to autoradiography. In vitro β-galactosidase (β-Gal) assays were performed as described previously (Liljeström and Garoff, 1991). For metabolic labeling of transiently transfected cultures, newly produced proteins were labeled directly by incubation in pulse medium for 1 h, with or without sodium arsenite as indicated. For direct quantitation of protein levels, EGFP was immunoprecipitated from lysates using green fluorescent protein (GFP) monoclonal antibody (BD Biosciences Clontech, Palo Alto, CA) and protein G-Sepharose (Amersham Biosciences). Unincorporated [35S]methionine was removed from lysates by Microcon YM3 filtration (Millipore, Billerica, MA). Samples of each were mixed with Emulsifier-Safe (PerkinElmer Life and Analytical Sciences, Boston, MA) and quantified by scintillation counting.
Immunofluorescence
Antibodies used for immunofluorescence include goat anti TIA-1 (Santa Cruz Biotechnology), mouse anti-TIA-1/R (3E6; Taupin et al., 1995), goat anti-eIF3 (Santa Cruz Biotechnology), mouse anti-bromouridine triphosphate (BrdUTP) (Stressgen Biotechnologies, Victoria, British Columbia, Canada), and mouse anti-digoxygenin (DIG) (Roche Diagnostics, Indianapolis, IN). Secondary antibodies were Cy2- and Cy3-conjugated donkey anti-goat sera and Cy3-conjugated donkey anti-mouse serum (all ML grade from Jackson ImmunoResearch Laboratories, West Grove, PA). Cells grown on coverslips were fixed by incubation in 4% paraformaldehyde (in PBS) for 8–10 min at room temperature followed by incubation in methanol for 8–10 min at –20°C. Coverslips were rinsed three times in PBS before incubation in blocking buffer (5% horse serum [Invitrogen], 0.02% sodium azide in PBS) for 1 h at room temperature. Primary antibodies were diluted in blocking buffer and incubated with the cells for 1–3 h. After rinsing in PBS, secondary antibodies were diluted in blocking buffer and incubated with the cells for 1–3 h. This secondary incubation contained 0.5 μg/ml Hoechst 33258 (Molecular Probes, Eugene, OR) for identification of cell nuclei. Washed coverslips were then mounted in Vinol mounting medium, and images were captured using a Leitz DM RB fluorescent microscope with a Hamamatsu cooled charged-coupled device camera C4880. Images were processed and compiled using Adobe Photoshop software.
RNA Quantitation by [14C]Uridine Labeling
Equal numbers of MEFs were infected with wt SFV at multiplicity of infection (MOI) 10 (as described above). Thirty minutes before each labeling period, 1 μg/ml actinomycin D (ActD) (Sigma) was added to the medium to block cellular RNA synthesis. Then, 1 μCi/ml [14C]uridine (Amersham Biosciences) was added so that all newly produced vRNA would incorporate this label. After the labeling period, total cell RNA was then isolated with TRIzol reagent (Invitrogen) according to the manufacturer's instructions and analyzed by agarose gel electrophoresis. The gel was then soaked for 30 min in 1 M salicylic acid (Sigma), dried, and exposed to x-ray film (Fujifilm, Tokyo, Japan).
Labeling of Newly Produced vRNA with BrUTP
For labeling of newly synthesized vRNA, MEFs grown on 13-mm-diameter glass coverslips, were infected with wt SFV at low MOI. Thirty minutes before and throughout each labeling period, ActD was added to the growth medium to 1 μg/ml. Lipofectamine (Invitrogen) was used to facilitate entry of the BrUTP label into the cells, according to the manufacturer's instructions. Briefly, growth medium was removed and after three PBS washes, it was replaced with Opti-MEM (Invitrogen) containing the appropriate volumes of Lipofectamine reagent and ActD and 10 mM BrUTP (Sigma). After the labeling period, cells were washed three times in PBS and fixed and processed for immunofluorescence as described above.
In Situ Hybridization (ISH)
The PCR DIG probe synthesis kit (Roche Diagnostics) was used to generate a virus-specific double-stranded DNA (dsDNA) probe labeled with DIG according to the manufacturer's instructions. The template for the PCR reaction was pSFVHelper1 (Liljeström and Garoff, 1991) and the primers were 5′-TATGCTCCCGGGCGACGAGCTGCAGTT-3′ (forward primer) and 5′-GTACACCCCGGGGTAAACCTTGCA-3′ (reverse), amplifying a 506-base pair region of the viral structural protein-coding region. ISH analysis for total vRNA (42S + 26S) was performed as follows. After fixing (as for immunofluorescence), cells were washed twice in 2× SSC. DIG containing dsDNA probes (denatured by boiling and diluted in hybridization buffer [0.5× formamide, 4× SSC, 1× Denhardt's reagent, 20 mM sodium phosphate, pH 7.0, 5% (wt/vol) dextran sulfate, 200 mM dithiothreitol, and 0.5 mg/ml denatured salmon sperm DNA] were hybridized to the cells at 44°C overnight in a humid chamber. Cells were then washed with 2× SSC and incubated with primary antibodies (typically goat anti TIA-1 and mouse anti-DIG) in 4× SSC containing 0.1% NP-40 for 1 h at room temperature. After washing in 4× SSC, cells were incubated with the requisite secondary antibodies and 0.5 μg/ml Hoechst 33258 (Molecular Probes) in 2× SSC containing 0.2% NP-40 for 1 h at room temperature. Cells were then washed sequentially in 4× SSC and 2× SSC and mounted and analyzed as for immunofluorescence. Controls performed with each experiment included mock-infected cells, mock-hybridized infected cells, and infected cells hybridized with non-DIG–containing probes. Furthermore, low MOI was used for easy comparison of background staining from noninfected cells.
RESULTS
Phosphorylation of eIF2α on Serine 51 Is Necessary for Shutoff in SFV-infected Cells
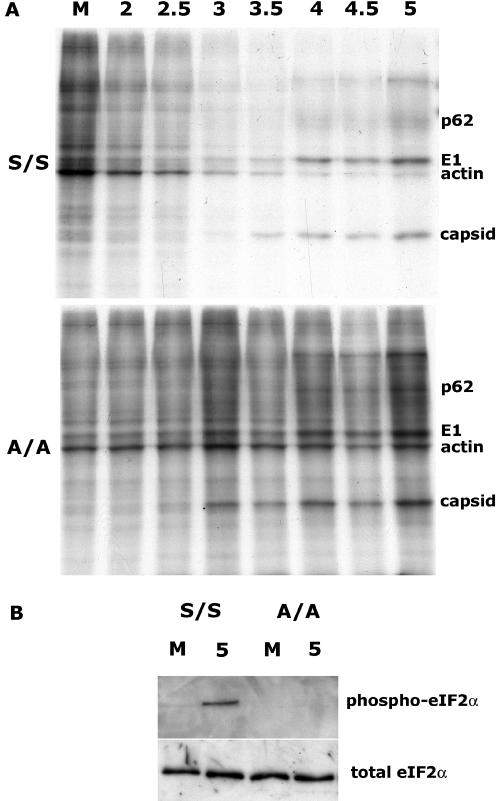
To determine whether the cellular stress response is involved in translation regulation during SFV infection, we studied translation in SV129 MEFs expressing a mutant form of eIF2α (Ser51Ala or A/A) that cannot be phosphorylated on serine 51 (Scheuner et al., 2001). Equal numbers of mutant and wt cells were infected with wt SFV at MOI 10. At different time points after infection, total protein production was analyzed by metabolic labeling. Figure 1A shows that in SV129 wt MEFs (S/S), efficient host cell translation inhibition was observed after ∼3 h of infection. Thereafter, newly produced proteins were predominantly viral, although a small residual level of cellular protein synthesis could be detected, probably from uninfected cells in the culture. In the MEFs expressing the nonphosphorylatable mutant (A/A), host shutoff did not occur at such efficiency, although the earliest production of viral proteins was not delayed relative to the S/S cells. Both sets of cells were infected at similar levels as determined by immunofluorescence (our unpublished data).
Figure 1.
Phosphorylation of eIF2α is necessary for efficient shutoff to occur in SFV-infected cells. (A) Total protein production at various times in infected wt eIF2α (S/S) and nonphosphorylatable mutant eIF2α (A/A)-expressing MEF cells was analyzed as indicated in Materials and Methods. Positions of the cellular protein actin and viral proteins p62, E1, and capsid are indicated. All lanes are labeled in hpi or as M, mock-infected cells. (B) The phosphorylation state of eIF2α in these cells 5 h postinfection or mock infection was determined by Western blotting for phospho-eIF2α (top) and related to total eIF2α levels by stripping and reprobing the membrane (bottom).
Western blotting was used to confirm the eIF2α phosphorylation status in these cells. Again, equal numbers of S/S and A/A cells were infected, and lysates were taken for analysis at 5 hpi. Figure 1B, top, shows that eIF2α was phosphorylated after infection with SFV but only in the S/S cells, as expected. Figure 1B, bottom, shows the same membrane that was stripped and reprobed for total eIF2α, showing that eIF2α protein expression was not altered in the mutant cells. Together, these results show that eIF2α becomes phosphorylated during infection with SFV and that this phosphorylation is important for shutoff of host mRNA translation.
The SFV Translational Enhancer Counteracts the Translation Block Imposed by eIF2α Phosphorylation
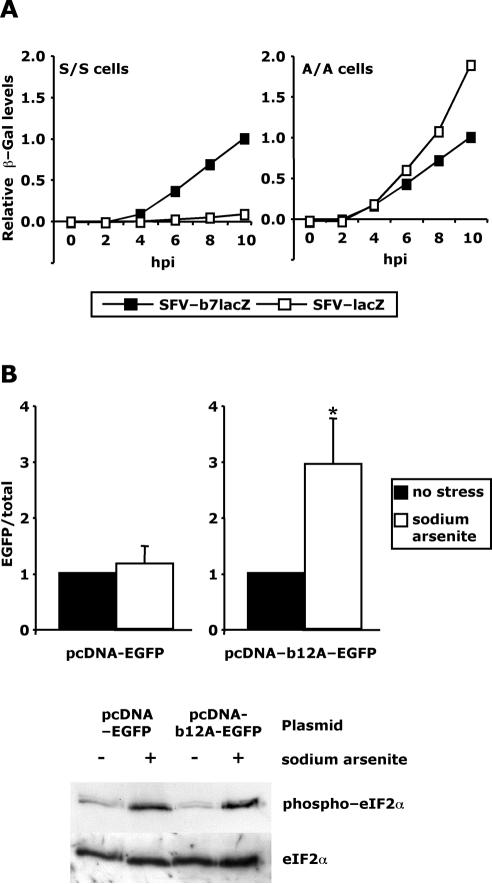
To explain the efficient translation of viral mRNAs in the presence of eIF2α phosphorylation, we initially studied the effect of the viral translational enhancer element in the S/S and A/A cell lines. As discussed, the enhancement of this element has been previously shown only to occur in infected cells and that it most likely functions at the initiation stage of translation. Analysis of protein production levels from vectors containing the enhancer in the A/A mutant cells therefore would indicate the relationship between the enhancer and eIF2α phosphorylation. Equal numbers of S/S and A/A cells were infected at MOI 10 with either SFV-lacZ or SFV-b7lacZ, lacking or containing the enhancer, respectively. In previous work, the replication of similar recombinant SFV genomes led to shutoff as efficiently as wt SFV (Liljeström and Garoff, 1991). Equal infection of each cell line in this experiment was verified by an in situ β-Gal assay (our unpublished data). Lysates were taken at different times postinfection and analyzed for β-gal protein levels by an in vitro assay. As expected, infection with the enhancer-containing vector led to greatly increased β-Gal levels in wt (S/S) MEFs (Figure 2A, left). 12-fold more β-Gal protein was detected in the S/S cells after infection with SFV-b7lacZ than after infection with SFV-lacZ, an increase consistent with other reports of alphavirus enhancer activity in infected wt cells (Frolov and Schlesinger, 1994b; Sjöberg et al., 1994). Interestingly, in the A/A mutant cells, both recombinant viruses expressed high levels of β-Gal with SFV-lacZ (lacking the enhancer), producing ∼50% more protein than the enhancer-containing virus (Figure 2B, right). Thus, the enhancer is necessary for efficient viral protein synthesis but only in the presence of eIF2α phosphorylation.
Figure 2.
The SFV translational enhancer counteracts the translation block imposed by eIF2α phosphorylation. (A) Equal numbers of S/S and A/A cells were infected with SFV-b7lacZ or SFV-lacZ at MOI 10. Lysates were taken at different times postinfection and analyzed by an in vitro β-Gal assay. For each cell line, all protein levels are presented relative to the level of protein present in cells infected for 10 h with enhancer-containing virus. In this way, the level of translational enhancement in the cells can easily be appreciated. The results are averages of three independent experiments with similar results. (B) BHK cells were transfected with either pcDNA-EGFP or pcDNA-b12A-EGFP. Twenty-four hours later, the cells were labeled with [35S]methionine for 1 h, in the presence or absence of sodium arsenite. EGFP levels were determined by immunoprecipitation and scintillation counting. Total protein levels were determined by scintillation counting after removal of unincorporated label. For each construct, the results are presented as EGFP/total and shown relative to that value for nonstressed cells. Thus, the efficiency of each transfected gene can be related to total cellular translation in each condition. The results are averages of four independent experiments with similar results. Error bars are SD, and * indicates p < 0.05 versus nonstressed cells (Student's t test). Shown in the lower panel is Western blot analysis for phospho- and total eIF2α in each set of cells.
In an alternative approach to study the relationship between the enhancer activity and eIF2α phosphorylation, we constructed plasmids for transient transfection of the EGFP gene, either alone (pcDNA-EGFP) or in fusion with the enhancer sequence (pcDNA-b12A-EGFP). We then assayed protein expression levels in the context of sodium arsenite-induced eIF2α phosphorylation. Baby hamster kidney (BHK) cells were transfected with either plasmid and at 24 h, the cells were metabolically labeled with [35S]methionine in the presence or absence of arsenite. A measure of the translational efficiency, under each condition, of the plasmid-derived transcripts relative to cellular transcripts was determined and related to that in the absence of stress for each plasmid. Although the levels of total protein expression were reduced ∼10-fold in all cells under stress, the relative efficiencies of the plasmid-derived mRNAs differed (Figure 2B). The relative expression of pcDNA-EGFP mRNA was not altered by stress treatment (left), and thus we conclude that it was inhibited by factor similar to that of the cellular mRNAs. In contrast however, it can be seen that the relative expression of the pcDNA-b12A-EGFP–derived mRNA was increased threefold in the presence of arsenite (right). Figure 2B, bottom, shows that the arsenite treatment in each case led to similar levels of eIF2α phosphorylation. The results in Figure 2 together demonstrate that the presence of the enhancer on the 26S mRNA allows the RNA to be efficiently translated in the absence of normal ternary complex levels as observed during infection or stress induced by other means.
Transient Assembly of SGs in SFV-infected Cells
Because it has been shown that the formation of SGs is a mechanism to down-regulate cellular housekeeping translation during stress and that phosphorylation of eIF2α is sufficient for their formation (Kedersha et al., 1999), we determined whether these structures were indeed formed during SFV infection. Accordingly, we analyzed the localization of TIA-1/R proteins in SFV-infected cells by immunofluorescence. Figure 3A shows a time-course experiment in which MEFs were infected at MOI 1 with SFV-b7EGFP, fixed at various time points, and stained for TIA-1. Low MOI was used so that noninfected cells would be visible in each field, serving as internal controls. Shortly after infection, TIA-1 completely relocated to the cytoplasm and was found in SGs. This change in localization corresponded in time to the change in protein production from host to viral proteins, as evidenced by the EGFP production visible in the same cells. As the infection proceeded, however, there was a marked inverse relationship between the intensity of EGFP signal and the number of SGs in each cell. This was evident in all cells viewed at 4 hpi or later. At the conclusion of these time-course experiments (8 hpi), all infected cells showed very high viral protein production (EGFP) and complete TIA-1 relocalization out of SGs, into a diffuse distribution throughout the cytoplasm. Cells that were mock infected or infected with UV-inactivated SFV-b7EGFP did not show alteration of TIA-1 localization after 8 h, demonstrating the requirement for vRNA replication for this to occur. Similar results were seen using 3E6, an antibody predominantly directed to TIAR (our unpublished data).
Figure 3.
Time course of SG assembly in SFV-infected MEFs. (A) wt MEFs were either infected at low MOI with SFV-b7EGFP, mock infected, or infected with UV-inactivated SFV-b7EGFP. They were then fixed at the indicated times postinfection and processed for immunofluorescence using anti TIA-1 according to Materials and Methods. Representative images are shown for each time point. (B) eIF3 is a component of SFV-induced SGs. wt MEFs were infected at low MOI with wt SFV, fixed 5 hpi, and stained for eIF3 and TIA-1. (C) wt MEFs were infected at low MOI with SFV1-EGFP, fixed at the indicated times postinfection, and processed for immunofluorescence using anti TIA-1. Representative images are shown for each time point. Cells containing SGs are indicated with arrowheads. Bar, 50 μm.
Confirming that this transient granular TIA-1/R staining does indeed represent SGs, we also observed eIF3 (a component of the translation preinitiation complex and another SG marker; Kedersha et al., 2002) colocalization with TIA-1 in SGs in cells infected at MOI 1 with wt SFV (Figure 3B). Furthermore, SFV-induced SGs were in dynamic equilibrium with polysomes as shown by their differential reactions to treatments with puromycin or cycloheximide (our unpublished data) as observed previously for arsenite-induced SGs (Kedersha et al., 2000). These experiments suggest that SG formation during SFV infection is transient and occurs at the time of host shutoff, i.e., when the profile of protein production changes from cellular to viral.
Because SG components are in dynamic equilibrium with polysomes (Kedersha et al., 2000), we investigated whether the SG disassembly seen at later times (Figure 3A) was a result of the efficient translation of the viral 26S mRNA competing with the SGs for the 48S complexes. Accordingly, we infected MEFs with SFV1-EGFP, which does not contain the translation enhancer element and thus does not direct as efficient translation of the 26S mRNA as wt SFV or SFV–b7EGFP, and analyzed the levels of SG assembly and disassembly (Figure 3C). Despite the very low level of translation of the 26S mRNA of this virus at these early times, the complete disassembly of SGs was observed at similar times for this virus as for the enhancer-containing viruses.
Protein Production during the Early Stages of SFV Infection of TIA-1-/- Cell Lines
We wanted to determine the rate and extent of shutoff in the absence of the TIA-1/R proteins. However, because total knockout of both TIA-1/R proteins is lethal (Le Guiner et al., 2003), it is technically impossible to assay SFV-induced effects in their absence. Thus, the TIA-1-/- MEF cells (Piecyk et al., 2000) constitute a significant knockdown (rather than knockout) of total TIA-1/R protein, and results regarding SFV-induced shutoff and SG formation in these cells should be analyzed with this in mind. Similar numbers of wt, and TIA-1-/- MEF cells were infected with SFV at MOI 10. Total protein production in the infected cells was analyzed at different times postinfection. Complete infection was determined by immunofluorescence. High MOI infection of the wt B6 MEFs resulted in very efficient shutoff (Figure 4A). It can be seen that host protein production was undetectable after 2.5 h of infection. Viral proteins, on the other hand, were first detected at this time and were efficiently expressed thereafter. In contrast, the TIA-1-/- B6 cells were capable of cellular protein production at much later times than the wt cells. For example, actin was clearly expressed at all times, indicating that shutoff was significantly delayed in these cells. As with the eIF2α A/A mutant MEFs (Figure 1A), viral protein production did not seem to be affected by the mutation. Similar results were observed with TIAR-/- cells (our unpublished data).
Figure 4.
Shutoff is delayed in TIA-1-/- MEFs. (A) Total protein production at various times in wt SFV-infected wt and TIA-1-/- MEF cell lines was analyzed as indicated in Materials and Methods. Positions of the cellular protein actin and viral proteins p62, E1, and capsid are indicated. (B) The number of SGs was quantified in wt and TIA-1-/- MEFs infected with SFV-b7EFGP at low MOI. The SGs in 20 cells per time point were counted, and the mean values are plotted. (C) The phosphorylation state of eIF2α in the TIA-1-/- cells 5 h postinfection or mock infection was determined by Western blotting (top) and related to total eIF2α levels by stripping and reprobing the membrane (bottom). (D) vRNA production at the indicated times during infection of wt and TIA-1-/- MEF cell lines was analyzed as indicated in Materials and Methods. Positions of the viral genomic 42S and subgenomic 26S RNAs are indicated. All lanes are labeled in hpi or as M, mock-infected cells.
Because it has recently been shown that the TIA-1-/- cells are impaired for SG assembly (Gilks et al., 2004), we determined the effect on SFV-induced SG formation by the TIA-1 deletion. Using immunofluorescence with the 3E6 antibody, we observed an overall reduction in SG numbers in TIA-1-/- relative to wt cells (Figure 4B), even though the time of appearance and disappearance did not differ. These SGs were probably formed by TIAR-mediated aggregation of the 48S* complexes. Figure 4C shows that the phosphorylation of eIF2α occurred in SFV-infected TIA-1-/- cells.
One possibility for the slower kinetics of shutoff in the TIA-1-/- cells is that there is a lower level of 26S mRNA available for competition with the cellular mRNA transcripts. Both TIA-1/R proteins are RNA binding proteins and TIAR has been shown to be important for West Nile virus RNA replication (Li et al., 2002). We have observed that at the time points at which the differential shutoff rates were detected, no major difference in the quantities of 42S genomic and 26S subgenomic RNA existed in wt and TIA-1-/- cells (Figure 4D).
SG Disassembly in SFV-infected Cells Correlates with Increasing vRNA Levels
Because SGs are sites for aggregation of unused translational components, including mRNAs (Kedersha et al., 1999), we analyzed the localization of both TIA-1 and vRNA in infected cells to determine whether vRNA was localized to SGs. An explanation for the viral translational supremacy could be that the 26S mRNA has a specific mechanism to avoid SG association (analogous, perhaps, to that of the hsp70 mRNA in heat shock; Kedersha and Anderson, 2002). Alternatively, vRNA may be SG associated but efficiently translated anyway due to its high level. We determined the relationship between vRNA and SGs by combining ISH analysis for total vRNA and immunofluorescence for TIA-1 protein. wt MEFs were infected at MOI 1 with wt SFV and fixed and processed for ISH. In all cells where a strong vRNA signal was detected, the complete dissolution of SGs was seen, although cytoplasmic localization of TIA-1 was maintained (Figure 5A). Interestingly, we observed that cells at earlier stages of infection showed weaker, more restricted vRNA signals, and generally only contained SGs in areas of little or no vRNA. Uninfected cells within the field again served as internal controls for the specificity of ISH staining. Mock-infected cells gave undetectable vRNA signals (our unpublished data).
Figure 5.
SGs in SFV-infected cells are disassembled as vRNA levels increase. (A) ISH analysis of total vRNA at 5 hpi. wt MEFs were infected at low MOI with wt SFV and fixed at 5 hpi. Hybridization and immunostaining was performed as indicated in Materials and Methods. (B) Time course of vRNA accumulation. wt MEFs were infected at low MOI with wt SFV. vRNA produced during the indicated intervals was labeled with BrUTP as indicated in Materials and Methods and processed for immunofluorescence using anti TIA-1 and anti BrUTP. Cells containing SGs are indicated with arrowheads. Bar, 50 μm.
We also determined the relationship between vRNA accumulation and SGs using BrUTP to label vRNA produced during 1-h labeling periods only. To specifically label vRNA, ActD was added to the growth medium 30 min before and during each labeling period. A representative time-course experiment is shown in Figure 5B. The results show that SGs assembled before vRNA was detectable by BrUTP staining. As the infection progressed, however, it was evident that the vRNA was not located in SGs. In agreement with the results in Figure 5A, it was observed that the establishment of vRNA replication seemed to occur only in areas where SGs were disassembled. TIA-1 protein was found in a more diffuse pattern in the vicinity of newly produced vRNA. Together, these results show that SFV-induced SGs are dissolved in the vicinity of vRNA as replication progresses.
At Late Times Postinfection, SFV-infected Cells Are No Longer Capable of SG Formation
SGs have been shown to be dissolved when stress inducers are removed and normal protein production is resumed (Kedersha et al., 1999). It is possible, therefore, that the dissolution of SGs at the later stages of SFV infection could reflect the fact that the stress signal only exists early in the infection. Alternatively the SGs may be disassembled even though the stress is still present. To distinguish between these possibilities, we conducted an experiment in which wt MEFs were infected at low MOI with SFV-b7EGFP and subjected to sodium arsenite stress at a late stage. The ability to generate a stress response to the new stress was then analyzed in infected and uninfected cells simultaneously. Figure 6A shows infected cells at 8 hpi that, as expected, exhibited cytoplasmic localization of TIA-1 but no SGs. Figure 6B shows similarly infected cells that were further stressed with sodium arsenite for 30 min before fixation. In this case, only uninfected cells showed SG assembly in response to the new stress. The infected cells displayed diffuse cytoplasmic TIA-1 staining, characteristic of the late stage of SFV infection (Figures 3 and 5). This result showed that SFV-infected cells lose their ability to respond to stress by assembling SGs. This suggested that the SG disassembly seen in the infected cells resulted from this stress resistance rather than the removal of stress signals.
Figure 6.
At late times postinfection, SFV-infected cells are no longer capable of arsenite-induced SG formation. wt MEFs were infected at low MOI with SFV-b7EGFP. After 8 h, cells were either fixed directly (A) or treated with sodium arsenite for 30 min and then fixed (B). Cells were processed for immunofluorescence using anti TIA-1. Cells containing SGs are indicated with arrowheads. Bar, 50 μm.
DISCUSSION
The inhibition of translation during viral infection is a well known phenomenon (Gale et al., 2000; Pe'ery and Mathews, 2000). Many mechanisms have been proposed by which viruses both inactivate components of the translation initiation complex and counteract the cell's own infection-induced translation inhibition pathways. In this work, we have investigated the cellular response to infection with SFV. We found that upon infection, host cell translation shutoff is achieved in large part via the phosphorylation of eIF2α on serine 51. In other situations, this event leads to a general translation inhibition as discussed above. However, the viral mRNAs have a unique element, the translation enhancer, which we show, confers resistance to these conditions. We also showed that early in the infection, TIA-1/R proteins are mobilized from the nucleus into cytoplasmic foci described previously as SGs, part of the cellular stress response. SGs have previously been shown to be involved in translation regulation under stress via the aggregation of 48S* mRNP complexes. We also show that later in the infection, the SGs are disassembled and TIA-1/R remain cytoplasmic in location. The cells remain unable to assemble SGs in response to another stress.
The initial events after viral entry are uncoating and the initiation of translation of the replicase coding region carried on the 5′ two-thirds of the 42S genome. After the switch from translation to replication, the replicase complex catalyzes the production of a complementary copy of the genome (negative strand). From this negative strand, new positive-strand molecules are made (both 42S genomic and 26S subgenomic RNAs). Genome replication of many viruses has been shown to activate PKR (Gale et al., 2000; Barber, 2001; Schneider and Mohr, 2003). A well studied substrate for the kinase activity of PKR is eIF2α. Some viruses have mechanisms to inhibit the reduction of ternary complex levels by phosphorylation of eIF2α (Gale et al., 2000; Schneider and Mohr, 2003). These include PKR inhibitors (adenovirus VA RNA [Reichel et al., 1985], vaccinia virus E3L and K3L proteins [Beattie et al., 1991; Chang et al., 1992], influenza virus NS1 protein [Lu et al., 1995], and the herpes simplex virus [HSV] Us11 protein [Mulvey et al., 1999, among others], and eIF2α phosphatase activators [HSV γ34.5 protein; He et al., 1997]). Because we have shown that eIF2α phosphorylation is important for shutoff (Figure 1), we conclude that SFV does not have an eIF2α kinase inhibitory activity. Rather, SFV seems to tolerate eIF2α phosphorylation due to the presence of the enhancer element that we have shown functions well under these conditions (Figure 2). Although the enhancer has previously been shown to function only in infected cells (Frolov and Schlesinger, 1994b; Sjöberg and Garoff, 1996), we have extended this to include cells under arsenite stress. Although the translational enhancement in this situation was less than that seen in infected cells, the enhancer significantly increased protein production during stress. This suggests that this element, present in other alphaviruses (Frolov and Schlesinger, 1994b), represents an evolutionary adaptation to the translational block imposed by PKR-mediated eIF2α phosphorylation. Recently, a novel internal ribosome entry site (IRES) was identified in the cricket paralysis virus (CrPV) genome that can initiate translation independently of the ternary complex (Wilson et al., 2000) and thus is not subject to the profound translation inhibition caused by activation of PKR. The translation of the capped SFV 26S mRNA is unlikely to be driven by a mechanism similar to that of the ternary complex-independent CrPV IRES. It has a very short 5′-untranslated region that is unlikely to contain an analogous RNA structural element. Furthermore, methionine is conserved as the N-terminal amino acid in all alphavirus capsid proteins that would be unnecessary if translation was ternary complex independent. Despite the obvious differences, the CrPV IRES and the alphavirus translational enhancer are both present to facilitate the efficient initiation of translation of the viral structural protein coding regions under conditions where normal cellular mRNAs are severely inhibited due to reduced availability of ternary complexes.
In the absence of ternary complexes under stress, stalled preinitiation complexes (48S*) are assembled (Kedersha et al., 2002). These complexes contain all components of the normal 48S complex except eIF2 and eIF5. The 48S* complexes are then incorporated into SGs by the TIA-1/R proteins. Because the SG components are in dynamic equilibrium with polysomes (Kedersha et al., 2000), constant monitoring is maintained for stress recovery. When charged ternary complexes become available, translation competent initiation complexes form on mRNAs, and the equilibrium will shift toward polysomes. The assembly and disassembly of SGs seems to be mediated by TIA-1/R (Kedersha et al., 1999). We have observed the transient appearance of SGs in SFV-infected cells (Figures 3 and 5). SGs occur in cells at a time when cellular translation is inhibited, but vRNA replication and viral structural protein production are not yet fully efficient. Later, as vRNA replication develops, the SGs are disassembled (Figure 5). It seems that SGs, initially distributed throughout the cytoplasm, become limited to regions away from sites of vRNA replication. Eventually, vRNA is distributed throughout the cell, and the SGs are no longer evident. Although SGs are disassembled in regions containing a high concentration of newly produced vRNA, the TIA-1 staining is not lost in these areas but seems evenly distributed. Interestingly, this occurs despite the elevated levels of phospho-eIF2α in the cells at these times (Figure 1). Furthermore, we showed that the cells, at these time points, have lost the ability to form SGs in response to arsenite (Figure 6). Due to the dynamic equilibrium between polysomes and SGs, the latter's disassembly has the consequence that the components are again made available for assembly into translation-active polysomes. Thus, we speculate that this occurs to give the 26S mRNA (by these times, present in increased numbers) access to the translation machinery. Cellular mRNAs will remain inhibited for translation due to the eIF2α phosphorylation, by which, we have shown, the enhancer-containing viral 26S mRNA is less affected (Figure 2).
A question raised by this work concerns the fate of the SG-associated cellular mRNAs. Why does cellular translation not resume after SG disassembly? On one hand, these mRNAs would not be expected to be translated efficiently due to the fact that eIF2α remains phosphorylated and ternary complex availability is low. Also, it is very possible that these mRNAs are selectively degraded in the SGs or in other cytoplasmic mRNA processing bodies. Furthermore, in alphavirus-infected cells there is a significant decrease in cellular mRNA and rRNA transcription (Frolova et al., 2002; our unpublished observations). This inhibition, which follows the translational inhibition, also would serve to reduce the cytoplasmic pools of cellular mRNAs that are available for translation and thus consolidate the shutoff at later times.
The delay in host translation inhibition in the TIA-1-/- MEFs (Figure 4) suggests an important role for SGs in the shutoff process at these early times postinfection. At times when viral structural protein synthesis is beginning, the TIA-1-/- cells are still efficiently producing cellular proteins. We have shown in Figure 3 that SGs are prevalent in infected wt cells at these times. Because there is a reduced ability to form SGs in the TIA-1-/- cells (Figure 4B; Gilks et al., 2004), we propose that in these cells, the cellular mRNAs are less efficiently removed from active polysomes and can now compete with the low initial levels of newly produced 26S mRNA for active initiation complexes. As replication develops, increasing quantities of viral transcripts (containing the enhancer) inevitably outcompete the cellular mRNAs for access to initiation complexes and translational advantage is gained. SG assembly by the TIA-1/R proteins is well described, but other cellular proteins have been shown to be involved. Recently G3BP, an endoribonuclease, has been shown to initiate SG formation in a phosphorylation-dependent manner (Tourrière et al., 2003). Also, the Fragile X Mental Retardation protein has been shown to form cytoplasmic mRNA and protein aggregates reminiscent of SGs (Mazroui et al., 2002). Whether these or other unidentified proteins play a role in SFV-induced SGs in wt or TIA-1-/- MEFs is not known.
Other activities of TIA-1/R have recently been described to be involved in various aspects of the life cycles of other viruses. Both TIA-1/R have been shown to interact with West Nile virus (WNV) RNA, and this interaction seems to be necessary for RNA replication (Li et al., 2002). Similarly, Sendai virus (SeV) RNA replication generates short transcripts that bind TIAR and modulate virus-induced apoptosis (Iseni et al., 2002). Interestingly wt SeV infection gave rise to SG formation in a small fraction of cells that was increased when a viral mutant was used containing a deletion in the TIAR binding site. Because neither WNV nor SeV induces efficient shutoff in infected cells, it may well be a common mechanism between these diverse viruses to bind TIA-1/R in the cytoplasm to avoid SG formation and consequent translational repression and perhaps apoptosis. Recently, it has been described that TIA-1/R is mobilized from the nucleus to the cytoplasm in HSV-1–infected cells (Esclatine et al., 2004), and a mutant lacking the virion shutoff protein gene UL41 led to assembly of SGs.
In conclusion, we have studied the role of the cellular stress response in the regulation of translation during SFV infection. Although there may be distinct but cooperating mechanisms for translational shutoff to be achieved in alphavirus-infected cells (as suggested by other investigators; Strauss and Strauss, 1994; Gorchakov et al., 2004), our results describe a major role for the phosphorylation of eIF2α in the inhibition of cellular gene expression during SFV infection. The enhancer containing 26S mRNA, however, is clearly less dependent on normal levels of the GTP-eIF2-tRNAiMet ternary complex and consequently SFV- and SFV-based vectors encoding this element, are capable of high levels of protein production under these conditions. We also have shown that SGs play a role, at least in the early stages, in inhibiting the translation of cellular mRNAs. We propose that the disassembly of SGs that occurs later in infection allows the viral 26S mRNA to avoid inclusion in these structures and thereby aids the establishment of translational superiority.
Acknowledgments
We thank Graham Belsham, Åsa Hidmark, and Gunilla Karlsson for stimulating discussions and critical reading of the manuscript, and Eivor Bonin, Henrik Claeson, and Håkan Samuelsson for technical assistance. This work was supported by the Swedish Research Council and National Institutes of Health Grants AI-50167 and AI-33600.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-02-0124) on June 1, 2005.
References
- Agapov, E. V., Frolov, I., Lindenbach, B. D., Pragai, B. M., Schlesinger, S., and Rice, C. M. (1998). Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95, 12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P., and Kedersha, N. (2002). Stressful initiations. J. Cell Sci. 115, 3227-3234. [DOI] [PubMed] [Google Scholar]
- Barber, G. N. (2001). Host defense, viruses and apoptosis. Cell Death Differ. 8, 113-126. [DOI] [PubMed] [Google Scholar]
- Beattie, E., Tartaglia, J., and Paoletti, E. (1991). Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology 183, 419-422. [DOI] [PubMed] [Google Scholar]
- Beck, A. R., Miller, I. J., Anderson, P., and Streuli, M. (1998). RNA-binding protein TIAR is essential for primordial germ cell development. Proc. Natl. Acad. Sci. USA 95, 2331-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham, G. J., and Jackson, R. J. (2000). Translation initiation on picornavirus RNA. In: Translational Control of Gene Expression, ed. N. Sonenberg, J.W.B. Hershey, and M. B. Mathews, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 869-900.
- Chang, H. W., Watson, J. C., and Jacobs, B. L. (1992). The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 89, 4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgizoli, M., Dai, Y., Kempf, C., Koblet, H., and Michel, M. R. (1989). Semliki Forest virus capsid protein acts as a pleiotropic regulator of host cellular protein synthesis. J. Virol. 63, 2921-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esclatine, A., Taddeo, B., and Roizman, B. (2004). Herpes Simplex virus 1 induces cytoplasmic accumulation of TIA–1/TIAR and both synthesis and cytoplasmic accumulation of Tristetraprolin, two proteins that bind and destabilize AU-rish RNAs. J. Virol. 78, 8582-8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förch, P., Puig, O., Kedersha, N., Martinez, C., Granneman, S., Seraphin, B., Anderson, P., and Valcarcel, J. (2000). The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6, 1089-1098. [DOI] [PubMed] [Google Scholar]
- Frolov, I., Agapov, E., Hoffman, T. A., Jr., Pragai, B. M., Lippa, M., Schlesinger, S., and Rice, C. M. (1999). Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73, 3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov, I., and Schlesinger, S. (1994a). Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J. Virol. 68, 1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov, I., and Schlesinger, S. (1994b). Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J. Virol. 68, 8111-8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov, I., and Schlesinger, S. (1996). Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 70, 1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova, E. I., Fayzulin, R. Z., Cook, S. H., Griffin, D. E., Rice, C. M., and Frolov, I. (2002). Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76, 11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M., Jr., Tan, S. L., and Katze, M. G. (2000). Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64, 239-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks, N., Kedersha, N., Ayodele, M., Shen, L., Stoecklin, G., Dember, L. M., and Anderson, P. (2004). Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov, R., Frolova, E., Williams, B.R.G., Rice, C. M., and Frolov, I. (2004). PKR–dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 78, 8455-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B., Gross, M., and Roizman, B. (1997). The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94, 843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseni, F., Garcin, D., Nishio, M., Kedersha, N., Anderson, P., and Kolakofsky, D. (2002). Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 21, 5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kääriäinen, L., and Ranki, M. (1984). Inhibition of cell functions by RNA-virus infections. Annu. Rev. Microbiol. 38, 91-109. [DOI] [PubMed] [Google Scholar]
- Kaufman, R. J. (2000). The double-stranded RNA-activated protein kinase PKR. In: Translational Control of Gene Expression, ed. N. Sonenberg, J.W.B. Hershey, and M. B. Mathews, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 503-527.
- Kedersha, N., and Anderson, P. (2002). Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, 963-969. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., Chen, S., Gilks, N., Li, W., Miller, I. J., Stahl, J., and Anderson, P. (2002). Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., Cho, M. R., Li, W., Yacono, P. W., Chen, S., Gilks, N., Golan, D. E., and Anderson, P. (2000). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. L., Gupta, M., Li, W., Miller, I., and Anderson, P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guiner, C., Gesnel, M. C., and Breathnach, R. (2003). TIA-1 or TIAR is required for DT40 cell viability. J. Biol. Chem. 278, 10465-10476. [DOI] [PubMed] [Google Scholar]
- Li, W., Li, Y., Kedersha, N., Anderson, P., Emara, M., Swiderek, K. M., Moreno, G. T., and Brinton, M. A. (2002). Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76, 11989-12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeström, P., and Garoff, H. (1991). A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology 9, 1356-1361. [DOI] [PubMed] [Google Scholar]
- Liljeström, P., Lusa, S., Huylebroeck, D., and Garoff, H. (1991). In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65, 4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., Wambach, M., Katze, M. G., and Krug, R. M. (1995). Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214, 222-228. [DOI] [PubMed] [Google Scholar]
- Mazroui, R., Huot, M. E., Tremblay, S., Filion, C., Labelle, Y., and Khandjian, E. W. (2002). Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11, 3007-3017. [DOI] [PubMed] [Google Scholar]
- Mulvey, M., Poppers, J., Ladd, A., and Mohr, I. (1999). A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J. Virol. 73, 3375-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe'ery, T., and Mathews, M. B. (2000). Viral translational strategies and host defense mechanisms. In: Translational control of gene expression, ed. N. Sonenberg, J.W.B. Hershey, and M. B. Mathews, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 371-424.
- Perri, S., Driver, D. A., Gardner, J. P., Sherrill, S., Belli, B. A., Dubensky, T. W., Jr., and Polo, J. M. (2000). Replicon vectors derived from Sindbis virus and Semliki Forest virus that establish persistent replication in host cells. J. Virol. 74, 9802-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk, M., et al. (2000). TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19, 4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushko, P., Parker, M., Ludwig, G. V., Davis, N. L., Johnston, R. E., and Smith, J. F. (1997). Replicon-helper systems from attenuated Venezuelan equine encephalitis virus-expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239, 389-401. [DOI] [PubMed] [Google Scholar]
- Reichel, P. A., Merrick, W. C., Siekierka, J., and Mathews, M. B. (1985). Regulation of a protein synthesis initiation factor by adenovirus virus-associated RNAi. Nature 313, 196-200. [DOI] [PubMed] [Google Scholar]
- Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S., and Kaufman, R. J. (2001). Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell. 7, 1165-1176. [DOI] [PubMed] [Google Scholar]
- Schneider, R. J., and Mohr, I. (2003). Translation initiation and viral tricks. Trends Biochem. Sci. 28, 130-136. [DOI] [PubMed] [Google Scholar]
- Sjöberg, E. M., and Garoff, H. (1996). The translation-enhancing region of the Semliki Forest virus subgenome is only functional in the virus-infected cell. J. Gen. Virol. 77, 1323-1327. [DOI] [PubMed] [Google Scholar]
- Sjöberg, E. M., Suomalainen, M., and Garoff, H. (1994). A significantly improved Semliki Forest virus expression system based on translational enhancer segments from the viral capsid gene. Biotechnology 12, 1127-1131. [DOI] [PubMed] [Google Scholar]
- Smerdou, C., and Liljeström, P. (1999). Two-helper RNA system for production of recombinant Semliki Forest virus particles. J. Virol. 73, 1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, J. H., and Strauss, E. G. (1994). The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58, 491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taupin, J. L., Tian, Q., Kedersha, N., Robertson, M., and Anderson, P. (1995). The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc. Natl. Acad. Sci. USA 92, 1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière, H., Chebli, K., Zekri, L., Courselaud, B., Blanchard, J. M., Bertrand, E., and Tazi, J. (2003). The RasGAP–associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823-831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- van Steeg, H., Kasperaitis, M., Voorma, H. O., and Benne, R. (1984). Infection of neuroblastoma cells by Semliki Forest virus. The interference of viral capsid protein with the binding of host messenger RNAs into initiation complexes is the cause of the shut-off of host protein synthesis. Eur. J. Biochem. 138, 473-478. [DOI] [PubMed] [Google Scholar]
- van Steeg, H., van Grinsven, M., van Mansfeld, F., Voorma, H. O., and Benne, R. (1981). Initiation of protein synthesis in neuroblastoma cells infected by Semliki Forest Virus. A decreased requirement of late viral mRNA for eIF-4B and cap binding protein. FEBS Lett. 129, 62-66. [DOI] [PubMed] [Google Scholar]
- Wengler, G. (1980). Effects of alphaviruses on host cell macromolecular synthesis. In: The Togaviruses: Biology Structure, Replication, ed. R. W. Schlesinger, New York: Academic Press, 459-472.
- Wilson, J. E., Pestova, T. V., Hellen, C. U., and Sarnow, P. (2000). Initiation of protein synthesis from the A site of the ribosome. Cell 102, 511-520. [DOI] [PubMed] [Google Scholar]
- Xiong, C., Levis, R., Shen, P., Schlesinger, S., Rice, C. M., and Huang, H. V. (1989). Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science 243, 1188-1191. [DOI] [PubMed] [Google Scholar]